Abstract
Sigma-H is an alternative RNA polymerase sigma factor that directs the transcription of many genes that function at the transition from exponential growth to stationary phase in Bacillus subtilis. Twenty-three promoters, which drive transcription of 33 genes, are known to be recognized by sigma-H-containing RNA polymerase. To identify additional genes under the control of sigma-H on a genome-wide basis, we carried out transcriptional profiling experiments using a DNA microarray containing >99% of the annotated B. subtilis open reading frames. In addition, we used a bioinformatics-based approach aimed at the identification of promoters recognized by RNA polymerase containing sigma-H. This combination of approaches was successful in confirming most of the previously described sigma-H-controlled genes. In addition, we identified 26 putative promoters that drive expression of 54 genes not previously known to be under the direct control of sigma-H. Based on the known or inferred function of most of these genes, we conclude that, in addition to its previously known roles in sporulation and competence, sigma-H controls genes involved in many physiological processes associated with the transition to stationary phase, including cytochrome biogenesis, generation of potential nutrient sources, transport, and cell wall metabolism.
Bacterial sigma factors are positive regulators of gene expression that interact with core RNA polymerase and direct the initiation of transcription from defined promoter sequences (22, 25). The major sigma factor in most bacteria, sigma-A, is required for expression of many of the so-called housekeeping functions and the bulk of the RNA during growth. Many bacteria have multiple alternative sigma factors, which are responsible for directing transcription of specialized gene sets. Bacillus subtilis has at least 17 alternative sigma factors which are involved in a variety of processes, including certain stress responses, chemotaxis, and motility (25, 30). One of the more dramatic examples of gene regulation by alternative sigma factors is the process of endospore formation (sporulation) in B. subtilis. The sporulation program of gene expression in B. subtilis is carried out under the direction of five alternative sigma factors whose activities are subject to spatial and temporal control (14, 51). Here we report the results of transcriptional profiling experiments aimed at identifying, on a genome-wide basis, genes under the control of one of these sigma factors, sigma-H.
Sigma-H, the sigH (spo0H) gene product, directs the transcription of several genes that function in the transition from exponential growth to stationary phase, including the initiation of spore formation and entry into the state of genetic competence (1, 7, 12, 20). Sigma-H is required at an early stage of sporulation and directly activates transcription of several sporulation genes including spo0A, spo0F, kinA, spo0M, spoVG, and spoVS and the spoIIA operon (2, 4, 28, 41, 42, 46, 54, 59, 61). Sigma-H also directs the transcription of several members of the phr family of genes, which encode secreted peptide pheromones (31, 37). Each Phr peptide likely inhibits the activity of a corresponding Rap phosphatase that modulates entry into genetic competence, sporulation, and perhaps other processes (32, 39).
Several of the genes that are transcribed by a sigma-H-recognized promoter have additional promoters that are recognized by other sigma factors. For example, spo0A (sporulation response regulator), ftsA (cell division), dnaG (DNA replication), sigA (encoding sigma-A, the major sigma factor), and citG (tricarboxylic acid cycle) are transcribed under sigma-H control but are also transcribed from sigma-A-dependent promoters.
In addition to genes that are under the direct control of sigma-H, there are many genes whose transcription is indirectly influenced by sigma-H. For example, during sporulation, sigma-H stimulates transcription of the master regulator of sporulation, spo0A, from a sigma-H-recognized promoter, Ps (41). Expression from Ps is essential for efficient sporulation (49). The product of spo0A (Spo0A), in turn, activates or represses a large number of genes, many of which are transcribed by RNA polymerase containing sigma-A. Thus, sigma-H contributes to gene expression during sporulation both directly and indirectly.
The regulation of sigma-H itself is complex. Transcription of sigH is controlled directly by the transcriptional repressor AbrB and indirectly by the phosphorylated form of Spo0A (Spo0A∼P), which represses abrB, and by sigma-H itself (6, 40, 52, 57). spo0A gene expression is driven in part by sigma-H (see above), which results in increased levels of Spo0A∼P (under the appropriate conditions). Increased levels of Spo0A∼P result in more repression of abrB and therefore increased levels of sigH transcription, thereby setting up a self-reinforcing cycle. Sigma-H activity is also controlled at the posttranscriptional, translational, and posttranslational levels and responds to a variety of external conditions including pH, carbon source, and availability of amino acids (3, 9, 10, 17, 34). The precise mechanisms of regulation are not completely understood.
We used a combination of DNA microarray analysis and a bioinformatics approach to identify genes of the sigma-H regulon. We performed two types of DNA microarray experiments, one comparing RNAs from wild-type cells to RNAs from a sigH-null mutant and the other identifying RNAs that were induced by the overexpression of sigma-H during growth. We found that this combined microarray approach, along with the use of a hidden Markov model (HMM) database of possible sigma-H promoters, was successful both in confirming the identification of genes previously known to be directly controlled by sigma-H and in assigning many additional genes to the sigma-H regulon. Our results indicate that, in addition to its previously known role in sporulation and genetic competence, sigma-H controls many genes that are involved in helping cells adapt to conditions of nutrient depletion. The products of these genes are involved in a variety of processes, including transport, cell wall metabolism, proteolysis, and cytochrome biogenesis. In combination with the several regulatory genes that it controls, sigma-H contributes significantly to the intertwined networks that influence physiological decisions during entry into stationary phase.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
All strains used in this study were constructed by standard procedures in the wild-type strain PY79 (60). RL1265 (ΔsigF::kan) (15) and AG665 (ΔsigH::cat) (24) were used to construct PE170 (ΔsigH::cat ΔsigF::kan). A mutation in sigF was included to eliminate sigma-F-dependent gene expression from the experiments. To compare the transcriptional profiles of sigH+ and sigH mutant cells, strains RL1265 and PE170 were grown in Difco sporulation medium at 37°C. Samples for RNA isolation were taken at T−1, T0, and T1. T0 refers to the time at which the culture proceeds from exponential growth to stationary phase; T−1 is 1 h before and T1 is 1 h after entry into stationary phase. Cells were allowed to double at least four times prior to the taking of samples for array analysis.
To induce ectopic expression of sigH, we constructed a strain with sigH under the control of the LacI-repressible, isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter Pspank-hy. pDR111 (gift from David Rudner and Federico Gueiros Filho, Harvard University) is a derivative of the Pspac-hy plasmid pJQ43 (43) that contains an additional lacO binding site to achieve better repression in the absence of the inducer IPTG. An EcoRI/BamHI fragment of pDR111 was cloned into pDG1727 (21) to generate plasmid pPE30. A 603-bp fragment, including the ribosome binding site and the N-terminal coding sequence of sigH, was amplified by PCR with primers PE193 and PE195 (sequences available on request) and chromosomal DNA from PY79 as template. This PCR fragment was digested with SalI and SphI and cloned into pPE30 to yield pPE31. This plasmid was integrated into PY79 by single-crossover recombination to generate strain EG232. To analyze the transcriptional profile of a strain in which sigH was overexpressed, cells were grown in Luria-Bertani medium at 37°C to mid-exponential phase (optical density at 600 nm = 0.5), at which time the culture was split in two and IPTG was added to one culture. Samples were taken for analysis immediately after and 15, 30, and 60 min after the addition of IPTG and compared to the same time points from the parallel culture without IPTG.
