Abstract
Proteins H-NS and Hha form a nucleoprotein complex that modulates expression of the thermoregulated hly operon of Escherichia coli. We have been able to identify two H-NS binding sites in the hly regulatory region. One of them partially overlaps the promoter region (site II), and the other is located about 2 kbp upstream (site I). In contrast, Hha protein did not show any preference for specific sequences. In vitro, temperature influences the affinity of H-NS for a DNA fragment containing both binding sites and H-NS-mediated repression of hly operon transcription. Deletion analysis of the hly regulatory region confirms the relevance of site I for thermoregulation of this operon. We present a model to explain the temperature-modulated repression of the hly operon, based on the experiments reported here and other, preexisting data.
Temperature changes represent signals sensed and processed by the biochemical machinery of the bacterial cell to favor the best possible adaptation and ultimately to allow the microorganism to survive when the environment changes (for a review, see reference 18). A good example of thermosensing is that of many pathogenic bacteria, which sense the temperature increase that they encounter when they enter the human host and respond by triggering the expression of different virulence factors (26). In fact, it is well established that expression of many virulence factors is regulated by environmental parameters, such as temperature, osmolarity, pH, and oxygen availability (11, 12, 27). In spite of the extensive studies carried out in recent years, many molecular aspects of the mechanisms that switch transcription of virulence genes in response to environmental changes remain to be elucidated.
The toxin α-hemolysin (Hly), produced by many uropathogenic Escherichia coli strains, is one of the many virulence factors whose expression is thermomodulated (30). The hly genes can be either chromosome or plasmid encoded. The hly operon harbored by plasmid pHly152 (36), one of the most extensively studied hly operons, displays many unusual features. Essential regulatory sequences are spread upstream from the promoter and include a 650-bp sequence, the so-called hlyR sequence (42), located more than 1.5 kbp upstream of hlyC, the first structural gene of the operon. An antiterminator element (the ops element), frequently found in long bacterial operons such as those encoding lipopolysaccharides and capsule synthesis or fertility, is located within the hlyR sequence (32). Furthermore, an IS2 insertion element separates hlyR from the promoter region (Fig. 1). Deletion of hlyR results in an unnatural repression of hemolysin expression (4, 42). This phenotype could be interpreted as being caused by the loss of the above mentioned ops element, but this has not yet been demonstrated. In addition to this as yet unclear antitermination mechanism, the hly operon is regulated by both temperature and osmolarity (29, 30). Studies focused on the regulatory features of the hly operon of plasmid pHly152 led to the identification of the Hha protein (4, 33) and to the finding that hha mutants are partially derepressed under conditions involving temperature- and osmolarity-mediated hemolysin repression. The Hha protein belongs to a new family of modulators, which includes, among others, the YmoA protein, a temperature-dependent modulator of the expression of different virulence factors in Yersinia enterocolitica (5). Both proteins display a high degree of conservation, with 82% of their amino acid sequences identical, and are functionally interchangeable (2, 7, 28).
FIG. 1.
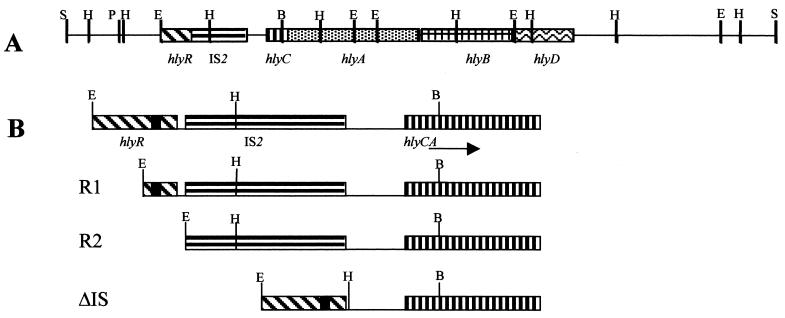
Schematic representation of the upstream and coding regions of the hly operon of plasmid pHly152. The hlyR sequence, the insertion element IS2, and sequences corresponding to the structural genes of the operon (hlyC, hlyA, hlyB, and hlyD) are indicated as bars with different patterns. The black box, in part B, indicates the ops sequence. E, H, S, P, and B correspond to restriction sites for EcoRI, HindIII, SalI, PstI, and BamHI, respectively. R1, R2, and ΔIS correspond to the deletions generated in this study.
Recently it has been found that Hha interacts with the nucleoid-associated protein H-NS (35). H-NS, a major component of the bacterial nucleoid, plays a central role as a modulator of gene expression in response to osmolarity and temperature changes (for reviews, see references 1, 41, and 43). In fact, H-NS, which binds preferentially to curved DNA (44) and is able to generate bends in noncurved DNA (38), affects the expression of a large number of genes (1, 16, 24) and is considered a general negative transcriptional regulator. Different models have been suggested to explain the mechanism by which H-NS represses gene expression. According to one model, termed “transcriptional silencing” (14), H-NS binding to DNA would occlude RNA polymerase access through a generalized effect on DNA compaction. Another model includes a direct competition between H-NS and the RNA polymerase for overlapping binding sites on or near the promoter (10, 40). Alternatively, H-NS would exert its repression through modifications of DNA supercoiling (17, 20, 39).
Interaction of H-NS with proteins other than Hha has been previously reported, but the significance of such interactions remains unclear (8, 22, 25, 31). In contrast, the phenotype of the hha/hns double mutants, which have lost both thermo- and osmoregulation (35), indicates a clear role of the Hha-H-NS complex in the thermo-osmotic regulation of the hly operon. For this work, we have further investigated the mechanism underlying Hha- and H-NS-mediated thermoregulation of the hly operon. We show that, rather than Hha, H-NS is the protein that binds at specific DNA sequences. In addition, we show that temperature is a critical factor in the modulation of the H-NS affinity for two distant DNA sequences of the regulatory region of the hly operon. A delicate balance between the moderate affinity of H-NS for its target sequences in this region and temperature-mediated changes in DNA flexibility is proposed as the factor allowing H-NS to generate a nucleoprotein complex (including Hha as well) that renders the hly operon repressed at low temperature.
