Abstract
Rob is regarded as a constitutively expressed protein, although little is known about how rob gene is regulated. We show here by reverse transcription-PCR that the transcriptional levels of rob are strongly down-regulated in response to the superoxide-generating agent paraquat (PQ). Repression reached a maximum of 20-fold after 10 min exposure at 10 μM PQ. The magnitude of rob repression was comparable to that of induction quantified for the most sensitive SoxS targets. β-Galactosidase expression with the rob2::lacZ transcriptional fusion indicates that down-regulation of rob expression takes place, at least in part, at the level of transcription initiation. Moreover, ca. 50% of the rob mRNA was degraded in <1 min after the addition of rifampin to inhibit transcription. This intrinsic short half-life, which is of obvious benefit for a rapid down-regulation after transcription ceases, was unaffected by the addition of PQ. No repression was observed in a soxR-null strain, indicating that the rob transcript level might be negatively modulated by the intracellular amounts of SoxS protein. Gel retardation assays support the idea that in vivo SoxS would block rob transcription directly.
Escherichia coli adapts to sublethal stress conditions by altering the transcription of a set of operons and regulons to restore homeostasis (15). The OxyR and SoxRS regulons deal with the threat of reactive oxygen species (27). Hydrogen peroxide oxidizes OxyR (encoded by oxyR) which, in turn, induces the transcription of a set of genes, including katG (catalase). The SoxRS response is a two-stage transcriptional process (29). SoxR is first activated by the univalent oxidation of its 2Fe-2S clusters in response to exposure to superoxide-generating reagents, such as paraquat (PQ). Oxidized SoxR stimulates the transcription of its only known target, soxS. SoxS is then produced in large amounts, leading to increased expression of genes of the SoxRS regulon, such as tolC, micF, and marA, that encode for an outer membrane protein, a regulatory RNA, and a positive transcriptional regulator, respectively.
Rob (rob) protein was first identified by its ability to bind the right border of the origin of the Escherichia coli chromosome (26). Rob is an abundant nucleoid-associated protein (up to 10,000 molecules per cell) (4), but its biological function remains unclear. At present, the only phenotype described for a rob-inactivated strain is increased susceptibility to organic solvents (30). Rob, MarA and SoxS are members of the same AraC/XylS family of transcriptional regulators (9, 28), and they are sufficiently similar to be able to activate, yet to different extents, a common subset of promoters (5, 19).
Most E. coli promoters are recognized by the rpoD-encoded σ70 factor, which is involved in the transcription of most of the genes expressed in unstressed exponentially growing cells (12). Multiple stresses, including entry into the stationary phase, trigger the synthesis of an alternative σS (or σ38) subunit (rpoS), and the RNA polymerase is directed to a specific set of promoters (the RpoS regulon) (11). σS expression is tightly regulated at the transcriptional, translational, and posttranslational levels (14). Two well-characterized RpoS-dependent genes are katE and osmY (osmotically inducible periplasmic protein).
We have recently designed and optimized a reverse transcription-multiplex PCR (RT-MPCR) procedure for the simultaneous detection and precise quantitation of both induction and repression of the in vivo transcript levels of well-defined sets of genes (24). We used this highly sensitive experimental approach here to quantify changes in the transcript levels of the 11 target genes outlined above in response to challenges posed by PQ. We emphasized that the finding of the SoxRS-mediated down-regulation of rob transcription upon PQ exposure might be relevant for the mechanism of transcriptional activation by SoxS regulator.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains are derived from E. coli K-12. Isogenic strains UC574 (arg56 nad113 araD81), UC1247 (ΔoxyR::kan), and UC1311 (rpoS::Tn10) have been previously described (1, 21). Strain UC1266 (ΔsoxR9::cat) was constructed by P1 transduction of the ΔsoxR9::cat (obtained from B. Weiss) mutant allele into strain UC574. Successful transfer was confirmed by screening for no DNA amplification with specific primers. It is known by complementation analysis that the ΔsoxR9::cat null mutation does not affect the expression of the nearby soxS gene (31). Strain M542 (λRS45:rob2::lacZ kan) was from J. L. Rosner (25). Bacteria were grown in M9 minimal medium as described previously (21). Overnight cultures were diluted into fresh medium (A600 = 0.03) and incubated at 37°C and 150 rpm to reach a turbidity (A600) of 0.2. At this stage, the bacteria were further grown in the absence or the presence of PQ for a fixed time period.