DNA microarray construction.
Our microarrays consist of >99% (4,074 of 4,106) of the annotated protein coding genes of the B. subtilis genome. Primers (B. subtilis ORFmers) to 4,100 of the 4,106 genes of the B. subtilis genome were purchased from Sigma-Genosys. Each gene was amplified by PCR from genomic DNA from B. subtilis strain JH642 by using Hot Start Taq Master mix (Qiagen). A second PCR was performed with a dilution of the first PCR as a template. Of the 4,100 genes, 180 were not successfully amplified with the B. subtilis ORFmer primers. We constructed new primer sets for these 180 genes, and all but 26 were successfully amplified for spotting on the microarrays. PCR products were purified with QIAquick 96-well PCR purification kits (Qiagen). The DNA was dried down and resuspended in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% Sarkosyl for spotting. DNA was spotted on CMT-GAPS slides (Corning) with the Affymetrix 417 arrayer. In addition to the B. subtilis genes, four control sequences from Escherichia coli (ybaS, yfiF, yciC, and ygjU) were amplified and spotted on the array. These were chosen because there are no homologous sequences in B. subtilis. Each is represented 15 to 20 times throughout the array. Slides were prepared for hybridization (postprocessed) as previously described (13). The quality of each set of printed arrays was checked by hybridizing to the arrays genomic DNA isolated from B. subtilis strains JH642 and PY79 that had been labeled with cyanine 3 (Cy3) or Cy5. Briefly, genomic DNA was digested with HpaII and labeled with Cy3-dUTP or Cy5-dUTP with random primers and the Klenow fragment of DNA polymerase I. Spots that did not show proper hybridization in the control experiment were excluded from further analysis.
Sample preparation and RNA isolation.
Samples of cells taken for RNA isolation were immediately mixed with an equal volume of methanol (−20°C) and kept at room temperature for 1 to 2 min. Cells were centrifuged for 5 min, and cell pellets were stored at −80°C. RNA was isolated by using RNeasy RNA isolation kits (Qiagen) or by a hot acid-phenol isolation procedure (15). RNA prepared by either method gave similar results. RNA was treated with DNase on Qiagen columns as described by the manufacturer. The quality of the RNA was checked by visualizing the integrity of the 23S and 16S rRNA bands on an agarose gel.
Labeling and hybridization conditions.
Labeled cDNA was generated from RNA samples by direct incorporation of Cy3- or Cy5-labeled dUTP into cDNA. Differentially labeled samples from two different conditions (strains) were mixed and hybridized to the DNA microarrays, and each experiment was done at least three times (see below). For labeling, RNA (10 to 50 μg) was incubated with 1 μg of random hexamers and E. coli control RNA at 70°C for 10 min and then placed on ice for 2 min. A labeling mix containing 2× reverse transcription buffer (Life Technologies), 5 mM MgCl2, 20 mM dithiothreitol, deoxynucleoside triphosphates (1 mM dATP, 1 mM dGTP, 1 mM dCTP, and 0.4 mM dTTP), and either Cy3-dUTP or Cy5-dUTP (Perkin-Elmer Life Sciences) was added to the RNA-primer mixture and incubated at 25°C for 5 min. Superscript II reverse transcriptase (300 U) (Life Technologies) was added, and the mixture was incubated at 25°C for 10 min and then at 42°C for 70 min. The reaction was stopped by heating the reaction mixture to 70°C for 15 min. RNA was digested by adding RNase A and RNase H and incubating the mixture at 37°C for 30 min. Unincorporated nucleotides were removed by using QiaQuick purification spin columns (Qiagen) or DyeEx spin columns (Qiagen). Labeled cDNA was dried and resuspended in hybridization buffer (25 mM HEPES [pH 8.0], 1 mM EDTA, 0.8 μg of yeast tRNA/μl, 3× SSC, 0.2% sodium dodecyl sulfate). Hybridizations were performed as described previously (13). Slides were scanned on a GenePix 4000B scanner (Axon Instruments, Inc.). E. coli control RNA corresponded to the genes ybaS, yfiF, yciC, and ygjU. These genes were amplified by PCR from E. coli with an upstream primer that contained a promoter recognized by T7 RNA polymerase. RNA was made by in vitro transcription with T7 RNA polymerase with the PCR products as template.
Data analysis. (i) Image analysis and normalization.
Images were processed and analyzed with GenePix 3.0 software (Axon Instruments, Inc.). To be considered a valid signal, 40% of the pixels in a spot had to be at least 1 standard deviation above the local background in at least one of the channels. Spots not making this cutoff were excluded from further analysis. Because many of the genes in the analysis are expressed under only one condition, we had to assign a value to the spots that did not contain a significant signal in one channel. This value was the lowest signal in a channel that met our criterion of being at least 1 standard deviation above background. Background signal was not subtracted from the signal intensity of the spots. Once spots with significant signals were identified, the two channels were normalized by making the total signal in each of the channels equal.
(ii) Determination of outliers.
Genes whose expression differed significantly between the two conditions being compared were determined by two independent methods. We analyzed our data by a method similar to the previously described iterative outlier analysis (35). Each time point was the average of at least three independent experiments (independently grown and prepared samples). In at least one hybridization, the fluorophores were swapped to help decrease bias introduced by the dyes. In cases where multiple hybridizations were done from the same RNA sample, the data were averaged and treated as a single value for the experiment. The ratios from the independent samples were log2 transformed, and then the data for each individual spot were averaged between the replicate experiments. We then calculated the geometric mean and standard deviation of the entire population. Any spot that had a ratio that was more than 2.5 standard deviations away from the mean was considered an outlier. Outliers were then removed from the population, and the means and standard deviations were recalculated. Once again, any spot more than 2.5 standard deviations away from the mean was considered an outlier. This process was repeated until few or no outliers were detected. In these experiments generally three iterations were needed to identify all outliers in the population.
Array data were also analyzed with the Rosetta Resolver application Axon error model (Rosetta Biosoftware). The lists of outliers from the two analysis methods were compared, and only those genes that were considered significantly changed in both were considered further. The range of ratios of the outliers was from a high of 12 to a low of 1.6.
HMM analysis.
The sigma-H promoter sequence was modeled by using the HMMER 2.1.1 suite of software packages to create a series of closely related HMMs (http://hmmer.wustl.edu). Known sigma-H promoter sequences from citG, ftsAZ, kinA, sigA-P3, spo0A, spo0F, spoIIA, spoVG, spoVS, ureaABC, dnaG/sigA, minC, and ytxG were manually aligned and used to hand specify models with the HMMBUILD module. The various models differed only in their accommodation of alternate direction of transcription and variation in the size of the spacer region. In all cases, a background nucleotide distribution consistent with that of B. subtilis was assigned to portions of the promoter sequences corresponding to the spacer region, while regions corresponding to the −10 and −35 boxes were assigned to an HMM “match” state. To enable detection of multiple occurrences of promoters within the genome, all HMMs were required to match globally with respect to the model but locally with respect to the B. subtilis genome. By using the HMMSEARCH module, B. subtilis genome release 14.2 was searched with each HMM, and the positions of hits were compiled and correlated with nearby reading frames.