MATERIALS AND METHODS
Bacterial strains, plasmids and media.
E. coli strains 5K (21), BSN26 (hha+ hns+) (19), BSN26H (hha hns+) (35), BSN27 (hha+ Δhns) (19), and BSN27H (hha Δhns) (35) and plasmids pHly152 (36) and pANN202312-R (13) have been previously described.
Plasmids pLGHlyR0, pLGHlyR1, pLGHlyR2, and pLGHlyΔIS are pLG338-30 (6) derivatives. Plasmid pLGHlyR0 was constructed by cloning the complete hly operon of plasmid pHly152, from the EcoRI site upstream of the hlyR sequence to the SalI site downstream of the hlyD sequence (Fig. 1). To do this, the EcoRI-BamHI fragment corresponding to the regulatory region of the hly operon was first cloned into pLG338-30, generating pLGR0. Upon BamHI-SalI digestion, the 11.7-kbp BamHI-SalI fragment of the hly operon from plasmid pHly152 was inserted, generating pLGHlyR0.
A similar strategy was used to construct plasmids pLGHlyR1, pLGHlyR2, and pLGHlyΔIS, which contain different deletions of the regulatory region of the hly operon (Fig. 1). The EcoRI-BamHI fragment of this plasmid was first modified to generate the deletions that were synthesized by PCR, using the primers described in Table 1. Primers HlyR1/HlyBam and HlyR2/HlyBam, containing EcoRI and BamHI restriction sites, were used to amplify the different EcoRI-BamHI fragments that were cloned into pLG338-30, generating plasmids pLGR1 and pLGR2, respectively. To construct the EcoRI-BamHI fragment containing the deletion of the IS2 element, we used primers HlyR0/ISA, with EcoRI and HindIII restriction sites, and ISB/HlyBam, with HindIII and BamHI restriction sites. These fragments were digested with HindIII, ligated, and then cloned as EcoRI-BamHI fragments into pLG338-30, obtaining plasmid pLGΔIS. We reconstructed the hly operon in all these plasmids by cloning the 11.7-kbp BamHI-SalI fragment of the hly operon of plasmid pHly152 to generate plasmids pLGHlyR1, pLGHlyR2, and pLGHlyΔIS.
TABLE 1.
Oligonucleotides used in this study to amplify DNA fragments corresponding to different deletions (HlyR1 and HlyBam to amplify the R1 fragment, HlyR2 and HlyBam to amplify the R2 fragment, and HlyRO/ISA and ISB/HlyBam to amplify the fragments corresponding to ΔIS)
| Oligonucleotide | Sequence |
|---|---|
| HlyRO | 5′-GGGGAATTCCAAGCGAAGTCCA-3′ |
| HlyBam | 5′-GTTTTGGGATCCACCCTGATGG-3′ |
| HlyR1 | 5′-ACTCCTTCAGAATTCTGGTCCC-3′ |
| HlyR2 | 5′-GGGAATTCTCCAGACAACCAA-3′ |
| ISA | 5′-ATCTCATAAAGCTTGATGC-3′ |
| ISB | 5′-TATCTGAAGCTTGACGAAGT-5′ |
Gel retardation assays.
Binding of H-NS to DNA was carried out in 20 μl of 40 mM Tris-HCl (pH 8.0)-100 mM KCl-1 mM dithiothreitol (DTT)-5 mM K phosphate-5% glycerol-50 mM EDTA, containing increasing concentrations of purified H-NS (0.12 to 0.48 μM) and 50 ng of DNA. Either linear DNA corresponding to the EcoRI-BamHI fragment of plasmid pLGR0 or the supercoiled DNA corresponding to plasmid pLGR0 was used. After incubation for 20 min at 25 or 37°C, the protein-DNA complexes were subjected to electrophoretic separation at 100 V at constant temperature (25 or 37°C) in 0.8% agarose gels in 0.5× Tris-borate-EDTA buffer. The DNA was visualized by ethidium bromide stain.
Gel competition retardation assay.
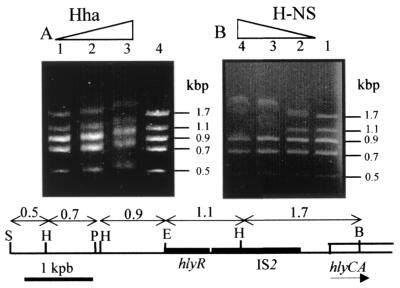
The DNA fragments used in this assay were obtained by digesting the SalI-BamHI fragment of the hly operon (Fig. 2). This fragment was first EcoRI digested, and the two resulting fragments were then purified. The SalI-EcoRI fragment was subsequently cut with HindIII and PstI, thus generating fragments of 0.1, 0.5, 0.7, and 0.9 kbp. The band corresponding to the 0.1-kbp PstI-HindIII fragment was not detected under the electrophoretic conditions used. The EcoRI-BamHI fragment was HindIII digested, generating fragments of 1.1 and 1.7 kbp. All these fragments were mixed either with Hha (19.12 to 76.48 μM) or with H-NS (0.24 to 0.97 μM) and incubated 10 min at 37°C under the conditions described above and then loaded in 1.5% agarose gels. Electrophoresis was carried out at room temperature, and the DNA was visualized by ethidium bromide staining.
FIG. 2.
Effect of increasing concentrations of Hha (A) and H-NS (B) proteins on the electrophoretic mobility of DNA fragments from the regulatory region of the hly operon. These fragments and their sizes are shown in the lower part of the figure, where the sequences corresponding to hlyR and IS2 (black boxes) and to the hlyC and hlyA genes (white boxes) and the restriction sites for SalI (S), PstI (P), HindIII (H), EcoRI (E), and BamHI (B) are indicated. Lanes 1, 2, 3, and 4 of panel A contained 19.12, 38.24, 76.48, and 0 μM Hha protein, respectively. Lanes 1, 2, 3 and 4 of panel B contained 0, 0.24, 0.48 and 0.97 μM H-NS protein, respectively.
In vitro transcription assay.