RNA purification and RT-MPCR.
Total RNA extraction and in vitro synthesis of external standard RNAs were as detailed (24). Bacterial RNA (0.5 μg) plus external standard RNAs (60 pg of gapA competitor and 0.1 pg of CYP1A noncompetitor) were retrotranscribed as described previously (21, 24). At least two independent RNA preparations were isolated for each experimental condition, with each RNA sample being retrotranscribed at least twice.
Primers were designed with Oligo 6.1.1/98 as detailed elsewhere (24). To obtain the highest specificity and acceptability for use in multiplex PCRs, primers were chosen to have a high Tm (≥81°C) and an optimal 3′ ΔG (≥−5.4°C) values. Primer sequences are available from the authors upon request. Primers for the regulatory RNA micF (Tm of 70°C) were in a separate set B, since its small size did not allow us to design primers with optimal characteristics for amplification of multiple target genes. Sets A and B also included primer pairs for amplification of gapA (internal standard and competitor external standard) and CYP1A (noncompetitor external standard). Forward primers were labeled with 6-carboxyfluorescein-N-hydroxysuccinimide ester, except micF with 6-carboxy-2′,4′,5′,7′,4,7-hexachloro-fluorescein.
The MPCR amplification was carried out in a mixture (25 μl [final volume]) containing MPCR buffer 3 (2.5 μl) supplemented with 1 mM MgCl2, a 250 μM concentration of each deoxynucleoside triphosphate, 0.2 μl of cDNA, 1.25 U of AmpliTaq Gold DNA polymerase, and primers at the following concentrations: (i) set A, 0.04 μM (katE), 0.04 μM (katG), 0.03 μM (rob), 0.04 μM (oxyR), 0.04 μM (soxS), 0.08 μM (rpoS), 0.04 μM (rpoD), 0.05 μM (tolC), 0.12 μM (marA), 0.11 μM (osmY), 0.03 μM (gapA), and 0.15 μM (CYP1A); and (ii) set B, 0.08 μM (micF), 0.01 μM (gapA), and 0.05 μM (CYP1A). Forward and reverse primers were used at identical concentrations. Twenty-eight cycles of PCR were performed with set A, with each cycle consisting of 1 min of denaturation at 94°C and 45 s of annealing and extension at 70°C. Twenty-seven cycles of 1 min of denaturation at 94°C, 15 s of annealing at 64°C, and 30 s of extension at 72°C were carried out with set B. These MPCR conditions were optimized as detailed elsewhere (24) to ensure that the amplifications were in the exponential phase and that the efficiencies remained constant in the course of the PCR.
After amplification, the fluorescent PCR fragments were separated and quantified in an ABI Prism 377 DNA Sequencer/GeneScan (Applied Biosystems) as detailed previously (24). Differences among PCR outcomes were normalized by dividing the fluorescent intensity of each band by that resulting from gapA amplification. As reported elsewhere (16, 23, 24), the potential variability of the reference gene was controlled by means of external standards. The levels of gapA in reference to the external standards remained essentially equal among the strains and experimental conditions investigated in this work. Consequently, changes detected with reference to the control gapA gene were accurately attributed to variations in the expression levels of the target genes under analysis. Samples for comparison of different experimental conditions or different bacterial strains were handled in parallel. Data are the means ± the standard errors of the means (SEM) from n (≥4) independent multiplexed PCR amplifications. Statistical comparisons were done by a hierarchical analysis of variance with SAS software (Statistical Analysis System v.6.03). The ratios between data from experimental and control samples represent the fold changes in gene expression.
Real-time PCR.
