Abstract
The dif site is located in the replication terminus region of bacterial chromosomes, having a function of resolving dimeric chromosomes formed during replication. We demonstrate that filamentous bacteriophages of vibrios, such as f237 (Vibrio parahaemolyticus) and CTXφ (V. cholerae), are integrated into the dif-like site of host chromosome.
Cholera toxin, the most important virulence factor of Vibrio cholerae, has long been believed to be encoded in the chromosome of the bacterium, but recently it was revealed to be encoded within the genome of a lysogenic filamentous bacteriophage, CTXφ (17). Integration of CTXφ into the host chromosome is a site-specific recombination event in which a 17-bp DNA sequence on the host chromosome, designated attRS1, has been demonstrated to be the target for the site-specific integration by the phage genome (3, 11). Although the role as a target for phage integration is well established, it is not clear whether the attRS1 sequence serves any function for the host bacterium.
Vibrio parahaemolyticus is another vibrio that also is recognized as a major, worldwide cause of acute gastroenteritis (8). In the last 6 years, V. parahaemolyticus strains belonging to a few specific serotypes, most likely derived from a common clonal ancestor, have caused a pandemic of gastroenteritis (1, 2). In previous studies, we reported on a filamentous bacteriophage f237 that is specifically associated with the recent pandemic V. parahaemolyticus strains (7, 10). f237 is a single-stranded DNA phage demonstrating a pattern of genomic organization similar to that of CTXφ (7, 10).
In this study, we demonstrate that f237 is integrated into the dif-like site. dif is located in the replication terminus region of bacterial chromosomes and has a function of resolving dimeric chromosomes that result from an uneven number of recombination events between sister chromosomes during the replication cycle of bacterial DNA (5, 13, 15). Close scrutiny of the whole genome sequence of V. cholerae N16961 suggests that CTXφ also utilizes the dif-like site for its integration into the host chromosome.
V. parahaemolyticus strains RIMD2210633 (referred to here as KX-V237) and RIMD2210587 (KX-V191) used in this study were isolates from patients (10). The DNA probe for the phage f237 genome was previously described (10). Sample preparation of bacterial genomic DNA for pulsed-field gel electrophoresis (PFGE) was according to a previously described method (6, 18). For the construction of the lambda-based library, the V. parahaemolyticus KX-V237 cells were grown in Luria-Bertani broth supplemented with 3% NaCl and the whole gnomic DNA was prepared by the standard method (14). Gigapack III XL packaging extracts (Stratagene) were used to package lambda phages. Shotgun sequencing was carried out as previously described (10).
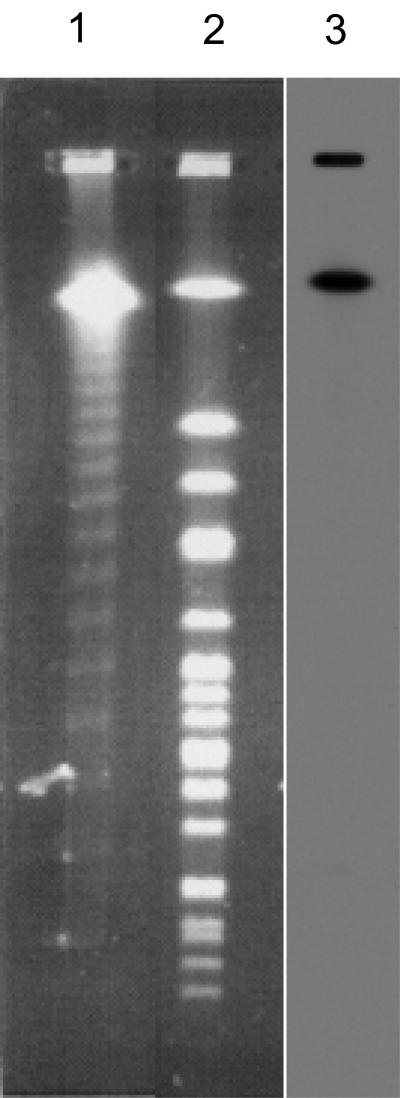
f237 was originally discovered as an episomal replicative form present in the host cytosol (10). Since the genome organization of f237 is similar to that of CTXφ, we examined whether the genome of f237 might be integrated into the host bacterial chromosome as CTXφ. Genomic DNA of V. parahaemolyticus KX-V237 cells known to have been infected with f237 was completely digested with the NotI restriction enzyme prior to separation by PFGE. Southern blot analysis using a DNA probe specific for the f237 genome demonstrated hybridization with the largest NotI fragment (1,080 kb in size) (Fig. 1). The results suggest that f237 is integrated into the host chromosome as a prophage.
FIG. 1.
Southern hybridization with probe for f237 phage. Lane 1, lambda ladder as size standards; lanes 2 and 3, NotI digests of KX-V237 genomic DNA. Lanes 1 and 2, ethidium bromide-stained gel; lane 3, Southern hybridization with a probe for f237 phage. The conditions for PFGE were on 1% agarose gel in 0.5× Tris-borate-EDTA buffer at 6 V/cm with pulse times of 1 to 60 s for 21 h at 14°C. The upper signal in lane 3 is the position of the well.