Transcription reactions containing 150 ng of supercoiled plasmid DNA (pANN202-312-R) and 1 U of E. coli RNA polymerase (USB) were performed in 40 mM Tris-HCl (pH 8.0)-10 mM MgCl2-5 mM DTT-200 μM ATP, CTP, GTP, and UTP-4 U of RNasin (Promega)-50 ng of BSA-150 mM KCl. The reaction mixtures were incubated for 30 min at 25°C or 37°C, with or without increasing amounts of H-NS protein (0.39 to 1.17 μM). The reactions were stopped by the addition of 1 μl of 0.5 M EDTA, 10 μg of carrier tRNA, 3 μl of 3 M sodium acetate (pH 5.4), and 90 μl of ethanol. RNA was precipitated and analyzed by primer extension.
Primer extension of RNAs.
The RNA was resuspended in a solution containing 40 mM Tris-HCl (pH 7.5)-20 mM MgCl2-50 mM NaCl-8 U of RNasin-5 pmol of the oligonucleotide used (5′-AACTTAAATTATATGATTAAA-3′), which hybridizes coordinates 2466 to 2486 (see Fig. 7). The mixture was heated for 5 min at 90°C and then allowed to cool slowly to 20°C. The solution was then put on ice for 5 min, and the RNA was precipitated by the addition of a 1/10 volume of 3 M sodium acetate (pH 5.4) and 2.5 volumes of ethanol. Precipitated RNA was resuspended in 5 μl of water, and the primer was extended in a solution containing 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 0.5 mM spermidine, 10 mM DTT, 8 U of RNasin (Promega), 0.2 mM each dNTP (except dATP, which was 100 μM), 2 μCi of [α-32P]dATP (3,000 Ci/mmol; Amersham Pharmacia Biotech), and 10 U of AMV-reverse transcriptase (Promega) in a total volume of 10 μl.
FIG. 7.
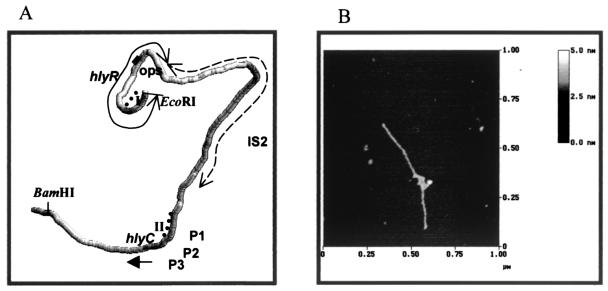
(A) Computer-generated prediction of the intrinsic curvature of the 3.1-kbp fragment covering the hly regulatory region and part of the structural genes. Dotted lines indicate the H-NS binding sites I and II. (B) AFM image in dry air of the 2.8-kbp EcoRI-BamHI fragment in HEPES-MgCl2 buffer adsorbed on bare mica (see Materials and Methods for details). The molecule shown is representative of many others observed.
The reaction mixture was incubated at 42°C for 45 min and then stopped by addition of 0.5 μl of 0.5 M EDTA and 30 μl of 10 mM Tris-HCl (pH 7.5)-1 mM EDTA. The sample was filtered through a 1-ml Sephadex G-50 spin column to eliminate the nonincorporated labeled nucleotide. The eluted cDNA was precipitated by the addition of 4 μl of 3 M sodium acetate (pH 5.4) and 200 μl of ethanol. The cDNA was analyzed in urea-6% polyacrylamide gel electrophoresis (PAGE) gels and visualized by autoradiography.
Measurement of hly operon transcription by RNase protection assay.
Total cellular RNA was isolated from mid-exponential-phase cultures (optical density at 600 nm = 0.5) using the acid phenol method (23). Plasmid pGEMhlyC (33) contains a fragment of the hlyC gene under the control of the T7 RNA polymerase promoter. Linearized (NdeI) pGEMhlyC was used as template to obtain antisense radiolabeled probes to the hlyC gene by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]UTP. Purity of the probe was checked by urea-6% PAGE.
For the RNase protection assay, 25 μg of total RNA was hybridized to an excess of radiolabeled probe and the nonhybridized RNA and probe were degraded with RNase-ONE (Promega). The protected probe was separated in urea-6% PAGE gels and visualized by autoradiography.
DNase I footprinting.
Supercoiled plasmid pLGHlyR0 (100 ng per sample) was preincubated for 20 min at 25°C with the appropriate concentrations of purified H-NS. After DNase I treatment, the partial-digestion DNA products were ethanol precipitated and subjected to 30 cycles of linear PCR using 5′-end 32P-labeled primer and TaqI polymerase (Polymed) essentially as previously described (11). The extension PCR products were separated on a 7% sequencing gel. The convergent oligonucleotide pair Hly543 (5′-CCAAGCGAAGTCCATCCCCCTCC-3′ [coordinates 1 to 28]) and Hly544 (5′-CTTGAAGGAGTGAGTTTGGATATGG-3′ [coordinates 473 to 448]) and the convergent oligonucleotide pair Hly545 (5′-CCTGGTCATTATCTGGAATTTGACG-3′ [coordinates 2032 to 2056]) and Hly546 (5′-CCCATAGCCAGGATACATGCCC-3′ [coordinates 2552 to 2531]) were used to detect H-NS protection on both DNA strands at sites I and II, respectively.
Atomic force microscopy.
Samples were prepared by placing a drop (2 μl) of DNA solution onto freshly cleaved green mica (Ashville-Schoonmaker Mica Co., Newport News, Va.). After adsorption for 5 min at room temperature, the samples were rinsed for 10 s in a jet of deionized water of 18 MΩ/cm from a Milli-Q water purification system (Millipore, Molsheim, France), directed onto the surface with a squeeze bottle. The samples were dried in an isolated chamber under pure N2 pressure for 30 min before imaging in the atomic force microscope (AFM) (37).
The samples were imaged in a Nanoscope III Multimode AFM (Digital Instruments Inc., Santa Barbara, Calif.), operating in tapping mode in air (15), at a scan rate of 1 to 2 Hz. The AFM probes were 125-μm-long monocrystal silicon cantilevers with integrated conical Si tips (Nanosensors GmbH, Norderfriedrichskoog, Germany) with an average resonance frequency of ≈300 kHz and a spring constant K of ≈35 N/m. The cantilever is rectangular, with a tip radius as given by the supplier of 5 to 15 nm, a cone angle of 35°, and a high aspect ratio. In general, the images were obtained at room temperature (23 ± 2°C), and the relative humidity was typically 55%.