Real-time PCRs were performed in triplicate by using 50 ng of cDNA template, 0.3 μM concentrations of each primer, 3 mM MgCl2, 250 μM concentrations of each deoxynucleoside triphosphate, 0.75 U of platinum Taq DNA polymerase, and a 1:100,000 concentration of SYBR Green I dye (Roche) in a volume of 25 μl. Reactions were analyzed on an iCycler iQ real-time PCR system (Bio-Rad). Cycling conditions were as follows: 2 min at 95°C for the platinum Taq activation and 40 cycles for the melting (15 s at 95°C) and annealing-extension (30 s at 70°C) steps. These conditions generate specific PCR products of the desired lengths. No primer dimers were present. Investigated transcripts showed optimal PCR efficiencies: 1.02 for rob, 1.00 for soxS, 1.01 for marA, and 1.03 for gapA (control gene) in the range from 0.3 to 100 ng of cDNA input with high linearity (correlation coefficient ≥ 0.97). An absolute standard curve was constructed with the gapA competitor in the range from 109 to 102 molecules. The number of copies of the experimental transcripts were calculated from the linear regression of the standard curve: y = −3.325x + 40.07 (r2 = 0.98).
β-Galactosidase assays.
M542 (λRS45::rob2::lacZ kan) cells in M9 minimal medium were diluted into fresh medium and incubated until an optical density at 600 nm of 0.2 was reached. At this stage, the bacteria were further grown in the absence or presence of PQ for a fixed time period. β-Galactosidase activity was then assayed in permeabilized cells as described previously (10, 22). Data are from triplicate cultures.
Gel retardation assays.
A fragment of 355 bp (containing a putative SoxS binding site in rob promoter) was prepared by PCR with the primers 5′-CGAACCAATCTCTTCTGCATGAGCCAAT and 5′-ACAGGGGCTGATCCAGATGACCTTCC. Nonspecific binding was excluded by using a non-promoter-associated DNA fragment. Binding reactions and protein-DNA complexes separations were as described previously (8, 10). Each reaction contained ∼150 fmol of DNA. After electrophoresis, the gel was stained with a 1:10,000 concentration of SYBR Green I in TGE (25 mM Tris, 190 mM glycine, 0.1 mM EDTA [pH 8.3]) buffer for 30 min. Fluorescence was detected with the FMBIO II System (Hitachi).
RESULTS AND DISCUSSION
Quantitation of gene expression profiles in response to PQ.
The in vivo expression profiles of the 11 target genes were first quantified by exposing wild-type cells to increasing concentrations of PQ for 10 min. These PQ treatments modified the transcript levels of six genes, with five genes (soxS, micF, tolC, marA, and katG) being activated and one gene (rob) being down-regulated. The transcript levels of the remaining five genes (oxyR, rpoS, katE, osmY, and rpoD) were unaffected by PQ.
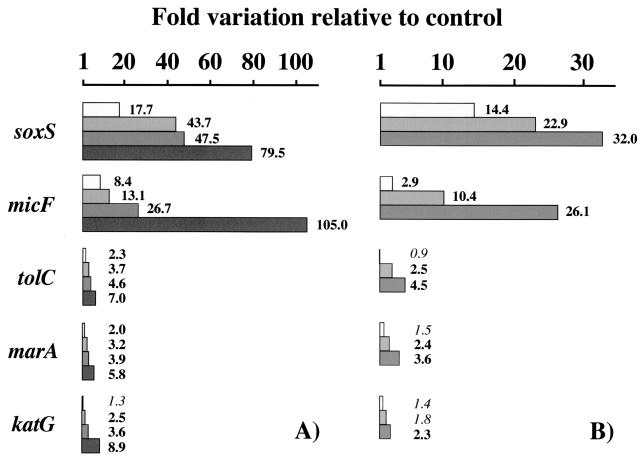
The genes whose expression was induced by PQ in wild-type cells are displayed in Fig. 1. The up-regulation of these genes was ablated by the mutational elimination of SoxR, but the activation of katG was strictly dependent on a functional OxyR (data not shown). Therefore, the PQ activation of the OxyR-regulated katG gene is attributed to the intracellular conversion of O2·− to H2O2. Previous studies have reported the PQ induction of soxS, micF, and mar transcription in a SoxRS-dependent manner (7, 20, 31). Our data are consistent with these previous studies, but they also provide new pieces of information.
FIG. 1.
