Abstract
The Salmonella pathogenicity island 2 (SPI2) type III secretion system (TTSS) promotes Salmonella enterica serovar Typhimurium virulence for mice and increased survival and replication within eukaryotic cells. After phagocytosis, Salmonella serovar Typhimurium assembles the SPI2 TTSS to translocate over a dozen effector proteins across the phagosome membrane. SpiC has been previously shown to be a translocated effector with a large contribution to virulence (K. Uchiya, M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman, EMBO J. 18:3924-3933, 1999). This report demonstrates by competitive index that the virulence phenotype of a spiC mutant is equivalent to that of a secretion component mutant. In addition, translocation of SPI2 effector proteins was shown to require SpiC. Thus, the severe virulence phenotype resulting from deletion of spiC is likely due to the inability to translocate all SPI2 effectors. SpiC was also required to secrete translocon proteins SseB and SseC but not translocated effector SseJ, indicating that lack of assembly of the translocon explains the spiC mutant phenotype.
Type III secretion systems (TTSS) promote the virulence of gram-negative bacteria for eukaryotic hosts by delivering bacterial proteins directly from the bacterial cytosol into eukaryotic cells (17). These secretion systems are composed of cytoplasmic, membrane-spanning, and secreted proteins. The cytoplasmic proteins consist of those that have homology to the flagellar energy apparatus, which powers the secretion of all substrate proteins, and chaperone proteins. Chaperones are typically small acidic proteins that are required for the secretion of specific subsets of TTSS substrate proteins. The TTSS membrane-spanning proteins have homology to the components of the flagellar basal body and collectively form the needle complex, which spans both the bacterial inner and outer membranes and which is required for the secretion of all substrate proteins. The TTSS secreted proteins can be subdivided into two classes, commonly known as translocon and effector proteins. Effector proteins are translocated into the eukaryotic cytoplasm, where they typically alter a specific function or pathway of the cell's physiology. In contrast, translocon proteins are hypothesized to connect the needle complex and the eukaryotic cell and to insert into the eukaryotic cell membrane to allow the delivery of effector proteins. Additionally, translocon proteins may exert effects on the eukaryotic cell themselves, possibly by interacting with membrane-associated proteins (31).
Despite the basic conservation of the structure and function of different TTSS, they can have remarkably distinct effects on eukaryotic cells. The differences in function between TTSS are presumed to be a result of the total activities of the effector and, to some extent, translocon proteins (5, 14, 16, 31). Since the translocon proteins are necessary for effector delivery, TTSS apparatus and translocon mutants typically display identical and significant defects in their virulence phenotypes. In contrast, bacteria with mutated single translocated effectors can have relatively minor measurable virulence defects, often due to redundancy in effector function (11, 13, 18-20, 24, 42).
Salmonellae are gram-negative bacteria that cause gastroenteritis and typhoid fever, a systemic illness characterized by fever and bacterial replication in lymphoid tissue rich in phagocytic cells (25). Salmonella enterica serovar Typhimurium is a broad-host-range pathogen that can infect a variety of animal hosts with different disease outcomes. In immunocompetent humans, Salmonella serovar Typhimurium infection typically results in self-limited gastroenteritis, whereas, in specific inbred mouse strains, Salmonella serovar Typhimurium can cause a systemic disease similar to human typhoid fever. While typhoid fever is caused by the human-specific pathogen S. enterica serovar Typhi, the limitation of experimental models for this pathogen has led to the widespread study of Salmonella serovar Typhimurium in Nramp mutant mice as a model for systemic disease. The ability of Salmonella serovar Typhimurium to cause systemic disease in inbred mice is correlated with its ability to survive and replicate within host cells, particularly macrophages. One important factor in this intracellular survival and replication is the TTSS encoded on Salmonella pathogenicity island 2 (SPI2) (28, 34) (Fig. 1). Most TTSS, including the Salmonella SPI1 system, which promotes invasion and enteric disease, translocate bacterial proteins across the eukaryotic cell plasma membrane (17). The SPI2 TTSS is relatively unique in that its expression is largely limited to intracellular bacteria (6, 39), and salmonellae use the SPI2 TTSS to translocate proteins across the phagosomal membrane (24).
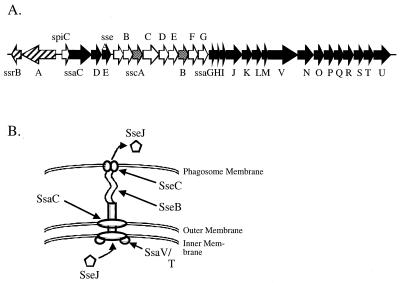
FIG. 1.
Genetic organization of SPI2 and putative model of the structure of the SPI2 TTSS with associated translocon. (A) Genes encoding the secretion apparatus (ssa), putative secreted substrate proteins (sse), putative chaperone proteins (ssc), and regulatory proteins (ssr) (black, white, shaded, and hatched arrows, respectively). (B) Predicted function and localization of various proteins of the SPI2 TTSS. Assignment is based predominantly on homology to other characterized TTSS proteins. SseB, SseC, and SseD are secreted by the SPI2 TTSS and form a complex on the bacterial cell surface required for the translocation of known SPI2 effector proteins such as SseJ (26). Based on its homology to EspA, SseB may form a filament connecting the SPI2 TTSS to the phagosomal membrane, allowing the insertion of SseC in the phagosomal membrane. SsaC is predicted to form the outer membrane ring of the SPI2 TTSS based on its homology to InvG and other related proteins, while SsaT and SsaV are predicted to function in a more proximal aspect of the SPI2 TTSS.