Preparation of H-NS and Hha proteins.
Hha and H-NS proteins were purified as described previously (see references 9 and 35, respectively).
Curvature predictions.
Computer-generated predictions of intrinsic curvature were obtained with 3D-WEDGE, a program developed by G. Micheli (11).
Nucleotide sequence accession numbers.
Accession numbers of DNA sequences (all previously published) used in this work are x07565 (hlyR sequence), m14107 (plasmid pHly152 hlyC, A, B and D genes), and m18426 (insertion element IS2). These sequences are partially overlapping and were used to create a composite of the upstream regulatory region of the hly operon.
RESULTS
Interaction between H-NS and the regulatory region of the hly operon.
We had previously tested binding of Hha and H-NS proteins to different DNA fragments of the hly regulatory region in the presence of competitor DNA. This experimental approach did not allow us to determine preferred binding sites for either protein (35). To find such sites, we have used a different approach: we performed competition gel retardation assays, selecting a mixture of DNA fragments corresponding to the hly regulatory region and increasing concentrations of either Hha or H-NS. As seen from Fig. 1 and the lower part of Fig. 2, the SalI-BamHI restriction fragment of plasmid pHly152 spans approximately 4.9 kb upstream of the BamHI site located at hlyC, the first structural gene of the hly operon. This fragment includes the 2.8-kbp region (the 1.1-kbp EcoRI-HindIII fragment plus the 1.7-kbp HindIII-BamHI fragment) that should contain all the regulatory elements controlling the expression of this operon. To study the interaction between Hha and H-NS proteins and this DNA region, competition gel retardation assays were performed using a mixture of five DNA restriction fragments derived from the SalI-BamHI fragment (Fig. 2). The mixture was incubated in the presence of increasing concentrations of either Hha or H-NS. We found that the Hha protein does not display a preferential binding for any one of the five DNA fragments present in the reaction mixture (Fig. 2A). In contrast, H-NS exhibits a preferential interaction with the two fragments (the 1.7- and 1.1-kbp fragments) that contain predicted regulatory sequences (Fig. 2B). The fact that both fragments are preferentially retarded by H-NS suggested that there is more than one binding site for this protein within the regulatory sequences of the hly operon. Furthermore, since H-NS participates in the thermoregulation of the hly operon expression, it could be envisaged that the interaction of H-NS with these sites could be modulated by temperature within the physiologically relevant range.
Temperature affects the H-NS affinity for the 2.8-kbp hly regulatory region.
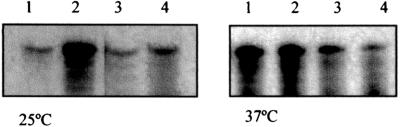
To determine whether the interaction of H-NS with the hly regulatory sequences could be affected by temperature, H-NS binding experiments were performed. Increasing concentrations of H-NS were mixed and incubated with the complete 2.8-kb EcoRI-BamHI linear DNA fragment at constant temperature (either 25°C or 37°C), and the interaction was determined by gel retardation assays, performed at the same temperature. The results obtained clearly indicate that H-NS shows more in vitro affinity to the DNA fragment at 25°C (Fig. 3A) than at 37°C (Fig. 3B). To investigate this temperature effect further, we performed experiments similar to those described above, replacing the linear DNA with supercoiled DNA. To this end, plasmid pLGR0, which contains the EcoRI-BamHI DNA fragment, was used to test the effect of temperature on H-NS affinity for the hly regulatory region. When pLGR0 was incubated in the presence of increasing concentrations of H-NS and the resulting protein-DNA complexes were separated in 0.8% agarose gels under conditions of constant temperature (either 25°C or 37°C), the results obtained clearly showed that the interaction of H-NS with pLGR0 is influenced by temperature and that the affinity is significantly higher at 25°C (Fig. 3C) than at 37°C (Fig. 3D). Taken together, the results determined so far indicate that unlike Hha, H-NS is able to select and bind specific regulatory sequences with the upstream region of the hly operon and that, at least in vitro, this binding is temperature dependent. Furthermore, taking into account the occurrence of an interaction between Hha and H-NS, these findings suggest that H-NS is the protein responsible for selecting and binding to specific DNA sites in the hly operon and that it thereby induces the temperature-dependent formation of a nucleoprotein complex, also containing Hha, that is responsible for the transcriptional repression of this operon.
FIG. 3.
Effect of increasing concentrations of H-NS on the electrophoretic mobility of the linear 2.8-kbp EcoRI-BamHI DNA fragment derived from the regulatory region of the hly operon (Fig. 2) (A and B) and on the mobility of plasmid pLGR0 (C and D). Binding reactions and electrophoretic separations were performed either at 25°C (A and C) or at 37°C (B and D). (A and B) Lane 1, molecular weight markers; lane 2, no protein added; lanes 3 to 5, 0.48, 0.24, and 0.12 μM H-NS protein, respectively. (C and D) Lanes 1 and 6, molecular weight markers; lane 2, no protein added; lanes 3 to 5, 0.97, 0.48, and 0.24 μM H-NS protein, respectively.
Temperature-mediated repression of hly operon transcription by H-NS.
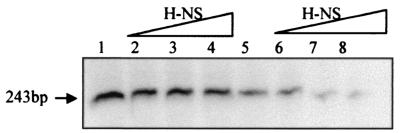
When analyzing the in vivo effect of an hns allele, we found that H-NS protein plays a major role in repressing hemolysin expression in E. coli cells growing at low temperature (35). In order to establish a correlation between the higher affinity that H-NS shows at 25°C for regulatory sequences of the hly operon (Fig. 3) and the observed role of H-NS in repressing transcription, we have analyzed the effect of H-NS on the in vitro transcription of the hly operon at different temperatures. Previous results indicated that transcription of the hly operon from plasmid pANN202-312R is more efficient when supercoiled DNA is used as a template (C. Madrid, unpublished data). Thus, supercoiled plasmid pANN202-312R was used as the template in transcriptional assays carried out at either 25 or 37°C in the presence of increasing amounts of purified H-NS. The data obtained (Fig. 4) indicate that H-NS-mediated repression of the hly operon is temperature dependent. We have quantified the results (using Molecular Analyst software by Bio-Rad) to determine the reduction in transcription levels that the presence of H-NS protein causes. At 37°C, the highest concentration of H-NS used accounts for a 20% reduction of transcription, whereas at 25°C, the reduction is about 60%. The finding that H-NS represses transcription of the hly operon more efficiently at 25°C than at 37°C correlates well with the fact that H-NS shows more affinity for regulatory sequences of the hly operon at low temperature and with the observed phenotype of hns mutants, which derepress hemolysin expression at low temperature (35).