Gene expression induction by PQ. Wild-type bacteria (UC574) were treated for 10 min with 1, 10, 100, and 500 μM PQ (A) or with 100 μM PQ for <1 min (immediately after the addition of PQ) and for 5 and 10 min (B). Fold variations in transcript levels were plotted for the genes whose expression was significantly induced by PQ. Boldface type indicates statistically significant increments relative to the untreated control bacteria.
First, significant induction was quantified for each SoxRS-regulated gene from the minimal dose assayed of 1 μM PQ (Fig. 1A). Prior inductions were detected under more acute treatment conditions. Second, genes displayed notable differences with respect to the extent of their individual in vivo activation; soxS (the specific target of oxidized SoxR) showed the highest induction levels, and marA (the regulator of Mar regulon) showed the lowest induction levels (e.g., 17.7- versus 2.0-fold induction at 1 μM PQ). Time course experiments at 100 μM PQ (Fig. 1B) confirmed these relative inducibilities; a large upregulation of 14.4-fold was quantified for the most highly induced soxS gene at a remarkably short time of exposure (<1 min after the addition of the oxidant), but the less-responsive genes (such as marA) required a minimum of 5 min of exposure to result in a significant variation of 2.4-fold. Third, our data show that tolC mRNA increases after the exposure of E. coli to PQ. The induction was visible over a range of PQ concentrations and exposure times; it was abolished in bacteria lacking SoxR, and its magnitude was similar to that quantified for the marA gene. Previous assignment of tolC to the SoxRS regulon was based on elevation of TolC protein levels in bacteria with multiple copies of soxS and on finding of a possible sox-mar-rob box sequence upstream of tolC (2). Fourth, PQ stress conditions (10 μM, 10-min exposure) that were much less stringent than those regularly used to induce the SoxRS regulon generate sufficient H2O2 to also induce the OxyR regulon.
Rob is regarded as a constitutively expressed protein, although little is known about how rob is regulated. We show here for the first time that rob transcript levels are strongly down-regulated in response to PQ. As shown in Tables 1 and 2, repression increased with PQ concentration and time of exposure to reach a maximum of 20-fold. Significant decreases of 2.1- and 9.9-fold were readily seen for rob mRNA immediately after the addition of 100 μM PQ or after 10 min of exposure to just a 1 μM concentration of this oxidant. The magnitude of rob repression was comparable to the level of induction quantified for the most sensitive SoxS targets such as micF (Fig. 1).
TABLE 1.
Down-regulation of rob in response to PQ: PQ dosea
| Dose (μM) | Mean rob/gapA ratio ± SEMb | Variationc (fold) |
|---|---|---|
| 0 | 1.58 ± 0.14 | 1.0 |
| 1 | 0.16 ± 0.02 | 9.9* |
| 10 | 0.08 ± 0.01 | 19.8* |
| 100 | 0.14 ± 0.04 | 11.3* |
The dose response results for wild-type bacteria treated for 10 min with PQ at the indicated concentrations are shown.
That is, the mean values of the fluorescence signal of the rob target sequence relative to that of gapA (internal standard) ± the SEM.
That is, the fold variations in rob transcript levels (for untreated versus treated bacteria). Statistically significant variations are indicated with an asterisk.
TABLE 2.
Down-regulation of rob in response to PQ: exposure timea
SoxRS dependence of down-regulation of rob.
To dissect the regulatory cascade of rob repression in response to PQ, strains carrying mutations that eliminate the SoxR, OxyR, or RpoS major global regulators were used in conjunction with wild-type bacteria (Table 3). It is clear that rob mRNA did not decrease in the ΔsoxR9::cat mutant strain, suggesting a negative regulation via the soxRS genes. In contrast, PQ elicited a profound repression in bacteria with null mutations in other regulators, such as oxyR or rpoS, indicating that SoxRS proteins might be the unique mediators of the response.
TABLE 3.