Several proteins have recently been demonstrated to be secreted by the SPI2 TTSS in vitro when Salmonella serovar Typhimurium is grown under SPI2-inducing conditions (1, 21, 26). These include SseB, SseC, and SseD, all of which are required for the translocation of known SPI2 effector proteins into infected mammalian cells. These proteins can be extracted from the bacterial cell surface and are believed to form a macromolecular structure that functions as the translocon (26). SseB has homology to EspA from enterohemorrhagic Escherichia coli and enteropathogenic E. coli (15). EspA forms a filament on the bacterial cell surface which connects the needle complex to the infected mammalian cell (7, 22, 32). Although SseB has not been shown to form such a filament, it has been visualized on the cell surface by immunofluorescence and is required for the surface association of SseC and SseD (26). SseC and SseD have homology to YopB and YopD, respectively, and thus probably function to insert into the phagosomal membrane to facilitate protein translocation (15).
In addition to the secreted translocon proteins, several other proteins have recently been shown to be translocated into infected mammalian cells. These include a family of TTSS effector proteins, such as SspH2 and SseJ, that share a conserved amino-terminal sequence that is required for translocation by SPI2 (3, 23, 24). All of the genes encoding these effector proteins are located independently outside of SPI2. One of these proteins, SifA, is required for the formation of lysosomal glycoprotein-containing filaments and for the maintenance of the phagosomal membrane (2, 37). SifA was shown to promote intracellular survival within macrophages and to be required for systemic infection in susceptible mice (2, 4). In addition to the effectors encoded outside of SPI2, SpiC (SsaB), a protein encoded within SPI2, has been shown to be translocated into mammalian cells (38). SpiC interferes with endosomal trafficking upon expression in mammalian cells and in in vitro assays. Furthermore, Salmonella serovar Typhimurium spiC mutants also demonstrate increased trafficking to vacuolar compartments containing lysosomal markers within infected macrophages (38). Strikingly, Salmonella serovar Typhimurium spiC mutants demonstrate significant defects in both virulence for mice and intracellular survival. Such a significant virulence phenotype is unusual for an effector and is actually similar to that resulting from mutation of the SPI2 TTSS.
Therefore, experiments were performed to further define the virulence phenotype of spiC mutants and the role of SpiC in type III secretion.
MATERIALS AND METHODS
Bacterial strains, eukaryotic cell lines, and growth conditions.
Bacterial strains and plasmids used are listed in Table 1. Salmonella serovar Typhimurium was grown to stationary phase in Luria broth (LB) with aeration for all infection and virulence assays. The composition of minimal media has been described previously (26). Murine monocyte cell line RAW264.7 was cultured in Dulbecco's modified Eagle medium (Gibco BRL) containing 10% fetal calf serum (FCS) (Gibco BRL) and 2 mM glutamine at 37°C in 5% CO2.
TABLE 1.
Strains and plasmids used in this study
| Designation | Description | Reference or source |
|---|---|---|
| Plasmids | ||
| pEM25 | sspH1-cyaA in pMJH20, Apr | 24 |
| pEM30 | sspH2-cyaA in pMJH20, Apr | 24 |
| pEM80 | sseI-cyaA in pMJH20, Apr | 23 |
| pEM82 | sseJ-cyaA in pMJH20, Apr | 23 |
| pKAS32 | rpsL suicide vector for allelic exchange | 35 |
| pJAF21 | ΔsifA in pKAS32 | This study |
| pWSK29 | Low-copy-number plasmid | 40 |
| pCAS61 | pWSK29 with HA epitope tag | This study |
| pJAF111 | sseJ-HA in pCAS61, Apr | This study |
| pWSK30 | Low-copy-number plasmid | 40 |
| p2062 | M45 epitope tag in pSK+ | 12 |
| p2129 | sseJ-M45 in pWSK29, Apr | 12 |
| p2127 | spiC-M45 in pWSK30, Apr | This study |
| pEG9127 | spiC in pBAC108L, Cmr | 38 |
| pKD46 | Plasmid expressing λ Red recombinase | 8 |
| pKD4 | Plasmid with FRT-flankeda kanamycin resistance gene | 8 |
| pCP20 | Plasmid expressing the FLP recombinase | 8 |
| Salmonella serovar Typhimurium strains | ||
| NCTC 12023 | Wild type, identical to ATCC 14028s | ATCC |
| P3F4 | NCTC 12023, ssrA::mTn5 Kmr | 34 |
| P7G2 | NCTC 12023, ssaC::mTn5 Kmr | 34 |
| HH109 | NCTC 12023, ssaV::aphT Kmr | 9 |
| EG10128 | ATCC 14028s, spiC::aphT Kmr | 38 |
| CS050 | ATCC 14028s, phoN::Tn10d-Kan | S. I. Miller laboratory |
| CS401 | ATCC 14028s, phoN2 zxx::6251 Tn10d-Cm, Strepr | 24 |
| CS600 | ATCC 14028s, phoN::Tn10d-Kan, Strepr | This study |
| EM232 | CS401, ssaT::mTn5 Kmr | 24 |
| JAF204 | CS401, ΔspiC | This study |
| JAF57 | CS401, ΔsifA | This study |
FRT, Flp recognition target.