FIG. 4.
Primer extension assay of the hly operon in the presence or absence of H-NS protein at 37°C (lanes 1 to 4) and at 25°C (lanes 5 to 8). Lanes 1 and 5, no protein added; lanes 2 and 6, 0.39 μM H-NS; lanes 3 and 7, 0.78 μM H-NS; lanes 4 and 8, 1.17 μM H-NS. Further experimental details are given in Materials and Methods.
Identification of H-NS binding sites by footprinting.
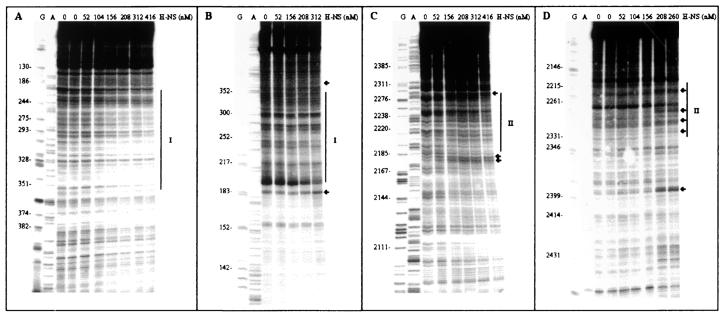
The above results prompted us to map the H-NS binding sites on the hly regulatory region by performing in vitro DNase I footprinting at 25°C in the presence and absence of this protein. These experiments were carried out on a supercoiled pLGHlyR0 plasmid carrying the entire hly operon, from the EcoRI site just upstream of hlyR to the SalI site downstream of hlyD (Fig. 1A). As seen from the results presented in Fig. 5A and B, H-NS influences the accessibility to DNase I digestion of two fairly extended sites, identified as site I and site II, for which H-NS seems to have, within the limits of accuracy of this type of analysis, a comparable affinity. Site I, which spans nucleotides 214 to 358 on one filament and nucleotides 193 to 348 on the other (Fig. 5A and B), clearly overlaps the hlyR sequence. Site II, on the other hand, extends approximately from nucleotide 2184 to 2285 on one filament and from nucleotide 2215 to 2329 on the other (Fig. 5B and C) and overlaps two of the three promoters described for the hly operon of plasmid pHly152 (23) (Fig. 6). Thus, site I and site II are flanking the IS2 insertion element such that the intrinsic DNA curvature predicted to occur within the IS2 element (Fig. 7A) can favor the interaction between the H-NS molecules located at both sites and thereby produce a stable H-NS-DNA complex. The hypothesis of the actual existence of the computer-predicted curvature is supported by an analysis of the EcoRI-BamHI fragment carried out by AFM, which revealed the presence of a strongly curved region approximately located in the central region of this fragment (Fig. 7B). This conclusion is also supported by additional data obtained by gel electrophoresis in agarose medium, which shows different levels of mobility of this particular EcoRI-BamHI fragment at 55°C and 4°C (our unpublished data). In addition to this intrinsic DNA bend, the appearance of several points (indicated by the arrows in Fig. 5) where H-NS induces an enhanced sensibility to DNase I cleavage, suggests that upon binding, this protein induces a structural deformation (i.e., bending and/or looping) of its target DNA.
FIG. 5.
DNase I footprinting of hlyR regulatory region by H-NS. Plasmid pLGHlyR0 was incubated at 25°C with or without the amounts of H-NS (expressed as nanomolar concentrations of dimers) indicated above each lane. The samples were processed using Hly544 (A), Hly543 (B), Hly545 (C), and Hly546 (D) as primers, as described in Materials and Methods. Lanes G and A represent TaqI polymerase sequencing reactions using the same primers. The H-NS-protected regions are indicated with vertical lines and labeled I and II; the sites hypersensitive to DNase I are indicated by arrows.
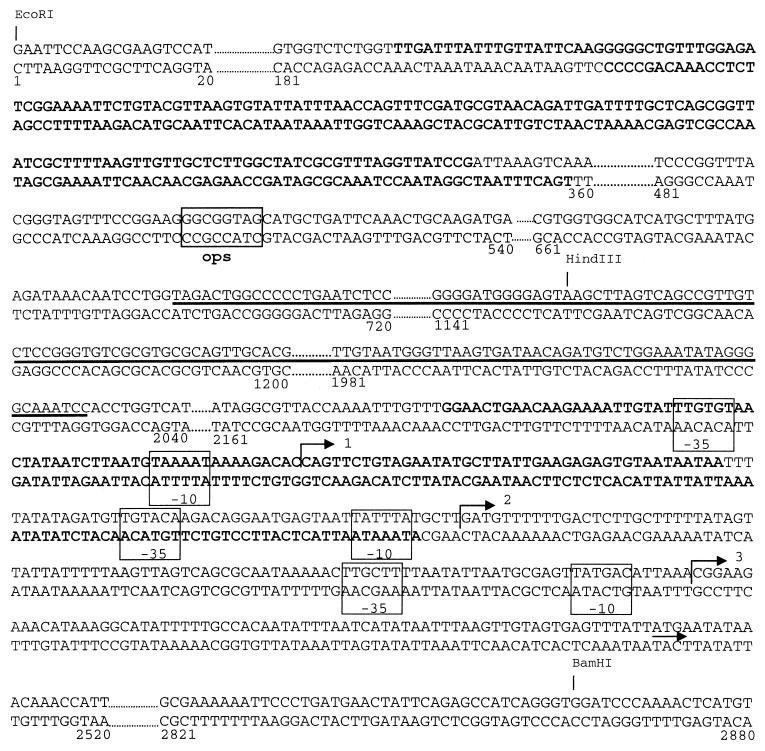
FIG. 6.