RESULTS AND DISCUSSION
Comparison of transcriptional profiles from sigH+ and sigH mutant cells.
We found significant differences between the transcriptional profiles (RNA levels) of sigH+ and sigH-null mutant cells. All strains in these studies contained a mutation in sigF, which encodes a sigma factor that is required for early-stage sporulation gene expression. By including a sigF mutation in all of the strain backgrounds, we eliminated most of the gene expression differences between sigH+ and sigH mutant cells that are due to downstream sporulation differences between the two strains. This allowed us to focus on changes in gene expression that are associated with the time that sigma-H is most active, the transition from exponential growth to stationary phase. sigH+ (RL1265) and sigH mutant (PE170) strains were grown in sporulation medium, and samples were taken during late exponential growth (approximately 1 h before the onset of stationary phase), at the onset of stationary phase (the end of exponential growth and the beginning of sporulation), and 1 h after entry into stationary phase. RNA was isolated from the samples, labeled, and hybridized to DNA microarrays containing 4,074 of the 4,106 protein coding genes in the B. subtilis genome.
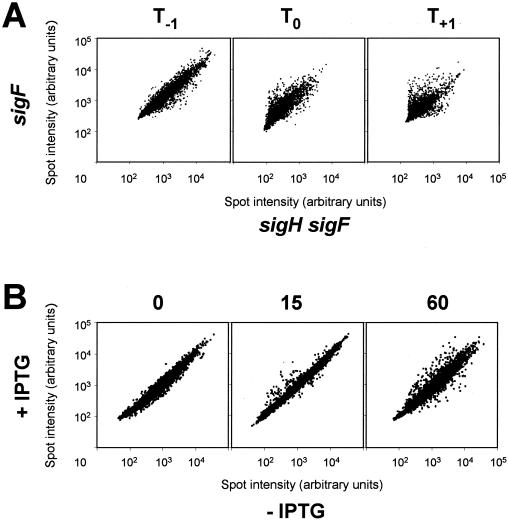
We found a total of 433 genes that had significantly different levels of RNA from at least one time point in the sigH mutant compared to the sigH+ strain. Of the genes altered, 245 were dependent on sigma-H for expression (that is, they were more highly expressed in wild-type cells) and 188 had higher expression levels in the sigH mutant. Together, over 10% of the genes in the B. subtilis genome were altered in the sigH mutant (discussed further below), demonstrating the important role of sigma-H in cellular physiology. Graphs comparing the relative abundance of RNA for each gene between the two strains are shown in Fig. 1A. Points that fall on or near the line with a slope of 1 are the majority of genes whose expression is not significantly affected in the sigH-null mutant. Points that are significantly off the line (outliers) are different between the two strains. As expected, most of the effects of the sigH mutation are seen an hour into stationary phase. We found 312 genes that had a significant change at this time point, 199 that were more highly expressed in the wild type, and 113 that had increased expression in the sigH mutant. Many of the genes dependent on sigma-H had previously been characterized as dependent on Spo0A (15).
FIG. 1.
Logarithmic scale plots of spot intensities (arbitrary units). (A) Transcriptional profiles from sigH+ and sigH mutant cells at T−1, T0, and T1 of sporulation. The intensity of each spot in one channel (sigH+) is plotted versus the intensity of that spot in the other channel (sigH mutant). (B) Overexpression of sigH during vegetative growth. Data are presented for RNA samples taken immediately after and 15 and 60 min after addition of IPTG to induce increased expression of sigH. The intensity of each spot in one channel (−IPTG) is plotted versus the intensity of that spot in the other channel (+IPTG). Data presented are from a representative experiment and are not normalized.
Overexpression of sigH during vegetative growth.
To identify many of the genes that are directly activated by sigma-H, we analyzed gene expression in response to overexpression of sigH during exponential growth in Luria-Bertani medium. Despite the complex posttranscriptional regulation of sigma-H, this strategy was quite useful. The sigH gene was placed under the control of the LacI-repressible, IPTG-inducible promoter Pspank-hy (Materials and Methods). Cells were grown to mid-exponential phase, at which time the cultures were split in two and sigH was induced by addition of IPTG to one of the cultures. Samples of cells for RNA extraction were taken just after addition of the inducer, IPTG, and 15, 30, and 60 min after induction and were compared to the same time points in the parallel cultures without IPTG. We found 160 genes that were significantly changed in response to overexpression of sigH; 110 of these had higher levels of expression at at least one of the time points after induction.
Data from a representative induction experiment are presented in Fig. 1B. Immediately after the addition of inducer, a single outlier is detected, sigH (Fig. 1B). This indicates how rapidly transcription from the promoter Pspank-hy is induced in response to addition of IPTG. Fifteen minutes after induction, most of the genes that are significantly changed are more highly expressed in the cells overproducing sigma-H, consistent with the function of sigma-H as an activator of transcription (Fig. 1B). That this approach is successful is underscored by the fact that many of these genes are known to be regulated by sigma-H, such as dnaG and citG. There is a more even distribution of genes that are up and down regulated 60 min after the overexpression of sigma-H (Fig. 1B). This is likely because we are observing some indirect effects on gene expression caused by increased levels of sigma-H and/or cells are beginning to proceed from exponential growth to stationary phase at this time. Interestingly, we observe known sigma-H promoters that additionally require the transcription factor Spo0A for activation (i.e., spoIIA operon) at 60 min after induction and not 15 min, most likely because Spo0A is more active at the transition to stationary phase, which is occurring about 60 min after addition of IPTG. Additional transcription factors that regulate gene expression at the transition from exponential growth to stationary phase such as AbrB and CodY are likely involved in the increase in genes regulated at the 60-min time point.
Identifying potential sigma-H binding sites in the genome by using an HMM approach.
HMMs of the promoter sequence recognized by RNA polymerase containing sigma-H were created (Materials and Methods) by using sequences from experimentally determined sigma-H promoters reported in the literature. Promoter sequences from the genes citG, ftsAZ, kinA, sigA-P3, spo0A, spo0F, spoIIA, spoVG, spoVS, ureABC, dnaG, minC, and ytxG were used to create HMMs of the sigma-H promoter. These models were used to search the B. subtilis genome to create a database of possible sigma-H promoters. These potential promoters were then compared to the microarray data described above. Genes that showed dependence on sigma-H for expression and had an HMM-predicted promoter were considered strong candidates for direct regulation by sigma-H.
We found that the models gave us a large number of false positives. This was probably because there were relatively few promoters known to be recognized directly by RNA polymerase containing sigma-H that could be used to make the HMMs and because the consensus promoter sequence is AT rich, as is the B. subtilis genome. Thus, without the transcriptional profiling data it would have been difficult to accurately predict sigma-H promoters based on sequence alone. Because we knew that the models were not ideal, we considered the possibility that some sigma-H promoters would be missed. Therefore, we additionally used a sequence based on the consensus to search all genes that were dependent on sigma-H for expression that did not already have a predicted HMM. The two approaches were then combined to give a final list of potential sigma-H promoters (discussed below).
Comparison of the microarray data with known sigma-H-regulated genes.