Construction of deletion mutants.
Deletion of sifA was accomplished by using an allelic-exchange plasmid. To construct this plasmid, DNA flanking both the 5′ and 3′ ends of sifA was amplified from Salmonella serovar Typhimurium chromosomal DNA by PCR using Pfu Turbo polymerase (Stratagene). The 5′ end was amplified with primers 5′-GGTTATCTCAATGAATTCCTGCTGTGG-3′ and 5′-GCGGGTACCGTCCGCTTTTGCTTTGCCAG-3′, and the 3′ end was amplified with primers 5′-GCGGGTACCCGCTCAGAACAACAAAGCGGC-3′ and 5′-GCGTCTAGACCAATAAAACGGTCGCCAGC-3′. The flanking ends were sequentially cloned into allelic-exchange vector pKAS32, yielding plasmid pJAF21. pJAF21 was then introduced into strain CS401 by conjugation and allelic exchange selecting for streptomycin resistance as previously described (29). The resulting deletion strain, JAF57, was verified by diagnostic PCR using Taq polymerase (Qiagen). The method described by Datsenko and Wanner (8) was used to delete spiC. Briefly, primers were designed to amplify a kanamycin resistance gene flanked by Flp recognition target sites from plasmid pKD4. The primers, designed to also be homologous to DNA flanking spiC, were 5′-TTGTGAGCGAATTTGATAGAAACTCCCATTTATGTCTGAGGTGTAGGCTGGAGCTGCTTC-3′ and5′-AATTAAGATTAAACGTTTATTTACTACCATTTTATACCCCCATATGAATATCCTCCTTAG-3′. Purified PCR product was then transformed into Salmonella serovar Typhimurium harboring the λ Red recombinase on plasmid pKD46, which contains a temperature-sensitive origin of replication, by selecting for kanamycin resistance. Kanamycin-resistant strains were then cured of pKD46 by incubation at 37°C. The inserted kanamycin resistance gene was subsequently removed by transforming the strain with plasmid pCP20, which expresses the FLP recombinase, resulting in an in-frame deletion of spiC, producing strain JAF204. The kanamycin-sensitive, spiC deletion strain was finally cured of the pCP20 plasmid.
Construction of HA-tagged SseJ.
Plasmid pCAS61 was constructed by annealing primers 5′-TCGACTATCCTTATGATGTTCCTGATTATGCATAAC-3′ and 5′-TCGAGTTATGCATAATCAGGAACATCATAAGGATAG-3′, which encode the hemagglutinin (HA) epitope tag sequence, and inserting the product into plasmid pWSK29 digested with SalI and XhoI. The sseJ open reading frame and promoter region were amplified with primers 5′-CGCGAATTCGTCAGATAATATGTACCAGGC-3′ and 5′-CGCGTCGACTTCAGTGGAATAATGATGAGC-3′ by PCR using Pfu Turbo. Plasmid pCAS61 and the PCR product were digested with EcoRI and SalI and ligated together, yielding plasmid pJAF111.
Generation of epitope-tagged SpiC.
PCR was performed using the Expand High Fidelity system (Roche) in order to minimize the error rate of the amplification procedure. Approximately 100 ng of genomic DNA of wild-type Salmonella serovar Typhimurium was used as a template for amplification with oligonucleotides ProA-For-EcoRI (5′-GCTGAATTCATCTCGGATAGAACGG-3′) and SsaB-Rev-SmaI (5′-ATGCCCGGGTACCCCACCCGAATAAAG-3′). The product was digested with EcoRI and SmaI and ligated with EcoRI- and EcoRV-digested plasmid p2062 to generate a gene fusion to the M45 coding sequence. The resulting construct was digested with EcoRI and XbaI, and the insert consisting of promoter ssaB and ssaB::M45 was subcloned in pWSK30 to generate plasmid p2127.
Bacterial infection of macrophages.
Salmonella serovar Typhimurium strains were grown to stationary phase in LB containing the necessary antibiotics, diluted in Dulbecco's modified Eagle medium containing FCS and glutamine, and added to RAW264.7 cells seeded in 24-well tissue culture plates. The bacteria were centrifuged onto the cells at 500 × g for 5 min and incubated for 25 min at 37°C in 5% CO2. After infection, the macrophages were washed three times with phosphate-buffered saline (PBS) and incubated for 1 h in medium containing FCS, glutamine, and 100 μg of gentamicin (Sigma)/ml. The cells were then incubated with medium containing FCS, glutamine, and 10 μg of gentamicin/ml for the remainder of the experiment.
Translocation assay.
Determination of SspH1 and SspH2 translocation by the SPI2 type III secretion system was performed as previously described (24). Briefly, wild-type and mutant Salmonella serovar Typhimurium cells expressing either sspH1-cyaA or sspH2-cyaA from a constitutive lac promoter on a low-copy-number plasmid were grown to stationary phase in LB. RAW264.7 cells were plated at 5 × 105 cells per well in 24-well dishes and infected with bacteria for 1 h at a multiplicity of infection of 10. Seven hours postinfection, the RAW264.7 cells were lysed in 0.1 N HCl and boiled for 5 min. Cellular cyclic AMP (cAMP) was then quantitated with the Direct cAMP Correlate enzyme immunoassay kit (Assay Designs, Ann Arbor, Mich.), and total protein concentration was quantitated with the Bradford assay. Cellular cAMP was then normalized for total protein content. All experiments were performed at least twice in triplicate.