Location of the H-NS binding sites within the nucleotide sequence of hly regulatory region. The sequences protected by H-NS protein are in bold letters. The IS2 sequence is underlined and the ops element is indicated by the corresponding box. The −35 and −10 elements of three promoters identified for the hly operon are indicated by the corresponding boxes. Bent arrows and the short arrow indicate the transcription start sites and the first codon of hlyC, respectively. Restriction sites for EcoRI, HindIII, and BamHI are also indicated.
Deletion analysis of the hly regulatory region.
The above finding that the H-NS binding sites are separated by an IS2 element suggests that this random insertion of the transposable element has been positively selected because it can contribute to a better regulation of the hly operon. To test the validity of this hypothesis as well as the relevance of the hlyR sequence in the thermoregulation of the hly operon, we generated a series of deletions that would either eliminate the hlyR sequence or fuse it directly to the promoter region, thereby removing the IS2 element. Since the hlyR sequence contains the ops element, as mentioned in the introduction, two different deletions of the hlyR sequence were created, one (R1) lacking H-NS binding site II but still containing the ops element and another (R2) that completely removes the hlyR sequence (Fig. 1). These deletions were generated by PCR (see Materials and Methods for details), and the resulting EcoRI-BamHI fragments containing the deletions were cloned in the low-copy-number plasmid pLG338-30, giving rise to pLGR1, pLGR2, and pLGΔIS. These plasmids were then used to reconstruct the hly operon (plasmids pLGHlyR0, pLGHlyR1, pLGHlyR2, and pLGHlyΔIS) (see Materials and Methods for details). In vivo expression of the hly operon was then tested, and the levels of the hly transcript in E. coli cells harboring plasmids pLGHlyR0, pLGHlyR1, pLGHlyR2, and pLGHlyΔIS were determined.
Deletion of the H-NS binding site I (deletion R1) causes a partial derepression of the transcription at low temperature (Fig. 8). On the other hand, transcription is strongly reduced at 37°C when the entire hlyR sequence, including the ops element, is deleted (R2). Deletion of the IS2 element moderately reduces transcription at both temperatures.
FIG. 8.
Effect of different deletions on transcription of the hly operon. RNase protection assay of hlyC transcripts from mid-exponential-phase cultures of E. coli 5K, grown at 25 or 37°C and carrying pLGHlyR0 (lanes 1), pLGHlyR1 (lanes 2), pLGHlyR2 (lanes 3), or pLGHlyΔIS (lanes 4). Representative results are shown.
DISCUSSION
In this work we have extended previous observations concerning the interaction between Hha and H-NS and the role of the complex formed by these proteins in the modulation of hly operon expression (35). A key aspect that has been examined here concerns the role played by each protein in the generation of a nucleoprotein complex with hly regulatory sequences capable of modulating the expression of the operon in a temperature-dependent manner. The competition gel retardation experiments described in this work provide additional information about the interaction of both Hha and H-NS with the upstream region of the hly operon. Hha was found to bind to this regulatory region at a high protein/DNA ratio only, without displaying a preference for any of the fragments covering more than 4 kbp upstream of the transcription start site(s) of the hly operon. In contrast, H-NS was found to bind preferentially to two of the five fragments used in the assay at a protein/DNA ratio significantly lower than that required for the nonspecific DNA binding of Hha. Further characterization by DNase I footprinting of the H-NS binding preference allowed us to identify two fairly extended binding sites (sites I and II), the first within the hlyR sequence (nucleotides 190 to 350) and the second (nucleotides 2180 to 2330) partially overlapping the promoter region of the hly operon of plasmid pHly152 (Fig. 6). The relevance of site I for thermoregulation is evidenced by the results of the deletion analysis of the hly operon that we have performed in the present study.
The results of two different approaches (computational prediction and AFM analysis) indicated the existence of an intrinsic DNA curvature in the region separating the two H-NS sites. The occurrence of an intrinsic curvature separating two H-NS binding sites has been previously reported in the upstream region of genes subjected to temperature-independent (hns and cspA) and temperature-dependent (virF) transcriptional repression by H-NS (3, 10, 11). A significant difference between the thermoregulated virF gene (11) and the hly operon is the presence in the latter of an IS2 sequence separating the two H-NS binding sites, thereby causing a more extensive separation between them. In any event, the role of the IS2 element in expression of the hly operon could not be clarified by removing it from its insertion site. As no hly promoter function has hitherto been assigned to this element, it is apparent that the moderate effect of IS2 deletion on hly transcription is most likely due to the fact that when the two H-NS binding sites are separated, topological modifications in the region are introduced.
It is apparent that temperature influences the affinity of H-NS for its target sequences within the regulatory region of the hly operon. In fact, the higher affinity of H-NS for the 2.8-kb EcoRI-BamHI fragment at low temperature correlates well with the more efficient transcriptional repression caused by H-NS at low temperature and with the hemolytic phenotype of the hns mutants. Taken together, these results indicate that H-NS is the protein that confers specificity to the regulatory loop by eliciting the temperature-dependent formation of a nucleoprotein complex comprising the regulatory sequences of hly.
With respect to Hha, different lines of experimental evidence suggest that it probably acts as a protein binder rather than as a DNA binder: (i) it shows strong affinity in vitro for H-NS (35) and (ii) a mutational analysis has shown that almost all of the protein present is required to bind H-NS (34). There is a certain degree of homology between the amino acid sequence of the described proteins from the Hha family and the oligomerization domain of the H-NS family of proteins (34). Considering the phenotype of hha mutants (partial derepression of temperature- and osmolarity-mediated hemolysin expression) and the role of this protein as an H-NS binder, it appears reasonable that the role of Hha in modulation of hly expression is accomplished by binding to H-NS to generate a hetero-oligomeric complex that represses hly transcription more efficiently than H-NS oligomers alone.