SoxR-mediated regulation of rob repression in response to PQ
| Genotype (strain) | PQ treatmenta | Mean rob/gapA ratio ± SEMb | Variationc (fold) |
|---|---|---|---|
| Wild type (UC574) | − | 1.06 ± 0.03 | 1.0 |
| + | 0.08 ± 0.01 | 13.3* | |
| ΔsoxR9::cat (UC1266) | − | 1.10 ± 0.09 | 1.0 |
| + | 0.94 ± 0.09 | 1.2 | |
| ΔoxyR::kan (UC1247) | − | 1.13 ± 0.06 | 1.0 |
| + | 0.06 ± 0.01 | 18.8* | |
| rpoS::Tn10 (UC1311) | − | 1.20 ± 0.12 | 1.0 |
| + | 0.10 ± 0.01 | 12.0* |
Bacteria carrying the indicated genetic marker were treated (+) or not (−) with PQ at 100 μM for 10 min.
Mean values of the fluorescence signal of the rob target sequence relative to that of gapA (internal standard) ± the SEM.
Fold variations in rob transcript levels (for untreated versus treated bacteria). Statistically significant variations are indicated with an asterisk.
Transcriptional regulation of rob.
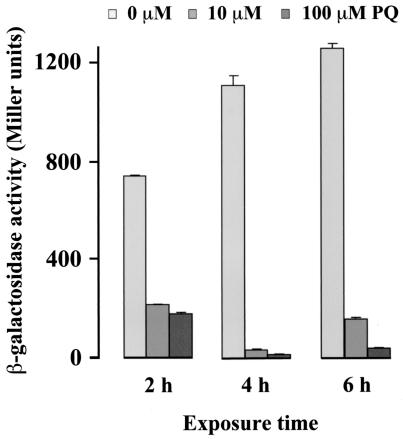
The steady-state abundance of every mRNA is dependent on both its rate of synthesis and its rate of decay. To address the contribution of transcription initiation to the regulation of rob, a strain with a rob2::lacZ transcriptional fusion (25) was tested. Treatments with PQ decreased the rob2::lacZ-directed β-galactosidase activity: minimal values of ≤30 Miller units were quantitated at 4 h of exposure to ≥10 μM PQ, which is <5% of the activity found in control cells (Fig. 2). This result indicates that the SoxRS-dependent down-regulation of rob expression takes place, at least in part, at the level of transcription initiation.
FIG. 2.
Effect of PQ on β-galactosidase activity of cells containing a rob2::lacZ fusion. Bacteria (M542) were treated for 2, 4, and 6 h with 10 or 100 μM PQ. Untreated bacteria were used as a control. Values are the means ± the SEM of β-galactosidase units. Bacteria with lacZ expressed from promoter insensitive to PQ treatments (nrdB::lacZ) were used to exclude the possibility that PQ might inactivate β-galactosidase protein.
Absolute quantitation and measurement of mRNA stability by real-time PCR.
A short half-life is of obvious benefit for a rapid down-regulation after transcription cease. To test whether intrinsic or PQ-induced mRNA stability makes a contribution to the down-regulation of rob, we investigated the effect of rifampin (a transcriptional inhibitor), alone or in combination with PQ treatment, on rob mRNA levels (Table 4). For comparison, three other mRNA species were also studied: those of the soxS and marA global regulators and that of the gapA housekeeping gene.
TABLE 4.
Absolute quantitation of transcript levels as determined by real-time PCRa
| Gene | Time (min) | No. of transcript molecules/pg of total RNA ± SEM (relative value)b
|
|||
|---|---|---|---|---|---|
| −Rif −PQ | −Rif +PQ | +Rif −PQ | +Rif +PQ | ||
| rob | <1 | 149 ± 1.0 (1.00) | 83 ± 3.0 (0.56)* | 88 ± 0.7 (0.59)* | 75 ± 4.3 (0.50)* |
| 6 | 150 ± 18.6 (1.01) | 11 ± 0.4 (0.07)* | 6 ± 0.7 (0.04)* | 6 ± 0.5 (0.04)* | |
| soxS | <1 | 66 ± 0.2 (1.00) | 768 ± 16.5 (11.64)* | 26 ± 4.1 (0.39)* | 163 ± 14.6 (2.47)* |
| 6 | 38 ± 1.4 (0.58)* | 1,924 ± 96.7 (29.15)* | 3 ± 0.1 (0.05)* | 10 ± 0.6 (0.15)* | |
| marA | <1 | 64 ± 5.7 (1.00) | 90 ± 3.7 (1.41)* | 32 ± 0.2 (0.50)* | 36 ± 2.2 (0.56)* |
| 6 | 62 ± 1.7 (0.97) | 269 ± 14.1 (4.62)* | 4 ± 0.1 (0.06)* | 7 ± 0.1 (0.11)* | |
| gapA | <1 | 3,233 ± 232.6 (1.00) | 2,671 ± 49.0 (0.83) | 3,003 ± 104.7 (0.93) | 3,010 ± 173.1 (0.93) |
| 6 | 3,340 ± 20.7 (1.03) | 3,273 ± 353.9 (1.01) | 2,367 ± 93.2 (0.73)* | 2,249 ± 170.5 (0.70)* | |
Time course results of the response of wild-type bacteria with (+) or without (−) 100 μM PQ in the absence (−) or presence (+) of 100 μg of rifampin (Rif)/ml.