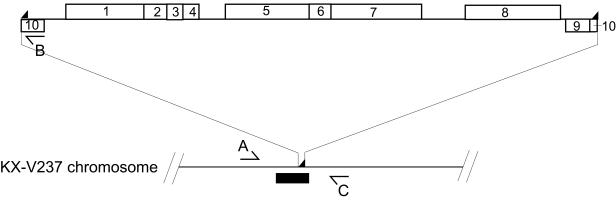
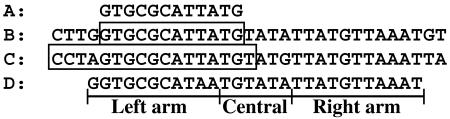
To identify the integration site of the f237 genome on the host chromosome, we constructed a lambda-based DNA library of the V. parahaemolyticus KX-V237 genome and screened for clones that reacted with the probe for the f237 genome. A lambda clone that hybridized with the probe was obtained (clone 5-7-H). Shotgun sequencing of the clone revealed that it contained the whole genome of f237 (Fig. 2). The sequence demonstrated that the f237 phage is integrated into an intergenic region on the KX-V237 chromosome without disrupting any open reading frame (ORF). By comparing the obtained sequence with the previously reported sequence of the f237 replicative form (10), we could identify the core of the attP sequence, which consisted of 12 bp (referred to as attPf237) (Fig. 3). This 12-bp sequence was directly repeated in the clone 5-7-H at both ends of the f237 phage genome (Fig. 2). We deduced the putative target sequence for integration of the f237 phage (attBf237) by subtracting the sequence of one set of the f237 phage genome sequence from the sequence of clone 5-7-H (Fig. 3). Unexpectedly, the 28-bp sequence containing the deduced attBf237 shared a high degree of homology (identity in 27 of the 28 bases) with the dif site of Escherichia coli (5) (Fig. 3). These results raised the possibility that f237 is integrated into the dif site on the chromosome of V. parahaemolyticus KX-V237.
FIG. 2.
Integration site of f237 phage on KX-V237 chromosome. The location of the integration site of f237 on clone 5-7-H is schematically presented. ◢, 12-bp direct repeats; black box, dif-like sequence; blocks, ORFs of the f237 genome. The numbers in the blocks are ORF numbers (10). Arrows (A, B, and C) show the positions and directions of primers for PCR.
FIG. 3.
Alignment of nucleotide sequences of attPf237, attBf237, attRS1, and E. coli dif. (A) attPf237; (B) attBf237 with flanking sequences (the sequence of attBf237 is boxed); (C) attRS1 with a flanking sequence (the sequence of attRS1 is boxed); (D) the dif sequence of E. coli. (C) The published sequence of V. cholerae N16961 (4) is the complement of the sequence. Base 1564104 corresponds to the end of the right arm of dif.
On the E. coli chromosome, the dif site is located 180° away from the replication origin (oriC) on the circular chromosome (5). To determine the location of the integration site of f237 on the chromosome of the KX-V237 strain, we localized the position of the f237 phage genome on the physical map (16) of the KX-V237 chromosome. The probe for the f237 genome hybridized with the NA and SA fragments when the NotI or SfiI digests of the KX-V237 genome were analyzed by PFGE (Fig. 4). Considering that dnaA, often found very close to oriC in many bacteria, is present on the NH fragment (16) (Fig. 4), the location of the f237 genome is speculated to be directly opposite oriC on the circular chromosome. Genomic sequencing by a Japanese consortium using the strain KX-V237 of V. parahaemolyticus is in progress. In the database of shotgun phase sequencing (7× coverage) of the whole genome of the strain, the attBf237 and its flanking region is the sequence most homologous to the E. coli dif among all the contigs (Makino et al., unpublished data). Furthermore, examination by PCR using primers A and B or A and C in Fig. 2 revealed that no DNA fragment is integrated in the attBf237 site in a V. parahaemolyticus strain KX-V191 (10) that is not infected with f237. These results suggest that f237 is integrated into the dif-like site of the large chromosome of KX-V237.
FIG. 4.
Location of the integration site of f237 on the large chromosome of V. parahaemolyticus KX-V237. The circle indicates a physical map of the KX-V237 large chromosome with restriction sites of NotI. The physical map is according to Tagomori et al. (16). NA and NH represent NotI fragments, and SA is an SfiI fragment. The probe for the f237 genome hybridized with the NA and SA fragments. dnaA is located on the NH fragment.
f237 is a closely related phage of CTXφ (7, 10, 17). The integration site on the host chromosome of CTXφ is a 17-bp element, attRS1 (3, 11). When compared with E. coli dif, the 28-bp sequence, including the majority of the attRS1 and its flanking region, was highly homologous to E. coli dif (25 out of 28 bases matched) (Fig. 3). In the whole genome sequence of V. cholerae N16961 (4), this 28-bp sequence corresponds to the position from the 1,564,104th base of chromosome 1, located nearly 180° away from the oriC on the circular chromosome. Also, the sequence is the most highly matched to E. coli dif in the whole genome sequence of the N16961 strain. All these results suggest that CTXφ also utilizes the dif-like site as a target for chromosomal integration.