Immunofluorescence detection and microscopy.
RAW264.7 cells, seeded on coverslips, were infected with Salmonella serovar Typhimurium at a multiplicity of infection of 2. Sixteen hours postinfection, the cells were fixed in 3% paraformaldehyde in PBS for 15 min and then washed three times with PBS. The coverslips were incubated with the appropriate antibodies diluted in a blocking solution consisting of 10% goat serum, 1% bovine serum albumin, and 0.1% saponin (Sigma) in PBS. After each incubation, the coverslips were washed three times with PBS. After staining, the coverslips were mounted on Fluoroprep (bioMèrieux) and sealed with Entellan (Merck). Samples were analyzed using a fluorescence microscope (Zeiss Axiophot) and the Metavue imaging software (Universal Imaging Corporation). The M45 epitope was detected with a mouse monoclonal antibody at a 1:5 dilution (27), and bacteria were detected with Bacto rabbit Salmonella anti-O test sera (Difco) at a 1:1,000 dilution. Secondary detection was performed with goat anti-mouse Cy3 (Jackson) at 1:500 and goat anti-rabbit Cy2 (Jackson) at 1:2,000.
Determination of mouse CIs.
Wild-type and mutant Salmonella serovar Typhimurium were grown to stationary phase in LB and used to infect BALB/c mice. Each mutant strain was marked with chloramphenicol resistance, while the wild type was marked with kanamycin resistance. Wild-type and mutant bacteria were diluted, and roughly equal numbers were mixed. An aliquot of the inoculum was then plated on selective media, and the numbers of wild-type and mutant bacteria were quantitated. A total of 105 bacteria in a volume of 0.2 ml were injected intraperitoneally into each mouse. In each experiment, four mice were infected with each mutant Salmonella serovar Typhimurium strain. Two days after infection, the mice were sacrificed and the infected spleens were removed and homogenized in sterile saline. Wild-type and mutant bacteria in each infected organ were then quantitated by serial dilution and plating on selective media. The competitive index (CI) was calculated by dividing the number of mutant bacteria isolated from infected animals by the number of wild-type bacteria recovered. This value was then corrected by the initial ratio of mutant to wild-type bacteria used to infect each animal. All experiments were performed at least twice (total of eight mice).
Preparation of surface-localized proteins.
Surface-localized proteins were prepared as described previously (12). Basically, for preparation of the detached fraction, bacteria were precultured in LB for 8 h at 37°C. Bacteria were washed once in minimal medium, and equal amounts of bacteria, as adjusted by optical density at 600 nm (OD600), were used to inoculate cultures in minimal media at neutral or acidic pH. Cultures (400 ml) in 1-liter glass flasks without baffles were incubated overnight with agitation of 200 rpm at 37°C. Bacteria were pelleted by centrifugation at 6,000 × g for 15 min and resuspended in 20 ml of PBS. This suspension was agitated at maximum speed in 50-ml centrifuge tubes (Falcon) on a vortex mixer (Vortex Genie 2; Scientific Industries) for 60 s. Bacterial cells were pelleted by centrifugation at 10,000 × g for 10 min, and the supernatant was passed through a 0.2-μm-pore-size filter to remove residual bacteria. Protein in the supernatant fraction was recovered by precipitation with trichloroacetic acid (10% [wt/vol], final concentration) on ice for 1 h and centrifugation for 1 h at 10,000 × g. The pellet was washed twice with 15 ml of acetone and recovered by centrifugation at 10,000 × g for 30 min. The final pellet was air dried and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris-HCl [pH 6.8], 4% SDS, 2% β-mercaptoethanol, 12.5% glycerol, 0.01% bromophenol blue). The final concentration of protein in the detached fraction was adjusted by the cell density of the original culture (OD600 of a 100-ml culture × 100 = volume of the sample buffer in microliters).
Preparation of secreted proteins.
Bacteria were pregrown for 12 h in LB at 37°C with shaking. Cells were washed once in sterile saline and inoculated into 40 ml of minimal medium at acidic pH at a 1/100 dilution. Cultures were grown at 37°C for 6 h. Bacteria were then removed by centrifugation at 125,000 × g for 90 min. Proteins in the culture supernatant were then precipitated in 10% trichloroacetic acid on ice for 1 h. Precipitated proteins were collected by centrifugation at 75,000 × g for 1 h. Protein pellets were washed twice in ice-cold acetone and allowed to dry. Samples were resuspended in SDS-PAGE sample buffer, of which 20 μl was analyzed by SDS-PAGE.
Subcellular fractionation.
A culture of Salmonella serovar Typhimurium carrying p2127 was grown in low-phosphate minimal medium at pH 5.8 for 16 h at 37°C in a glass flask without baffles. The culture was centrifuged, washed in PBS, and resuspended in PBS. In the presence of a protease inhibitor cocktail (Sigma) the suspension was passed two times through a French press (82.7 MPa). To remove nonlysed bacteria, the solution was centrifuged at 400 × g for 30 min at 4°C. After centrifugation at 100,000 × g for 1 h at 4°C, the pellet (membrane fraction) was washed with double-distilled H2O, and the supernatant (soluble fraction) was centrifuged again at 100,000 × g. The membrane pellet was resuspended in 10 mM Tris-HCl, pH 8.0, with a syringe needle (0.45 by 25 mm) and solubilized with 1% N-lauroylsarcosine (Sarkosyl) in 10 mM EDTA, pH 8.0, for 10 min on ice. The Sarkosyl-soluble and Sarkosyl-insoluble fractions were separated by centrifugation for 2 h at 100,000 × g. The pellet containing the Sarkosyl-insoluble outer membrane fraction was resuspended in 10 mM Tris-HCl, pH 8.0, and centrifuged again to remove the excess of Sarkosyl. The proteins of the soluble fraction and the Sarkosyl-soluble inner membrane fraction were precipitated with acetone.