Statistical significance is indicated by an asterisk.
As quantitated by real-time PCR, bacteria undergoing early exponential growth in minimal medium produced ca. 150 rob mRNA molecules per pg of total RNA. This transcript level was ∼2-fold higher than those of soxS and marA, but >20-fold lower than that of gapA (clearly, the most abundant of the four mRNA species). PQ treatments decreased the yield of rob mRNA molecules and concomitantly increased the yields of soxS and marA transcripts; the number of gapA mRNA molecules being unaffected.
Intrinsic rob mRNA stability was very low, since ∼50% of the mRNA was degraded in <1 min after the addition of transcription inhibitor. This short half-life is typically beneficial for early response genes (such as soxS or marA), whereas long half-lives are of benefit for ubiquitous housekeeping genes (such as gapA) that do not require rapid induction or repression (reviewed in reference 6). According to the data presented in Table 4, rob mRNA decay was apparently not affected by PQ.
Biological significance of rob down-regulation in response to superoxide stress.
In vivo and in vitro studies have shown that Rob can activate the transcription of a subset of target genes of the SoxRS and Mar regulons (3, 5, 13). On the other hand, Rob has a high basal level of expression. A recent calculation estimates the intracellular level of Rob in exponentially growing E. coli in 10,000 molecules per cell (4). The high basal amounts of Rob and the finding that Rob binds many promoter sites more tightly than do SoxS and MarA (17) leave an important question unanswered. If the basal expression of Rob is high enough to saturate some or most of the SoxS- and MarA-regulated promoters, how can SoxS and MarA exert their effect in response to specific stress conditions? The down-regulation of rob expression described here provides for the first time a consistent and experimentally verified explanation. Therefore, the induction of soxS and marA concomitantly with the repression of rob might contribute synergistically to the transcriptional response of the dozen or more promoters with a common “marbox” (18).
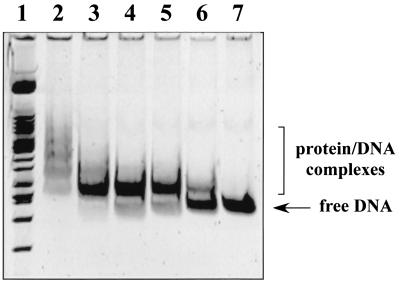
Besides, we have shown that the lack of SoxR eliminates the possibility of rob repression, suggesting that the rob transcript level might be negatively modulated by the intracellular amounts of SoxS protein. Therefore, we addressed, by performing gel retardation assays with a DNA fragment of rob (positions −288 to +67, relative to the initiation codon), the possibility that SoxS would block rob transcription directly. As shown in Fig. 3, clear retarded bands were found with increasing amounts of SoxS. Similar results were obtained with micF promoter (data not presented).
FIG. 3.
In vitro SoxS binding to rob DNA sequence as assessed by gel retardation assays. A DNA fragment containing the segment from positions −288 to +67 of rob (relative to the initiation codon) was incubated with purified His6-SoxS protein (10). The amounts of SoxS in the 25-μl reactions were as follows: 0.38 pmol (lane 6), 0.75 pmol (lane 5), 1.5 pmol (lane 4), 3 pmol (lane 3), and 6 pmol (lane 2). Control without SoxS (lane 7) and molecular weight markers (lane 1) are also shown.
Final remarks.