To validate our experimental approach, we analyzed the behavior of known sigma-H-regulated genes in the two types of DNA microarray experiments. The experiments reliably detected genes previously known to be activated by sigma-H. We were aware of 23 known sigma-H-regulated promoters driving the expression of 33 genes (Table 1 and Fig. 2 ). Of these 23 promoters we found sigma-H-dependent gene expression for 18 of the promoters in at least one of the microarray experiments (sigH+ versus sigH mutant or sigH overexpression). Classical sigma-H-regulated genes such as citG, spoVG, and kinA showed the correct regulation in both DNA microarray experiments.
TABLE 1.
Genes previously known to be regulated directly by sigma-H
| Gene or operona | sigH+/sigHb | Overexpressionc | −35 box | Spacing | −10 box | Function and/or comments | Reference(s) |
|---|---|---|---|---|---|---|---|
| citG (P2) | 3.5 | 2.6 | AAAGGATTT | 11 | GGCGAATTA | Fumarate hydratase (TCAi cycle), P1 recognized by sigma-A | 42, 54 |
| dnaG (sigA-P4) | 3.4 | 3.0 | GAAGGGATT | 12 | ATCGAATAA | DNA primase | 28, 61 |
| ftsAZ (P2) | 2.8 | 1.6 | AGAGGATAT | 11 | AACGAATAT | Cell division, P1 recognized by sigma-A | 19 |
| glgBCDAPd | 4.0 | 2.1 | AAAGGGCTT | 11 | TTCGAATAA | Glycogen biosynthesis | 29 |
| kinA | 3.9 | 2.8 | GAAGGAGAA | 12 | AGCGAATCA | Sporulation (phosphorelay) | 2, 41 |
| phrC | 1.6 | 1.6 | AGAGGATTT | 11 | GTAGCAAAA | Phosphatase regulator (promoter within rapC) | 31 |
| phrE (P1-2) | 1.6 | NOe | TTAGGAGGC | 11 | TTATAATGG | Phosphatase regulator (promoter within rapE) | 37 |
| phrE (P3) | 1.6 | NO | AGAGGATAG | 12 | CAAGAAAAT | See above | 37 |
| phrF | 2.4 | NO | TGAAGATTT | 13 | GGCAAATAA | Phosphatase regulator (promoter within rapF) | 37 |
| phrG | NO | NOf | GAAGGAAAA | 12 | GCCGAATAT | Phosphatase regulator (promoter within rapG) | 37 |
| phrl (P2) | NO | NOf | CAAGGAAAT | 12 | AATGAATAT | Phosphatase regulator (promoter within rapI) | 37 |
| phrK | NO | NOf | ACAGGAAAG | 12 | GGAGAATAA | Phosphatase regulator (promoter within rapK) | 37 |
| sigA (P3) | 1.6 | 2.1 | GCAGGAGTT | 12 | GGAGAATTA | Major sigma factor (promoter within yqxD), P1 recognized by sigma-A | 8 |
| spo0A (Ps) | NO | NOg | AGAGGGTAT | 11 | GTCGAATGT | Transcriptional regulation of sporulation, Pv recognized by sigma-A | 41 |
| spo0F (P2) | 3.2 | 2.3 | AAAGGAAAT | 11 | ACAGAATAC | Sporulation (phosphorelay), Spo0A regulated, P1 recognized by sigma-A | 4, 41 |
| spo0M (ygaI) | NO | NO | ATAGGAAAA | 12 | AACGAATCT | Sporulation | 23 |
| spoIIAA-AB-sigF | 9.0 | 3.2 | GAAGGAATT | 12 | ATCGAAACA | Forespore gene expression, Spo0A regulated | 59 |
| spoVG | 3.0 | 2.8 | GCAGGATTT | 11 | GTGGAATTG | Sporulation (peptidoglycan hydrolysis), AbrB regulated | 28, 61 |
| spoVS | 3.1 | 2.9 | GCAGGAATA | 12 | AGTGAATAT | Sporulation | 46 |
| ureABC (P2) | 1.6 | 2.1 | GAAGGAATT | 12 | GTCGAACTA | Urease, P3 recognized by sigma-A and repressed by CodY | 58 |
| yoxA-dacC (pbp) | 5.3 | 5.7 | GGAGGAAAT | 12 | ATTGAATTC | Unknown-d-alanyl-d-alanine carboxypeptidase | 38 |
| ytxG (P2) | (3.4)h | 1.6 | AAAGGATTT | 11 | GGAGAATAG | Stress, P1 recognized by sigma-B | 56 |
| yvyD (P2) | (2.2)h | 1.5 | GCAGGAATT | 12 | AGAGAAATA | Putative modulator of sigma-L, P1 recognized by sigma-B | 11 |
| Consensus (24)j | R-AGGAwWW | 11-12 | R--GAATww | R is A or G, W is A or T | 25 |
Gene or genes of an operon that are regulated by sigma-H. If the sigma-H promoter is a secondary promoter, its designation is listed in parentheses. lytE (cwlF), which had been proposed to be under sigma-H control (26), was not included in the table because its suggested promoter matches poorly with the proposed consensus.
Ratios of relative RNA levels in sigH+ versus sigH-null mutant. Ratios are presented from the time point at which the difference was largest (Fig. 2). In cases where multiple genes from an operon are listed, data are presented from the gene with the largest detected effect. Ratios are the averages of at least three independent experiments.
Ratios of relative RNA from sigH overexpression (+IPTG) compared to comparable time points without overexpression (−IPTG). Ratios are presented from the time point where the largest effect was detected (Fig. 2). In cases where multiple genes from an operon are listed, data are presented from the gene with the largest detected effect. Ratios are the average of at least three independent experiments.
Sigma-H-dependent regulation has been demonstrated only for Bacillus stearothermophilus (29).
NO, not determined to be an outlier in our analysis.
This phr gene was not detected as an outlier most likely because of its small size (<200 bp).
Expression of spo0A from Ps (the sigma-H-dependent promoter) is modest relative to the overall level of expression and is probably the reason that we did not detect it.
Parentheses denote that ytxG and yvyD were more highly produced in the sigH mutant strain, most likely due to activation of a sigma-B-dependent promoter in the absence of sigma-H.
TCA, tricarboxylic acid.
Boldface indicates a match with the consensus promoter sequence.
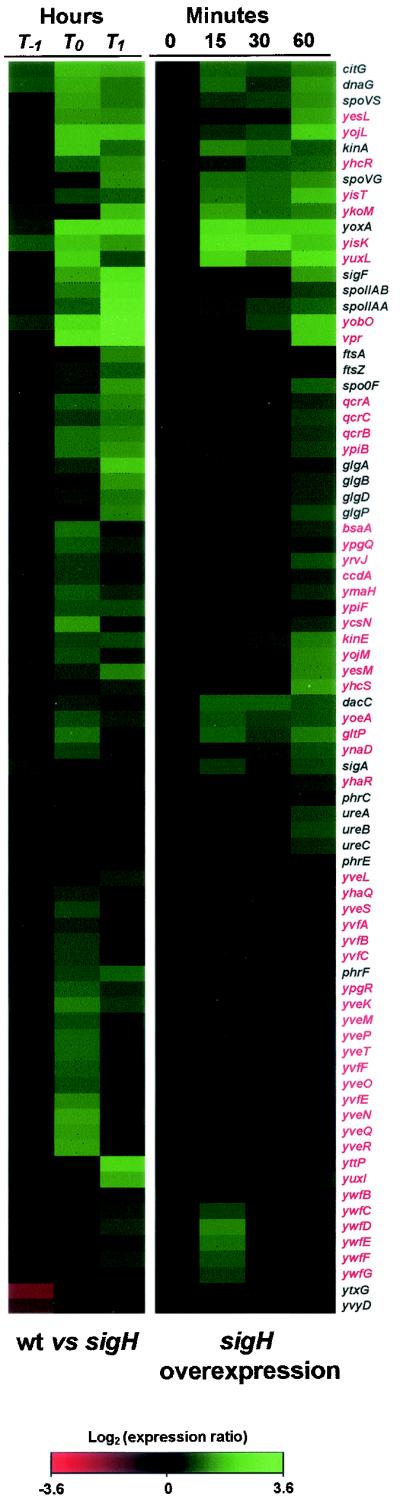
FIG. 2.