Production of antisera.
Polyclonal antiserum against OmpA of S. enterica serovar Typhimurium was raised in BALB/c mice. Outer membrane proteins were prepared by Sarkosyl extraction as described above. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes. A protein of 33 kDa corresponding to the molecular weight of OmpA was recovered and confirmed by N-terminal amino acid sequencing. The nitrocellulose fragment with OmpA was dissolved in dimethyl sulfoxide and used for immunization according to standard procedures (13a).
Protein analysis.
Cell-associated- and secreted-protein profiles of various Salmonella serovar Typhimurium strains were routinely analyzed by SDS-PAGE on Tricine gels (8 or 12%) as described by Schägger and von Jagow (30). For analysis by Western blotting, proteins were transferred onto nitrocellulose membranes (BA85; Schleicher and Schuell) by the discontinuous semidry blotting procedure. SseB and SseC were detected with polyclonal antibodies raised against recombinant proteins (1, 26). SseJ-HA was detected with a monoclonal anti-HA antibody (Covance), and SpiC-M45 was detected with a monoclonal anti-M45 antibody (27).
RESULTS
SpiC and SsaT mutants compete equally after intraperitoneal injection of BALB/c mice.
Uchiya et al. demonstrated that a spiC::aphT allele resulted in dramatic attenuation of Salmonella serovar Typhimurium virulence for BALB/c mice, with an 50% lethal dose of >3.6 × 106 (38). This level of attenuation is similar to that observed previously for SPI2 TTSS apparatus mutants (34). Uchiya et al. also demonstrated that the strain with spiC::aphT was equivalent to a SPI2 TTSS apparatus mutant in ability to survive and replicate within macrophages (38). These results suggest that spiC mutants could be altered in the delivery of SPI2 effectors to eukaryotic cells. To further establish that spiC mutants display virulence defects identical to those of SPI2 TTSS apparatus mutants within mouse tissues, a strain with an in-frame nonpolar deletion of spiC was constructed. The resulting strain, JAF204, and SPI2 TTSS mutant strain EM232 were then tested for the ability to compete with an isogenic wild-type strain upon intraperitoneal injection of BALB/c mice (33).
Table 2 reports the mean CIs resulting from the infection of at least eight mice for each strain. The CI represents the ratio of the number of mutant to wild-type bacteria recovered from infected spleens 48 h after the intraperitoneal inoculation of 105 bacteria. Like other SPI2 TTSS mutants, ssaT mutant strain EM232 was significantly outcompeted by the wild-type strain (2, 33). Based on homology to other TTSS and flagellar proteins, SsaT should be a component of the cytoplasmic energy apparatus that is required for type III secretion (17). spiC deletion strain JAF204 exhibited the same CI as the ssaT mutant, suggesting that the spiC mutant was attenuated to the same degree as the secretion mutant. To verify this result, the spiC deletion mutant and the ssaT mutant were assayed for competition against each other. Consistent with the previous results, equal numbers of spiC and ssaT mutants were recovered from infected spleens. These results suggested that SpiC could function, like SsaT, as part of the TTSS delivery apparatus.
TABLE 2.
Virulence of SPI2 mutant Salmonella serovar Typhimurium for mice
| Genotype of: | Log10 CIa | |
|---|---|---|
| Strain 1 | Strain 2 | |
| Wild typeb | Wild typec | 0.00 ± 0.09 |
| ssaT | Wild typec | −2.32 ± 0.18 |
| ΔspiC | Wild typec | −2.31 ± 0.22 |
| ΔspiC | ssaT | −0.04 ± 0.04 |
Ratio of the number of strain 1 to strain 2 bacteria recovered from infected spleens removed 2 days after the intraperitoneal injection of 105 total bacteria into mice. The mean log10 CIs from the infection of at least eight mice are reported with corresponding standard deviations.
Wild-type chloramphenicol-resistant strain CS401, parent strain for both the ssaT and ΔspiC strains.
Wild-type kanamycin-resistant strain CS600, isogenic to CS401.
SpiC is required for the translocation of SPI2 effector proteins into infected mammalian cells.
A variety of SPI2 effector proteins have been routinely assayed for translocation by construction of gene fusions to Bordetella pertussis cyaA, which encodes a toxin that catalyzes the formation of cAMP only in the presence of eukaryotic cytoplasmic protein calmodulin (36). To determine if SpiC is required for the delivery of SPI2 effector proteins into the cytoplasm of infected mammalian cells, plasmids pEM25, pEM30, and pEM82 encoding CyaA fusions to SspH1, SspH2, and SseJ, respectively, were introduced into spiC deletion strain JAF204 and into sifA deletion strain JAF57. SifA is another SPI2 effector protein which has previously been shown to play a significant role in Salmonella serovar Typhimurium virulence, and its mutation should not necessarily alter the translocation of other effectors (2).