The data reported here suggest that Rob levels might be tightly controlled, like those of SoxS and MarA. Briefly, superoxide stress upon PQ exposure activates SoxR, thereby increasing the intracellular amounts of SoxS. SoxS in turn represses rob transcription, thereby assisting SoxS and MarA to distinguish bona fide sites from non-promoter-associated sites. Therefore, the rob promoter itself might have a “soxbox” sequence (10), one degenerated enough to be functional in transcription activation although still an effective repressor site.
Acknowledgments
We are deeply grateful to J. L. Rosner and R. W. Wolf, Jr., for providing us with strain M542 and purified SoxS protein, respectively.
This work was supported by grant PB98-1627 (DGES). M.M. was the recipient of a predoctoral fellowship from the Spanish Ministry of Education and Culture.
C.M. and M.M. contributed equally to this study.
REFERENCES
- 1.Abril, N., T. Roldan-Arjona, M. J. Prieto-Alamo, A. A. van Zeeland, and C. Pueyo. 1992. Mutagenesis and DNA repair for alkylation damages in Escherichia coli K-12. Environ. Mol. Mutagen. 19:288-296. [DOI] [PubMed] [Google Scholar]
- 2.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariza, R. R., Z. Li, N. Ringstad, and B. Demple. 1995. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 177:1655-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennik, M. H., P. J. Pomposiello, D. F. Thorne, and B. Demple. 2000. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J. Bacteriol. 182:3794-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey, M., and S. T. Smale. 1999. Modes of regulating mRNA abudance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Chou, J. H., J. T. Greenberg, and B. Demple. 1993. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J. Bacteriol. 175:1026-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fawcett, W. P., and R. E. Wolf, Jr. 1994. Purification of a MalE-SoxS fusion protein and identification of the control sites of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 14:669-679. [DOI] [PubMed] [Google Scholar]
- 9.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffith, K. L., and R. E. Wolf, Jr. 2001. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 40:1141-1154. [DOI] [PubMed] [Google Scholar]
- 11.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 12.Ishihama, A. 1999. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells 4:135-143. [DOI] [PubMed] [Google Scholar]
- 13.Jair, K. W., X. Yu, K. Skarstad, B. Thöny, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 15.Lin, E. C. C., and A. S. Lynch. 1996. Regulation of gene expression in Escherichia coli. Chapman and Hall, New York, N.Y.
- 16.Manchado, M., C. Michán, and C. Pueyo. 2000. Hydrogen peroxide activates the SoxRS regulon in vivo. J. Bacteriol. 182:6842-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, R. G., W. K. Gillette, M. N. I., and J. L. Rosner. 2002. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol. Microbiol. 43:355-370. [DOI] [PubMed] [Google Scholar]
- 18.Martin, R. G., W. K. Gillette, S. Rhee, and J. L. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 19.Martin, R. G., W. K. Gillette, and J. L. Rosner. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 35:623-634. [DOI] [PubMed] [Google Scholar]
- 20.Martin, R. G., K. W. Jair, R. E. Wolf, and J. L. Rosner. 1996. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178:2216-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michán, C., M. Manchado, G. Dorado, and C. Pueyo. 1999. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J. Bacteriol. 181:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Prieto-Álamo, M. J., J. Jurado, R. Gallardo-Madueño, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 24.Pueyo, C., J. Jurado, M. J. Prieto-Alamo, F. Monje-Casas, and J. Lopez-Barea. 2002. Multiplex reverse transcription-polymerase chain reaction for determining transcriptional regulation of thioredoxin and glutaredoxin pathways. Methods Enzymol. 347:441-451. [DOI] [PubMed] [Google Scholar]
- 25.Rosner, J. L., B. Dangi, A. M. Gronenborn, and R. G. Martin. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skarstad, K., B. Thöny, D. S. Hwang, and A. Kornberg. 1993. A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268:5365-5370. [PubMed] [Google Scholar]
- 27.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 28.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touati, D. 2000. Sensing and protecting against superoxide stress in Escherichia coli: how many ways are there to trigger soxRS response? Redox Rep. 5:287-293. [DOI] [PubMed] [Google Scholar]
- 30.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, J., and B. Weiss. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]