Heat map indicating the expression profiles of 79 sigma-H-regulated genes. These genes were ordered by using a hierarchical clustering algorithm (J-Express v.2.1 application from MolMine AS), so that those with similar expression patterns were grouped together. The first three columns (left) display expression ratios for the wild typeversus the sigH mutant at T−1, T0, and T1. The next four columns display the expression ratios for cells harboring Pspank-hy-sigH immediately (0) and 15, 30, and 60 min after induction with IPTG. Hybridization ratios are displayed colorimetrically: shades of green indicate that a gene had a higher RNA level in wild-type cells (first three columns) or when sigH was overproduced (last four columns); shades of red indicate that a gene had less RNA in wild-type cells (first three columns) or when sigH was overexpressed. Genes identified in this work as sigma-H regulated are indicated in red. The ratios for each gene are from averages of at least three independent experiments.
Five promoters known to be regulated by sigma-H were not identified in our microarray analysis, and biological or technical reasons account for the discrepancy with previous reports. Three of the genes not found in our experiments were phr genes (phrG, phrI, and phrK), which are reading frames of under 200 nucleotides. Although we successfully detected several small open reading frames on our arrays (other phr genes, spoVG, and spoVS), DNA fragments under 200 bp tended to give lower signals with higher variability. Thus, it is not surprising that some of the small genes were missed (47). One of the other two genes (spo0A) would likely not have been detected under the experimental conditions used. The sigma-H-dependent promoter that drives expression of spo0A is a secondary promoter and has a less-than-twofold effect on the overall expression of spo0A (41), and therefore, we did not expect to find this gene in our experiments. The last gene, spo0M, is expressed at a low level during sporulation (23), and we did not detect a significant signal for this gene on our arrays.
The analysis of how the known sigma-H genes behaved in the microarray experiments was important in interpreting the data from the full-genome arrays. First, some genes that appeared to be dependent on sigma-H for expression are likely detected because the promoter for the downstream gene resides in their coding region. Examples of this are the promoters for the phr genes that reside in the coding sequence of the rap gene located immediately upstream. In these cases only a portion of the rap gene is transcribed and would result in a positive signal on the microarrays although the entire gene is not being expressed. Also, we found that promoters that require an additional transcription factor for activity (such as the requirement of Spo0A for the expression of the spoIIA operon) were detected only at time points when the transcription factor is active. Thus, in the sigH overexpression experiment, genes that were expressed at the 60-min time point, as cells were entering stationary phase, are excellent candidates for those that are regulated by additional transcription factors.
In some cases it is possible that a gene that appears to be independent of sigma-H (in the sigH+ versus sigH mutant experiment) is actually regulated by sigma-H. For example, ytxG is known to be under the control of sigma-H and is also directly controlled by the general stress response sigma factor sigma-B (56). We found that sigma-B activity was increased at the T−1 time point in the sigH mutant background, consistent with previous findings that expression of sigma-B-dependent genes is increased in sigH mutants (45, 56). RNA from ytxG was higher in the sigH mutant due to the fact that the promoter controlled by sigma-B was more active in the absence of sigma-H (56). We detected this regulation in the two types of microarray experiments: ytxG was induced by overexpression of sigma-H but showed a higher level of expression in a sigH mutant strain (Table 1). This example shows that one cannot necessarily assume that a gene that is regulated by sigma-H will behave as expected in the sigH+ versus sigH mutant experiment and demonstrates the importance of performing both types of microarray experiments. yvyD is also known to be controlled by both sigma-H and sigma-B (11) and had a pattern of expression similar to that of ytxG (Table 1).
Identification of genes that are strong candidates to be regulated directly by sigma-H.
In addition to the previously described genes activated by sigma-H, we found 26 operons containing 54 genes that showed dependence on sigma-H for expression in the microarray experiments and had a potential sigma-H promoter within 200 bp of the start codon (except yojLM, for which the promoter was 292 bp upstream) (Table 2 and Fig. 2). In 23 of the operons there was a promoter predicted by an HMM. Promoters for the other three operons were uncovered by using the pattern search algorithm on the SubtiList website (http://genolist.pasteur.fr/SubtiList/). In our analysis of genes that were differentially affected in the microarrays, we inferred direct regulation by sigma-H if a good sigma-H binding site was located upstream of the gene. It seems likely that most of the other genes are indirectly regulated by sigma-H. Many of the known or putative functions of the 54 genes that may be directly controlled by sigma-H highlight the critical role of sigma-H in adaptation to nutrient deprivation. Several genes under direct sigma-H control appear to be involved in adaptation to nutrient deficiency. Many of the proteins in this class are known or predicted to be secreted and could be used to modify the extracellular environment (Table 2). Proteins such as Vpr (extracellular serine protease) (50) could be used to scavenge for food in the extracellular environment by degrading proteins. In addition to Vpr there are other secreted proteases that are indirectly regulated by sigma-H, AprE and NprE, which also could be used to digest extracellular material. In addition there is a putative secreted nuclease, YhcR, that could be used to degrade nucleic acid that could also be used for food. (A recent study of E. coli has shown that DNA can be used as a sole source of carbon for the cell [16].) Another group of genes that could provide alternative nutrients to the cell are transporters. For example, gltP encodes a glutamate transporter and is predicted to be directly regulated by sigma-H. The yhaQ gene product has similarity to ABC transporter ATP binding proteins, likely has a role in transport, and appears to be directly regulated by sigma-H. Lastly, genes such as ccdA and the qcr operon encode proteins that are involved in the synthesis of cytochrome c and the cytochrome bc complex, respectively. These proteins are involved in the electron transport chain and may be up regulated by sigma-H in response to nutrient-limiting conditions in an attempt to generate energy. We also find the expression of resABC, which is also required for cytochrome c synthesis, to be indirectly regulated by sigma-H. Expression of resABC was previously shown to be induced upon entry into stationary phase by a putative sigma-A promoter (53).
TABLE 2.