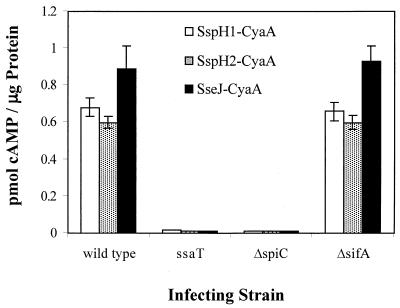
Production of cAMP was assayed 8 h after infection of cultured RAW264.7 macrophages and is shown in Fig. 2. As previously demonstrated, wild-type strain CS401 harboring pEM25, pEM30, and pEM82 induces production of cAMP but the ssaT mutant strain does not (24). The lack of cAMP production in the ssaT mutant-infected cells is not attributable to decreased viability of this mutant, for no intracellular replication difference can be detected within 8 h of infection (data not shown). Infection with sifA deletion strain JAF57 resulted in nearly wild-type levels of cAMP production, indicating that SifA is not required for the translocation of SspH1, SspH2, and SseJ into macrophages. SpiC, however, was shown to be required for the translocation of SspH1, SspH2, and SseJ, because infection of macrophages with strain JAF204, harboring pEM25, pEM30, and pEM82, did not result in production of cAMP. SpiC was also shown to be required for the translocation of a CyaA fusion to SPI2 effector protein SseI (data not shown).
FIG. 2.
Effect of deletion of spiC and sifA on the SPI2-dependent translocation of SspH1-CyaA and SspH2-CyaA. Wild-type Salmonella serovar Typhimurium strain CS401, ssaT mutant strain EM232, ΔspiC mutant strain JAF204, and ΔsifA mutant strain JAF57, each expressing sspH1-cyaA (pEM25), sspH2-cyaA (pEM30), and sseJ-cyaA (pEM82), were grown to stationary phase and used to infect RAW264.7 cells. Eight hours postinfection, infected macrophages were lysed and cellular cAMP levels were quantitated. Cellular cAMP content was normalized for total protein concentration. Error bars represent 1 standard deviation above and below the mean of experiments performed in triplicate.
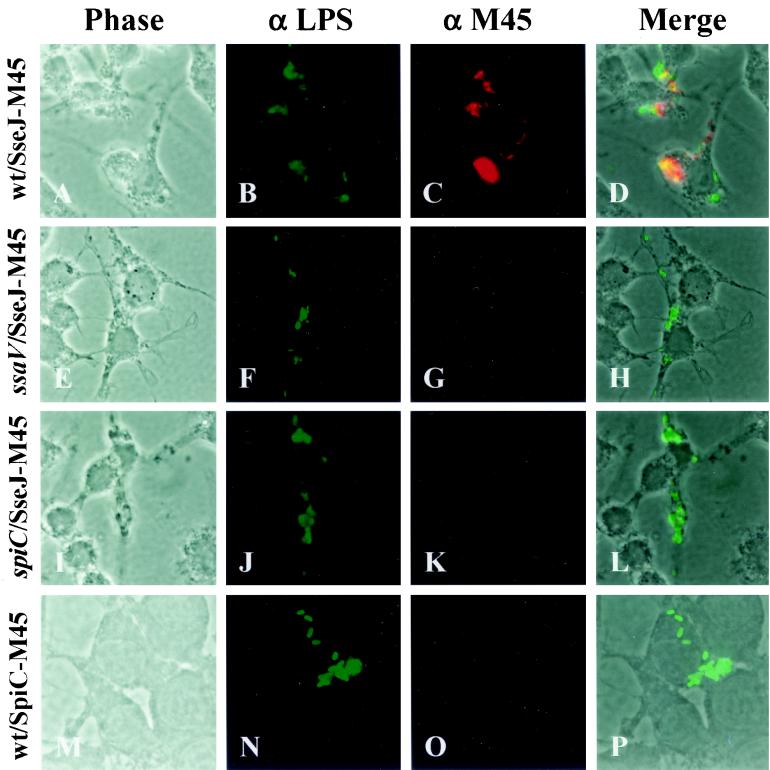
Further experiments were performed to establish that SpiC is essential for translocation of SPI2 effectors. Translocation of SPI2 effector SseJ was assayed by indirect immunofluorescence after bacterial infection of RAW264.7 cells. SseJ containing a carboxy-terminal M45 epitope of the adenovirus E4-6/7 protein has previously been shown to be expressed and secreted by the SPI2 TTSS under certain in vitro growth conditions (12). To determine if M45 epitope-tagged SseJ could be visualized upon translocation, the plasmid encoding SseJ-M45, p2129, was introduced into wild-type strain NCTC 12023, ssaV mutant strain HH109, and spiC mutant strain EG10128. SsaV encodes a component of the SPI2 TTSS apparatus and has previously been shown to be required for TTSS function (26). RAW264.7 cells were infected with the transformed Salmonella serovar Typhimurium strains and were fixed and stained for Salmonella and SseJ-M45 16 h postinfection. Because the Salmonella cells are not permeabilized in the procedure, SseJ retained within the bacteria is not detected and any intracellular M45 staining indicates SseJ-M45 translocation from the bacteria. SseJ-M45 was demonstrated to be translocated into infected RAW264.7 cells from intracellular wild-type bacteria, as shown by the diffuse red staining in Fig. 3. In cells infected with the ssaV and spiC mutants, however, no SseJ-M45 was detected, indicating that these two strains are defective in SseJ translocation. Therefore, SpiC is required for the translocation of at least several, and likely all, SPI2 effectors, suggesting that a lack of translocation of a number of SPI2 effectors contributes to the severe virulence phenotype associated with deletion of spiC.