The mechanism underlying temperature-mediated repression of hly operon can be modeled from the data reported in this paper. At low temperature, the increased affinity of H-NS for the two identified binding sites generates nucleoprotein complexes that include Hha as well. Because of the increased flexibility of DNA at low temperature (the increased flexibility being a consequence of a higher degree of DNA supercoiling), H-NS molecules located at both sites may contact, thereby allowing the generation of a larger nucleoprotein complex which includes H-NS and Hha. That complex would occlude both the ops sequence and the promoter region, thus repressing transcription and abolishing the antitermination effect of ops. An increase in the growth temperature may have two different effects: (i) reduction of the affinity of H-NS for its target sequences and (ii) reduction of the flexibility of the DNA. As a possible consequence, H-NS molecules colliding with their target sequences are not able to generate nucleoprotein complexes large enough to extend between them and the decrease in DNA flexibility does not allow it to stabilize a nucleoprotein complex which includes both sites. Under the latter circumstances, transcriptional repression is at least partially alleviated. A delicate balance between the moderate affinity of H-NS for its target sites and the effect of temperature on DNA structure and flexibility probably underlies thermoregulation of the hly operon. It is relevant to mention here that growth at 37°C does not result in complete derepression of hly expression. H-NS still partially represses transcription, as demonstrated by the significant increase in hemolysin expression that is evidenced in hns mutants growing at 37°C (35). This means that H-NS- and Hha-mediated repression of the hly operon is not an on-off process but, rather, a gradual one, in which different environmental factors that cause variations in the physicochemical parameters of DNA or sequestering or degradation of H-NS modify hemolysin expression.
The model presented here is strongly reminiscent of that previously reported for the thermoregulation of the virF operon (11) and hence supports a general model for H-NS-mediated thermoregulation of virulence gene expression. In addition, it is completely compatible with the previous reported models for H-NS-mediated repression of transcription. H-NS molecules generating a nucleoprotein complex that represses transcription fit with the promoter-occlusion model, and the interaction of H-NS molecules with DNA to modify the level of supercoiling and hence DNA flexibility fits with the model supporting a role for H-NS on DNA topology (11, 39, 40). This means that H-NS could play a dual role, both as a protein specifically modifying DNA structure and as a repressor of transcription.
Acknowledgments
We are grateful to G. Micheli for computer analysis of DNA curvature, to B. E. Uhlin for providing strains BSN26 and BSN27, and to the Nanometric Technics Unit of the Scientific-Technic Resources of the University of Barcelona for handling AFM.
This work was supported by grants from the Ministerio de Ciencia y Tecnología (PB97-0950) and the CIRIT from the Generalitat de Catalunya (2000 SGR 00038). The financial support of a MURST-PRIN 2001 grant (Transcriptional Response to Environmental Changes in Pathogenic Bacteria) to M.F. is gratefully acknowledged.
REFERENCES
- 1.Atlung, T., and A. Ingmer. 1997. H-NS: a modulator of environmental regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre, C., A. Juárez, C. Madrid, M. Mouriño, A. Prenafeta, and F. J. Muñoa. 1996. Complementation of the hha mutation in Escherichia coli by the ymoA gene from Yersinia enterocolitica: dependence of the gene dosage. Microbiology 142:1841-1846. [DOI] [PubMed] [Google Scholar]
- 3.Brandi, A., R. Spurio, C. O. Gualerzi, and C. L. Pon. 1999. Massive presence of the Escherichia coli major cold-shock protein CspA under non-stress conditions. EMBO J. 18:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmona, M., C. Balsalobre, F. J. Muñoa, M. Mouriño, Y. Jubete, F. De la Cruz, and A. Juárez. 1993. Escherichia coli hha mutants, DNA supercoiling and expression of haemolysin genes from the recombinant plasmid pANN202-312. Mol. Microbiol. 9:1011-1018. [DOI] [PubMed] [Google Scholar]
- 5.Cornelis, G. R., C. Sluiters, I. Delor, D. Gelb, K. Kaninga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michaelis. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, T. P., R. C. Montelaro, and K. E. Rushlow. 1993. Lentivirus envelope sequence and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vector. Gene 9:93-98. [DOI] [PubMed] [Google Scholar]
- 7.De la Cruz, F., M. Carmona, and A. Juárez. 1992. The Hha protein from Escherichia coli is highly homologous to the YmoA protein from Yersinia enterocolitica. Mol. Microbiol. 6:3451-3452. [DOI] [PubMed] [Google Scholar]
- 8.Donato, G. M., and T. H. Kawuala. 1998. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J. Biol. Chem. 11:24030-24036. [DOI] [PubMed] [Google Scholar]
- 9.Falconi, M., M. T. Gualtieri, A. La Teana, M. A. Losso, and C. L. Pon. 1988. Proteins from the prokaryotic nucleoid: primary and quaternary structure of the 15-kDa Escherichia coli DNA binding protein H-NS. Mol. Microbiol. 2:323-329. [DOI] [PubMed] [Google Scholar]
- 10.Falconi, M., N. P. Higgins, R. Spurio, C. L. Pon, and C. O. Gualerzi. 1993. Expression of the gene encoding the major bacterial nucleoid protein H-NS is subject to transcriptional auto-repression. Mol. Microbiol. 10:273-282. [DOI] [PubMed] [Google Scholar]
- 11.Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17:7033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardel, C. L., and J. J. Mekalanos. 1994. Regulation of cholera toxin by temperature, pH and osmolarity. Methods Enzymol. 235:517-526. [DOI] [PubMed] [Google Scholar]
- 13.Godessart, N., F. J. Muñoa, M. Regué, and A. Juárez. 1988. Chromosomal mutations that increase the production of plasmid-encoded haemolysin in Escherichia coli. J. Gen. Microbiol. 134:2779-2787. [DOI] [PubMed] [Google Scholar]