Newly identified sigma-H-regulated geneslegend
| Gene or operona | sigH+/sigHb | Over- expressionc | −35 box | Spacing | −10 box | Distanced | Methode | Putative function and/or comments | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bsaA-ypgQR | 2.6 | 1.8 | AGAGGGAAT | 12 | ATAGAATGA | 147 | PM | Putative glutathione peroxidase-unknowns | ||||||||
| ccdA | 2.0 | 1.7 | GAAGGAGTA | 12 | GTCAAATTA | 109 | HMM | Late step of cytochrome c synthesis (33) | ||||||||
| gltP | 2.6 | 2.7 | AAAGGTGTT | 11 | CACGAATGA | 40 | HMM | H+/Glu symport protein | ||||||||
| kinE (ykrQ) | 2.2 | 2.7 | GAAGGAAGT | 11 | GTAGAAATA | 35 | HMM | Two-component sensor His kinase (27) | ||||||||
| vpr | 9.5 | 4.1 | CAAGGATTT | 11 | CAAGAAATA | 47 | HMM | Minor extracellular serine protease (secretion sequenceg) (50) | ||||||||
| ycsN | 3.1 | 1.8 | AAAGGAAAA | 12 | GCCGAATGA | 50 | HMM | Similar to aryl-alcohol dehydrogenase | ||||||||
| yesL-yesM | 3.0 | 3.2 | GCAGGAATT | 11 | GGAGAAATA | 43 | HMM | Unknown—similar to two-component sensor histidine kinase | ||||||||
| yhaRQ | 1.8 | 1.8 | AAAGGTTTA | 12 | GGGGAATGT | 44 | HMM | Similar to enoyl CoAj hydratase-ABC transporter | ||||||||
| yhcRSf | 2.9 | 3.0 | AAAGGAATT | 12 | GTCGAAATG | 45 | HMM | Similar to 5′-nucleotidase (secretion sequenceg)-unknown | ||||||||
| yisK | 3.9 | 6.5 | AAAGGGATT | 12 | AGAGAATAC | 41 | HMM | Similar to 5-oxo-1,2,5-TCAk-3-penten acid decarboxylase | ||||||||
| yisT | 2.3 | 3.7 | GAAGGAGAA | 12 | AACGAATTT | 21 | HMM | Similar to nuclease inhibitor | ||||||||
| ykoM | 3.7 | 3.4 | GAAGGAATT | 11 | AGCGAATAC | 79 | HMM | Similar to transcriptional regulator (MarR family) | ||||||||
| ymaH | 2.3 | 1.9 | GCAGGAAAA | 11 | ATCGAAACT | 31 | HMM | Similar to host factor 1 protein, sporulation (MICADO database) | ||||||||
| ynaD | 2.1 | 2.2 | GAAGGATAG | 12 | GGAGAATCA | 143 | HMM | Similar to ribosomal protein alanine N-acetyltransferase | ||||||||
| yobO | 12.4 | 4.2 | AAAGGAATT | 11 | ACAGAATTG | 84 | HMM | Similar to phage-related preneck appendage | ||||||||
| yoeA | 2.2 | 2.3 | GAAGGGTTT | 12 | GATGAATAA | 78 | HMM | Similar to multidrug efflux (Clostridium acetobutylicum), sporulation (MICADO databaseh) | ||||||||
| yojLM | 3.9 | 4.0 | GAAGGGATT | 12 | AGAGAATTG | 292 | HMM | Cell wall binding protein (secretion sequenceg)—similar to superoxide dismutase | ||||||||
| ypiBF-qcrABC | 3.2 | 2.0 | AAACGATTT | 11 | GATGAATTT | 80 | HMM | Unknown-unknown-menaquinol:cytochrome c reductase | ||||||||
| ypiBF-qcrABC | 3.2 | 2.0 | GCAGGAATA | 12 | GTGGAACAT | 40 | HMM | Alternative promoter; for function see above | ||||||||
| yrvJ | 2.0 | 2.2 | AAAGGACTT | 12 | CAAGAATGC | 30 | PM | Putative cell wall binding protein (secretion sequenceg); similar to N-acetylmuramoyl-l-alanine amidase | ||||||||
| yttP | 4.9 | NOi | ACAGGAACA | 11 | CTTGAATAG | 24 | HMM | Similar to transcriptional regulator (AcrR/TetR family) | ||||||||
| yuxl | 3.5 | 1.6 | AAAGGAAAA | 12 | CACGAATTA | 33 | HMM | Unknown | ||||||||
| yuxL | 4.4 | 5.4 | AAAGGAGTT | 12 | AACGAAATA | 23 | HMM | Similar to acylaminoacyl-peptidase | ||||||||
| yveKLMNOPQRST- yvfABCDEF | 3.4 | NOi | TAAGGAATT | 11 | ATAAAATTT | 173 | PM | Exopolysaccharide biosynthesis-fruiting body formation | ||||||||
| ywfBCDE (FG) | 1.8 | 2.8 | AAAGGGTTT | 11 | TGGGAATAA | 86 | HMM | Unknown-unknown-similar to Glc-1-dehydrogenase-unknown | ||||||||
| ywfFG | 1.8 | 2.8 | GCAGTAATT | 11 | GCTGAATCA | 53 | HMM | Similar to efflux protein-similar to Asp aminotransferase | ||||||||
| Consensus (24) l | R-AGGAwWW | 11-12 | R--GAATww | R is A or G, W is A or T |
Gene or genes of a potential operon that are regulated by sigma-H.
Ratios of relative RNA levels in sigH+ versus sigH-null mutant. Ratios are presented from the time point at which the difference was largest (Fig. 2). In cases where multiple genes from an operon are listed, data are presented from the gene with the largest detected effect. Ratios are the averages of at least three independent experiments.
Ratios of relative RNA levels from sigH overexpression (+IPTG) compared to comparable time points without overexpression (−IPTG). Ratios are presented from the time point where the largest effect was detected (Fig. 2). In cases where multiple genes from an operon are listed, data are presented from the gene with the largest detected effect. Ratios are the average of at least three independent experiments.
The distance is measured from the last base pair of the −10 region to the first base pair of the start codon.
PM, pattern match. HMM, hidden Markov model.
The predicted sigma-H promoter for yhcR overlaps with predicted AUG (SubtiList), but a second AUG is present 60 bp downstream (a possible ribosome binding site is also found just upstream of this second AUG).
Secretion sequence is from reference 55.
MICADO database: http://locus.jouy.inra.fr/cgi-bin/genmic/madbase/progs/madbase.operl.
NO, not determined to be an outlier in our analysis.
CoA, coenzyme A.
TCA, tricarboxylic acid.
Boldface indicates a match with the consensus promoter sequence.
Expression of the ccdA operon was previously known to coincide with the time that sigma-H is fully active (33). The ccdA operon transcript has been mapped by primer extension, and the authors indicated that sigma-A was likely to drive transcription of the ccdA operon. We have identified a potential sigma-H promoter that overlaps the putative sigma-A promoter. Thus, sigma-H could be responsible for additional regulation of ccdA. ccdA mutant strains are deficient in sporulation at a very late stage (48), similar to what is observed with spoVS, a gene controlled by sigma-H.