FIG. 3.
Translocation of epitope-tagged SPI2 effector proteins. RAW264.7 cells were infected with wild-type Salmonella serovar Typhimurium strain NCTC 12023 expressing SseJ-M45 (A to D), ssaV mutant strain HH109 expressing SseJ-M45 (E to H), spiC mutant strain EG10128 expressing SseJ-M45 (I to L), and wild-type strain NCTC 12023 expressing SpiC-M45 (M to P). Sixteen hours postinfection, cells were fixed, permeabilized, and stained for bacteria (α LPS, green) and epitopes (α M45, red). Stained cells were then examined by fluorescence microscopy and photomicrographs were taken of phase contrast, green fluorescence, and red fluorescence. Merged images, in which yellow represents colocalization of green and red fluorescence, were generated with Metavue imaging software (Universal Imaging Corporation).
SpiC-M45 is not translocated into macrophages by intracellular Salmonella serovar Typhimurium.
The observation that SpiC is required for the translocation of SPI2 effectors suggests that SpiC itself may not, in fact, be an effector protein. To independently assess SpiC translocation, a spiC fusion to the M45 epitope tag coding sequence was generated on a low-copy-number plasmid. RAW267.4 cells were infected with wild-type Salmonella serovar Typhimurium strain NCTC 12023 harboring SpiC-M45 plasmid p2127, and translocation was assayed by indirect immunofluorescence 16 h after infection. As shown in Fig. 3, although SseJ-M45 was detected within macrophages infected with wild-type Salmonella serovar Typhimurium harboring p2129, SpiC-M45 was not observed within cells infected with wild-type bacteria harboring p2127. This result indicates that SpiC is likely not translocated by the SPI2 TTSS, though it is possible that it is translocated in an amount that cannot be detected or that, despite its small size, it is processed.
SpiC is limited to the cytoplasm under in vitro growth conditions that result in the secretion of translocon component SseB to the cell surface.
The facts that SpiC has previously been demonstrated to be translocated into infected macrophages and that it is required for the translocation of other effectors suggest that it may be a component of the translocon. Some translocon proteins have been shown to be translocated in small amounts into eukaryotic cells under certain conditions (10). SPI2 translocon proteins SseB, SseC, and SseD have previously been shown to localize to the bacterial cell surface in vitro when Salmonella serovar Typhimurium is grown under SPI2-inducing conditions (1, 21, 26). Presumably, these proteins are secreted by the SPI2 secretion apparatus and subsequently form a macromolecular structure on the cell surface to facilitate the translocation of SPI2 effector proteins across the phagosomal membrane of an infected host cell.
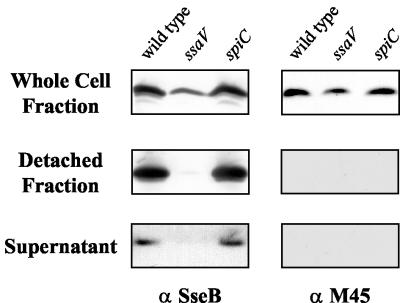
To determine if SpiC also localizes to the cell surface, plasmid p2127, encoding SpiC-M45, was introduced into wild-type and mutant Salmonella serovar Typhimurium and the resultant strains were grown in SPI2-inducing conditions. After growth to early stationary phase, both the surface-associated and secreted fractions were collected and examined. As shown in Fig. 4, translocon component SseB could be detected both on the cell surface (detached fraction) and in the culture supernatant of wild-type Salmonella serovar Typhimurium. This localization was dependent on a functional SPI2 secretion apparatus, for SseB was not secreted or localized to the cell surface by the ssaV mutant. SpiC, however, was not detectable either in the culture supernatant or on the cell surface of wild-type Salmonella serovar Typhimurium although SpiC was detected in the whole-cell fraction.
FIG. 4.
Analysis of secretion of SpiC-M45. Wild-type Salmonella serovar Typhimurium strain NCTC 12023, ssaV mutant strain HH109, and spiC mutant strain EG10128, each harboring plasmid p2127, were grown in minimal medium at pH 5.8 to induce expression and secretion of SPI2 substrate proteins. Total cell fractions and secreted protein from the culture supernatant or detached from the cell surface were prepared as described in Materials and Methods. Equal amounts of bacterial cells as adjusted according to the OD600 were analyzed for the total-cell fraction. Detached-protein and supernatant fractions were prepared from equal volumes of cultures grown to similar cell densities. SseB and SpiC-M45 were detected with a polyclonal antiserum raised against recombinant SseB (1) and monoclonal antibodies against the M45 epitope (27).
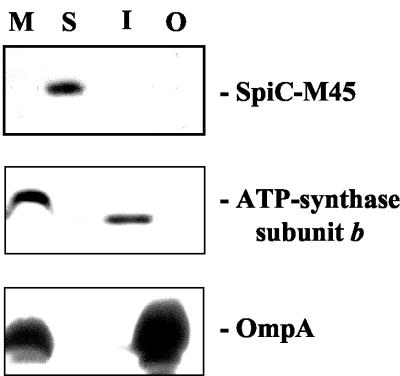
Since SpiC was not detected on either the cell surface or in the culture supernatant, the subcellular localization of SpiC was further addressed. Wild-type Salmonella serovar Typhimurium harboring p2127 was grown under SPI2-inducing conditions and subsequently fractionated into total membrane, soluble, inner membrane, and outer membrane fractions. As shown in Fig. 5, SpiC-M45 could be detected only in the soluble fraction. The purity of the soluble and two membrane fractions was demonstrated by probing for the β subunit of ATP synthase and OmpA. Taken together, these results indicate that SpiC is present in the cytoplasm when SseB is localized to the cell surface. This suggests that SpiC is limited to the cytoplasm though it remains possible that it could be secreted under different bacterial growth conditions.