Resolution of dimeric chromosomes at the dif site is a site-specific recombination event mediated by the tyrosine recombinases, XerCD (5, 13). Since the integration of phage genomes into the host chromosome is also a site-specific recombination event, there is a possibility that the chromosomal integration of vibrio phages is also mediated by host XerCD. However, considering that the overlap of the attPf237 or attRS1 sequence with the dif sequence is only partial (Fig. 3), it is also likely that the chromosomal integration of the vibrio phage is independent of XerCD, and the phages just use the dif-like site as one of the conserved sequences in bacterial genomes. To clarify whether the XerCD recombinases are involved in the chromosomal integration by vibrio phages, further studies including construction of xerCD-disrupted mutant strains of vibrios would be needed.
Because of the positioning of the duplicated region (attPf237) at one end of dif rather than in the center (Fig. 3), the consequence of integration of the f237 genome is that the whole dif site is regenerated at one end of the prophage. Thus, the integration should not destroy the dif site nor should it create two functional dif sites, both of which might be predicted to be bad for the host. dif (or its coordinate) has been found in a wide range of bacterial species (12). Recently, it was reported that Xanthomonas bacteriophage φLf integrated into a dif-like sequence (22 out of 28 bp identical to E. coli dif) (9). Likewise, it is possible that bacteriophages infecting other bacteria also use the dif site for chromosomal integration.
ADDENDUM IN PROOF
After acceptance of this article, a paper (K. E. Huber and M. K. Waldor, Nature 417:656-659, 2002) was published which reports that the CTXφ integration site overlaps with the dif site and that the integration requires XerC and XerD.
Acknowledgments
We thank the staff at the Kansai International Airport quarantine station for providing the V. parahaemolyticus strains used in this study.
This work was supported by the Research for the Future Programs (97L00101 and 97L00704) of the Japan Society for the Promotion of Science and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Chowdhury, N. R., S. Chakraborty, T. Ramamurthy, M. Nishibuchi, S. Yamasaki, Y. Takeda, and G. B. Nair. 2000. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 6:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniels, N. A., L. MacKinnon, R. Bishop, S. Altekruse, B. Ray, R. M. Hammond, S. Thompson, S. Wilson, N. H. Bean, P. M. Griffin, and L. Slutsker. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 3.Davis, B. M., and M. K. Waldor. 2000. CTXφ contains a hybrid genome derived from tandemly integrated elements. Proc. Natl. Acad. Sci. USA 97:8572-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidelberg, J. F., et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill, T. M. 1996. Features of the chromosomal terminus region, p. 1602-1614. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 6.Iida, T., O. Suthienkul, K.-S. Park, G.-Q. Tang, R. K. Yamamoto, M. Ishibashi, K. Yamamoto, and T. Honda. 1997. Evidence for genetic linkage between the ure and trh genes in Vibrio parahaemolyticus. J. Med. Microbiol. 46:639-645. [DOI] [PubMed] [Google Scholar]
- 7.Iida, T., A. Hattori, K. Tagomori, H. Nasu, R. Naim, and T. Honda. 2001. Filamentous phage associated with recent pandemic strains of Vibrio parahaemolyticus. Emerg. Infect. Dis. 7:477-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph, S. W., R. R. Colwell, and J. B. Kaper. 1982. Vibrio parahaemolyticus and related halophilic vibrios. Crit. Rev. Microbiol. 10:77-124. [DOI] [PubMed] [Google Scholar]
- 9.Lin, N.-T., R.-Y. Chang, S.-J. Lee, and Y.-H. Tseng. 2001. Plasmids carrying cloned fragments of RF DNA from the filamentous phage øLF can be integrated into the host chromosome via site-specific integration and homologous recombination. Mol. Genet. Genomics 266:425-435. [DOI] [PubMed] [Google Scholar]
- 10.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K.-S. Park, K. Yokoyama, K., Makino, H. Shinagawa, and T. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson, G. D., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 90:3750-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sciochetti, S. A., P. J. Piggot, and G. W. Blakely. 2001. Identification and characterization of the dif site from Bacillus subtilis. J. Bacteriol. 183:1058-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherratt, D. J., I. F. Lau, and F.-X. Barre. 2001. Chromosome segregation. Curr. Opin. Microbiol. 4:653-659. [DOI] [PubMed] [Google Scholar]
- 14.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusion. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Steiner, W. W., and P. L. Kuempel. 1998. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol. Microbiol. 27:257-268. [DOI] [PubMed] [Google Scholar]
- 16.Tagomori, K., T. Iida, and T. Honda. 2002. Comparison of genome structures of vibrios, bacteria possessing two chromosomes. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 17.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 18.Yamaichi, Y., T. Iida, K.-S. Park, K. Yamamoto, and T. Honda. 1999. Physical and genetic map of the genome of Vibrio parahaemolyticus: presence of two chromosomes in Vibrio species. Mol. Microbiol. 31:1513-1521. [DOI] [PubMed] [Google Scholar]