The best-known role of sigma-H is to activate sporulation. Many of the known sigma-H-controlled genes are involved in the signal transduction pathway involved in the initiation of sporulation (kinA, spo0F, and spo0A) and in the early stages of sporulation (spoIIAA, spoIIAB, and sigF). We find that an additional histidine kinase implicated in controlling the initiation of sporulation, kinE, is controlled by sigma-H (27). We also searched the MICADO (http://locus.jouy.inra.fr/cgi-bin/genmic/madbase/progs/madbase.operl) and JAFAN (http://bacillus.genome.ad.jp) databases to determine if any of the newly identified direct sigma-H targets display a sporulation defect when mutated. Two genes, ymaH and yoeA, are reported to be defective in endospore formation when mutated.
Recently, it was shown that natural isolates of B. subtilis can form multicellular aerial structures that may be important for the dispersal of spores and that sigH is required for the formation of these structures (5). Two genes required for the formation of these aerial structures, yveQ and yveR, are thought to be part of a large operon responsible for exopolysaccharide production. We find that genes that are members of the yve operon that is involved in fruiting body formation, yveKLMNOPQRST-yvfABCDEF, are dependent on sigma-H.
Three additional groups of newly identified sigma-H-controlled genes are worth mentioning: transcription factors, cell wall binding proteins/autolysins, and proteins involved in detoxification. Sigma-H was already known to regulate three transcription factors, sigma-A, Spo0A, and sigma-F. Two additional putative transcription factors show sigma-H-dependent gene expression, ykoM (MarR family transcriptional regulator) and yttP (TetR/AcrR family). How these putative transcriptional regulators contribute to gene expression will be an interesting avenue of future investigation.
The cell wall binding proteins affected by sigma-H are of interest because sigH mutants are unable to form the asymmetric septum during sporulation. With the exception of ftsA and ftsZ, we do not find that the expression of any known cell division genes is affected. We do find three genes that show dependence on sigma-H, yojL (similar to major autolysins lytE and lytF), yrvJ (similar to N-acetylmuramoyl-l-alanine amidase), and yuxL (similar to acylaminoacyl-peptidase), which are likely to be involved in the modification of the cell wall and possibly in the formation of the asymmetric septum. Interestingly two of the previously identified sigma-H-controlled genes, dacC and spoVG, may also be involved in cell wall modification (36, 38). Alternatively, these gene products may be involved in generating nutrients for the cell by digesting cell wall material. Conversely, the major autolysins lytC, lytD, and lytF (yhdD) appear to be indirectly controlled by sigma-H and are up regulated in the sigH mutant presumably via activation of sigma-D in the sigH mutant cells (see below).
We found four genes that are predicted to be involved in adapting to changing environmental conditions. They are yoeA (similar to multidrug efflux), yojM (similar to superoxide dismutase), ywfF (similar to efflux protein), and bsaA (putative glutathione peroxidase). These proteins likely provide protective properties to the cell and could contribute to the effects of sigma-H on survival in stationary phase at high and low pH and in the presence of ethanol (18).
Global changes in cell physiology.
As mentioned above, we found that the expression of over 10% of the genes is significantly different between sigH+ and sigH mutant cells. The majority of these genes are almost certainly not under the direct control of sigma-H. Many of the genes whose expression is altered are involved in metabolism, transport of nutrients, and antibiotic production. The large numbers of genes that appear to be indirectly controlled by sigma-H are listed in supplementary material (http://mcb.harvard.edu/losick). Many of these indirect effects could be due to increased association of alternate sigma factors, especially sigma-B and sigma-D, with core RNA polymerase in the absence of sigma-H.
Many of the changes that we observed in the sigH (spo0H) mutant were also found in spo0A mutants (15). The regulation of three transcription factors, Spo0A, AbrB, and sigma-H, is intertwined, and mutations in one gene affect the others (see the introduction). AbrB represses transcription of sigH, and this repression is relieved via Spo0A∼P repressing transcription of abrB. Thus, a spo0A mutant lacks full sigma-H activity. In turn, sigma-H is partially responsible for the activation of transcription of spo0A and expression of genes needed for (kinA and spo0F) or influencing (phr genes) phosphorylation of Spo0A. Thus, in a sigma-H mutant Spo0A activity is greatly reduced. The result is that mutations in either gene have significant effects on the activity of the other, and it is not surprising that their transcriptional profiles are similar.
An example of how a sigH mutation can have profound effects on a large number of genes via its role in activating Spo0A is highlighted by the expression of genes under the control of sigma-D. Sigma-D regulates expression of genes involved in motility, chemotaxis, and autolysin production (25). We found that expression of many of these genes was increased in the sigH mutant. This effect is almost certainly due to the role of sigma-H in activation of Spo0A; a spo0A mutation also causes increased expression of genes dependent on sigma-D via regulation of sinI and sinR (15, 44).
Although many of the same genes are affected in spo0A and sigH mutants, the genes directly regulated by Spo0A and sigma-H are mostly different. The only promoters that have binding sites for both Spo0A and sigma-H are involved in the initial stages of sporulation, spo0F, spoIIA (sigF), and spo0A itself.
Conclusion.
Global analysis of gene expression has provided insights into the role of sigma-H in the transition from exponential growth to stationary phase and sporulation. We have identified 54 genes expressed by 26 newly identified putative sigma-H promoters. This brings the total regulon of sigma-H to potentially 49 promoters controlling the expression of at least 87 genes. Many of these are controlled by additional transcription factors, and several of these genes encode bona fide or putative transcription factors. In addition, there are many other genes that are indirectly affected by sigma-H. Identifying most of the genes on a particular pathway controlled by a specific transcription factor is a critical step in beginning to dissect how the various regulatory pathways are interconnected to form complex networks.
Acknowledgments
We thank Rachel Erlich, Keith Morneau, Tyler Aldredge, and Paul Grosu from the Bauer Center for Genomics Research at Harvard University for advice in the construction of the DNA microarrays and in the analysis of the data. We thank Natalia Comella and Emily Cornell Ruiz for assistance in the production of the microarrays, Nick Warner for assistance in data analysis, and the MIT BioMicro Center for providing equipment and technical support.
This work was supported in part by Public Health Service grant GM50895 (A.D.G.), by the Merck/MIT Collaborative program (A.D.G.), and by NIH grant GM18568 (R.L.). R.A.B. was supported in part by a postdoctoral fellowship from the NIH, P.E. was a postdoctoral fellow of the Human Frontier Science Program and of the Swiss National Science Foundation, and J.E.G.-P. was supported by the Ministerio de Educacion y Ciencia Postdoctoral Fellowship (Spain).
R.A.B., P.E., and J.E.G.-P. contributed equally to the work.