- 14.Göransson, M., B. Sondén, P. Nilsson, B. Dagberg, K. Forsman, K. Emanuelsson, and B. E. Uhlin. 1990. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature 344:682-685. [DOI] [PubMed] [Google Scholar]
- 15.Hansma, H. G., R. L. Sinsheimer, J. Groppe, T. C. Bruice, V. Elings, G. Gurley, M. Bezanilla, I. A. Mastrangelo, P. V. C. Hough, and P. K. Hansma. 1993. Recent advances in atomic force microscopy of DNA. Scanning 15:296-299. [DOI] [PubMed] [Google Scholar]
- 16.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. LeCaer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 17.Hulton, C. S. J., A. Seirafi, J. C. D. Hinton, J. A. M. Sidebotham, L. Waddell, G. D. Pavitt, T. Owen-Hughes, A. Spassky, H. Buc, and C. F. Higgins. 1990. Histone-like protein H1 (H-NS), DNA supercoiling and gene expression in bacteria. Cell 63:631-642. [DOI] [PubMed] [Google Scholar]
- 18.Hurme, R., and M. Rhen. 1998. Temperature sensing in bacterial regulation—what it all boils down to. Mol. Microbiol. 30:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Johansson, J., B. Dagberg, E. Richet, and B. E. Uhlin. 1988. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 180:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordi, B., T. A. Owen-Hughes, C. Hulton, and C. F. Higgins. 1995. DNA twist, flexibility and transcription of the osmoregulated proU promoter of Salmonella typhimurium. EMBO J. 14:5690-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juárez, A., M. Hartlein, and W. Goebel. 1984. Study of regulation and transport of hemolysin by using fusion of the β-galactosidase gene (lacZ) to hemolysin genes. J. Bacteriol. 160:161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajitani, M., and A. Ishahima. 1991. Identification and sequence determination of the host factor gene for bacteriophage Qβ. Nucleic Acids Res. 19:1063-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koronakis, V., and C. Hughes. 1988. Identification of the promoters directing in vivo expression of hemolysin genes in Proteus vulgaris and Escherichia coli. Mol. Gen. Genet. 213:99-104. [DOI] [PubMed] [Google Scholar]
- 24.Laurent-Winter, C., S. Ngo, A. Danchin, and P. Bertin. 1997. Role of Escherichia coli histone-like nucleotide-structuring protein in bacterial metabolism and stress response: identification of targets by two-dimensional electrophoresis. Eur. J. Biochem. 244:767-773. [DOI] [PubMed] [Google Scholar]
- 25.Marykwas, D. L., S. A. Schmidt, and H. C. Berg. 1996. Interacting components of flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol. 256:564-576. [DOI] [PubMed] [Google Scholar]
- 26.Maurelli, A. T., and P. J. Sansonetti. 1988. Identification of a chromosomal gene controlling temperature regulated expression of Shigella virulence. Proc. Natl. Acad. Sci. USA 85:2820-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikulskis, A. V., and G. R. Cornelis. 1994. A new class of proteins regulating gene expression in enterobacteria. Mol. Microbiol. 11:77-86. [DOI] [PubMed] [Google Scholar]
- 29.Mouriño, M., C. Madrid, C. Balsalobre, A. Prenafeta, F. J. Muñoa, J. Blanco, M. Blanco, J. E. Blanco, and A. Juárez. 1996. The Hha protein as a modulator of expression of virulence factors in Escherichia coli. Infect. Immun. 64:2881-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouriño, M., F. J. Muñoa, C. Balsalobre, P. Díaz, C. Madrid, and A. Juárez. 1994. Environmental regulation of α-hemolysin expression in Escherichia coli. Microb. Pathog. 16:249-259. [DOI] [PubMed] [Google Scholar]
- 31.Muffler, A., D. Fisher, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10:1143-1151. [DOI] [PubMed] [Google Scholar]
- 32.Nieto, J. M., M. J. A. Bailey, C. Hughes, and V. Koronakis. 1996. Suppression of transcription polarity in the Escherichia coli haemolysin operon by a short upstream element shared by polysaccharide and DNA transfer determinants. Mol. Microbiol. 19:705-713. [DOI] [PubMed] [Google Scholar]
- 33.Nieto, J. M., M. Carmona, S. Bolland, Y. Jubete, F. de la Cruz, and A. Juárez. 1991. The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285-1293. [DOI] [PubMed] [Google Scholar]
- 34.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodriguez, and A. Juárez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juárez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263:349-358. [DOI] [PubMed] [Google Scholar]
- 36.Noegel, A., U. Rdest, and W. Goebel. 1981. Determination of the functions of hemolytic plasmid pHly152 of Escherichia coli. J. Bacteriol. 145:233-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onoa, G. B., G. Cervantes, V. Moreno, and M. J. Prieto. 1998. Study of the interaction of DNA with cisplatin and other Pd(II) and Pt(II) complexes by atomic force microscopy. Nucleic Acids Res. 26:1473-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for bending. EMBO J. 16:1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tupper, A. E., T. A. Owen-Hughes, D. W. Ussery, D. S. Santos, D. J. P. Ferguson, J. M. Sidebotham, J. C. D. Hinton, and C. F. Higgins. 1994. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 13:258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueguchi, C., and T. Mizuno. 1993. The Escherichia coli nucleoid protein H-NS functions as a transcriptional repressor. EMBO J. 12:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ussery, D. W., J. C. D. Hinton, B. J. A. M. Jordi, P. E. Granum, A. Seirafi, R. J. Stephen, A. E. Tupper, G. Berridge, J. M. Sidebotham, and C. F. Higgins. 1994. The chromatin-associated protein H-NS. Biochimie 76:968-980. [DOI] [PubMed] [Google Scholar]
- 42.Vogel, M., J. Hess, I. Then, A. Juárez, and W. Goebel. 1988. Characterization of a sequence (hlyR) which enhances synthesis and secretion of hemolysin in Escherichia coli. Mol. Gen. Genet. 212:76-84. [DOI] [PubMed] [Google Scholar]
- 43.Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the E. coli nucleoid protein H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175-185. [DOI] [PubMed] [Google Scholar]
- 44.Yamada, H., T. Yoshida, K. Tanaka, C. Sasakawa, and T. Mizuno. 1991. Molecular analysis of the Escherichia coli hns gene encoding a DNA binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 230:332-336. [DOI] [PubMed] [Google Scholar]