REFERENCES
- 1.Albano, M., J. Hahn, and D. Dubnau. 1987. Expression of competence genes in Bacillus subtilis. J. Bacteriol. 169:3110-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniewski, C., B. Savelli, and P. Stragier. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J. Bacteriol. 172:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai, K., F. Kawamura, H. Yoshikawa, and H. Takahashi. 1995. Expression of kinA and accumulation of σH at the onset of sporulation in Bacillus subtilis. J. Bacteriol. 177:6679-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, U., M. Lewandoski, E. Dubnau, and I. Smith. 1990. Temporal regulation of the Bacillus subtilis early sporulation gene spo0F. J. Bacteriol. 172:5432-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkholder, W. F., and A. D. Grossman. 2000. Regulation of the initiation of endospore formation in Bacillus subtilis, p. 151-166. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 7.Carter, H. L., III, and C. P. Moran, Jr. 1986. New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 83:9438-9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, H. L., III, L. F. Wang, R. H. Doi, and C. P. Moran, Jr. 1988. rpoD operon promoter used by σH-RNA polymerase in Bacillus subtilis. J. Bacteriol. 170:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosby, W. M., and P. Zuber. 1997. Regulation of Bacillus subtilis σH (Spo0H) and AbrB in response to changes in external pH. J. Bacteriol. 179:6778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon, L. G., S. Seredick, M. Richer, and G. B. Spiegelman. 2001. Developmental gene expression in Bacillus subtilis crsA47 mutants reveals glucose-activated control of the gene for the minor sigma factor σH. J. Bacteriol. 183:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drzewiecki, K., C. Eymann, G. Mittenhuber, and M. Hecker. 1998. The yvyD gene of Bacillus subtilis is under dual control of σB and σH. J. Bacteriol. 180:6674-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubnau, E., J. Weir, G. Nair, L. Carter III, C. Moran, Jr., and I. Smith. 1988. Bacillus sporulation gene spo0H codes for σ30 (σH). J. Bacteriol. 170:1054-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 14.Errington, J. 1996. Determination of cell fate in Bacillus subtilis. Trends Genet. 12:31-34. [DOI] [PubMed] [Google Scholar]
- 15.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisby, D., and P. Zuber. 1994. Mutations in pts cause catabolite-resistant sporulation and altered regulation of spo0H in Bacillus subtilis. J. Bacteriol. 176:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzy-Treboul, G., C. Karmazyn-Campelli, and P. Stragier. 1992. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J. Mol. Biol. 224:967-979. [DOI] [PubMed] [Google Scholar]
- 20.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 21.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 22.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, W. D., S. Kawamoto, Y. Hosoya, M. Fujita, Y. Sadaie, K. Suzuki, Y. Ohashi, F. Kawamura, and K. Ochi. 1998. A novel sporulation-control gene (spo0M) of Bacillus subtilis with a σH-regulated promoter. Gene 217:31-40. [DOI] [PubMed] [Google Scholar]
- 24.Healy, J., J. Weir, I. Smith, and R. Losick. 1991. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor σH in Bacillus subtilis. Mol. Microbiol. 5:477-487. [DOI] [PubMed] [Google Scholar]
- 25.Helmann, J. D., and C. P. Moran, Jr. 2001. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 26.Ishikawa, S., Y. Hara, R. Ohnishi, and J. Sekiguchi. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol. 180:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, W. C., C. P. Moran, Jr., and R. Losick. 1983. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature 302:800-804. [DOI] [PubMed] [Google Scholar]
- 29.Kiel, J. A., J. M. Boels, G. Beldman, and G. Venema. 1991. Molecular cloning and nucleotide sequence of the glycogen branching enzyme gene (glgB) from Bacillus stearothermophilus and expression in Escherichia coli and Bacillus subtilis. Mol. Gen. Genet. 230:136-144. [DOI] [PubMed] [Google Scholar]
- 30.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 31.Lazazzera, B. A., I. G. Kurtser, R. S. McQuade, and A. D. Grossman. 1999. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J. Bacteriol. 181:5193-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazazzera, B. A., J. M. Solomon, and A. D. Grossman. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89:917-925. [DOI] [PubMed] [Google Scholar]
- 33.Le Brun, N. E., J. Bengtsson, and L. Hederstedt. 2000. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 36:638-650. [DOI] [PubMed] [Google Scholar]
- 34.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol. Microbiol. 33:415-428. [DOI] [PubMed] [Google Scholar]
- 35.Loos, A., C. Glanemann, L. B. Willis, X. M. O'Brien, P. A. Lessard, R. Gerstmeir, S. Guillouet, and A. J. Sinskey. 2001. Development and validation of Corynebacterium DNA microarrays. Appl. Environ. Microbiol. 67:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuno, K., and A. L. Sonenshein. 1999. Role of SpoVG in asymmetric septation in Bacillus subtilis. J. Bacteriol. 181:3392-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McQuade, R. S., N. Comella, and A. D. Grossman. 2001. Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma-H of Bacillus subtilis. J. Bacteriol. 183:4905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen, L. B., T. Murray, D. L. Popham, and P. Setlow. 1998. Characterization of dacC, which encodes a new low-molecular-weight penicillin-binding protein in Bacillus subtilis. J. Bacteriol. 180:4967-4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perego, M., and J. A. Brannigan. 2001. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22:1541-1547. [DOI] [PubMed] [Google Scholar]
- 40.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 41.Predich, M., G. Nair, and I. Smith. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing σH. J. Bacteriol. 174:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, V. A., I. M. Feavers, and A. Moir. 1989. Role of σH in expression of the fumarase gene (citG) in vegetative cells of Bacillus subtilis 168. J. Bacteriol. 171:5933-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashid, M. H., and J. Sekiguchi. 1996. flaD (sinR) mutations affect SigD-dependent functions at multiple points in Bacillus subtilis. J. Bacteriol. 178:6640-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray, C., M. Igo, W. Shafer, R. Losick, and C. P. Moran, Jr. 1988. Suppression of ctc promoter mutations in Bacillus subtilis. J. Bacteriol. 170:900-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Resnekov, O., A. Driks, and R. Losick. 1995. Identification and characterization of sporulation gene spoVS from Bacillus subtilis. J. Bacteriol. 177:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiott, T., and L. Hederstedt. 2000. Efficient spore synthesis in Bacillus subtilis depends on the CcdA protein. J. Bacteriol. 182:2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siranosian, K. J., and A. D. Grossman. 1994. Activation of spo0A transcription by σH is necessary for sporulation but not for competence in Bacillus subtilis. J. Bacteriol. 176:3812-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sloma, A., G. A. Rufo, Jr., K. A. Theriault, M. Dwyer, S. W. Wilson, and J. Pero. 1991. Cloning and characterization of the gene for an additional extracellular serine protease of Bacillus subtilis. J. Bacteriol. 173:6889-6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 52.Strauch, M. A. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J. Bacteriol. 177:6999-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatti, K. M., H. L. Carter III, A. Moir, and C. P. Moran, Jr. 1989. Sigma H-directed transcription of citG in Bacillus subtilis. J. Bacteriol. 171:5928-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dijl, J. M., A. Bolhuis, H. Tjalsma, J. D. H. Jongbloed, A. De Jong, and S. Bron. 2001. Protein transport pathways in Bacillus subtilis: a genome-based road map, p. 337-355. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 56.Varon, D., M. S. Brody, and C. W. Price. 1996. Bacillus subtilis operon under the dual control of the general stress transcription factor σB and the sporulation transcription factor σH. Mol. Microbiol. 20:339-350. [DOI] [PubMed] [Google Scholar]
- 57.Weir, J., M. Predich, E. Dubnau, G. Nair, and I. Smith. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis σH factor. J. Bacteriol. 173:521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, J. J., P. J. Piggot, K. M. Tatti, and C. P. Moran, Jr. 1991. Transcription of the Bacillus subtilis spoIIA locus. Gene 101:113-116. [DOI] [PubMed] [Google Scholar]
- 60.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
- 61.Zuber, P., and R. Losick. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]