FIG. 5.
Subcellular localization of SpiC-M45. Wild-type Salmonella serovar Typhimurium strain NCTC 12023 carrying p2127 was grown under conditions inducing the expression and secretion of SPI2 substrate proteins. Bacteria were harvested and subjected to subcellular fractionation as described in Materials and Methods. The various fractions (M, total membrane fraction; S, soluble fraction; I, inner membrane fraction; O, outer membrane fraction) were subjected to SDS-PAGE on 12% Tricine gels; protein was transferred onto nitrocellulose membranes, and the M45 epitope was detected with monoclonal antibodies and an ECL kit (Amersham). The purity of the various fractions was confirmed by Western blotting using antibodies against subunit β of ATP synthase and OmpA as tracers for the inner membrane and outer membrane fractions, respectively.
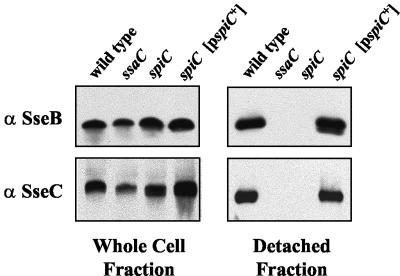
SpiC is required for the secretion and surface localization of translocon components SseB and SseC.
Since spiC mutants are defective in translocation and since SpiC is a cytoplasmic protein, it could function as part of the SPI2 type III secretion apparatus. To test this hypothesis, the spiC mutant was analyzed for the surface localization of SseB and SseC, translocon components whose surface localization is dependent on the SPI2 TTSS apparatus. Bacteria were grown in SPI2-inducing conditions, and the surface-associated proteins were extracted and subjected to analysis. As a control, wild-type bacteria and ssaC mutant bacteria were also grown similarly and analyzed. SsaC has homology to InvG from the SPI1 TTSS, which forms the outer membrane ring of the needle complex, and such mutants are completely defective in type III secretion (17). As shown in Fig. 6, SseB and SseC were detected in the detached fraction from the wild-type strain but not in the ssaC mutant strain. Although wild-type levels of SseB and SseC were produced in the cytoplasm, neither SseB nor SseC could be detected from the detached fraction of the spiC mutant, indicating that SpiC is required for the surface association of these translocon proteins. This phenotype was specifically due to the loss of SpiC, for SseB and SseC were detected on the cell surface of the spiC mutant complemented by a plasmid-borne wild-type allele of spiC. This indicated that SpiC functions in the SPI2 TTSS to promote translocon assembly, and the absence of a functional translocon explains the inability of the spiC mutant to translocate SPI2 effectors.
FIG. 6.
Effect of mutation of SPI2 genes on the surface association of SPI2 TTSS translocon proteins SseB and SseC. Wild-type Salmonella serovar Typhimurium strain NCTC 12023, ssaC mutant strain P7G2, spiC mutant strain EG10128, and spiC mutant strain EG10128 harboring pEG9127 were grown in minimal media at pH 5.8 to induce the expression of the SPI2 TTSS and the secretion of substrate proteins. Total-cell fractions and detached-protein fractions were prepared and analyzed as described in Materials and Methods.
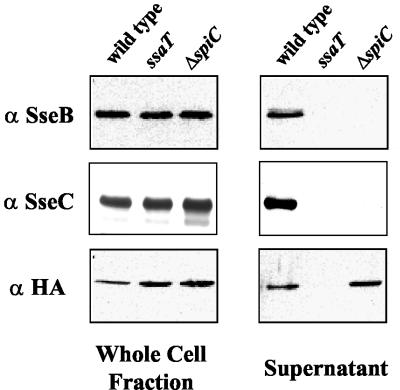
SpiC could function to maintain stability of the translocon structure on the bacterial surface. A lack of stability of the translocon structure could result in the release of translocon components into the bacterial supernatant. Therefore, to address this possibility, secretion of translocon proteins SseB and SseC into the culture supernatant in the spiC deletion mutant was assessed. Upon growth of wild-type and mutant Salmonella serovar Typhimurium in SPI2-inducing conditions, secreted proteins were precipitated from the culture supernatant. As shown in Fig. 7, SseB and SseC are secreted into the culture supernatant from wild-type, but not ssaT mutant, bacteria. Although spiC deletion strain JAF204 produces wild-type levels of SseB and SseC proteins, none are secreted into the culture supernatant. Therefore, the lack of surface localization of SseB and SseC in the spiC mutant is specifically due to a lack of secretion of SseB and SseC and not to their release into the supernatant.
FIG. 7.
Secretion of SPI2 substrate proteins. Wild-type Salmonella serovar Typhimurium strain CS401, ssaT mutant strain EM232, and ΔspiC mutant strain JAF204, each harboring HA epitope-tagged SseJ (pJAF111), were grown to early stationary phase in minimal media at pH 5.8 to induce the synthesis and secretion of SPI2 substrate proteins. Equal amounts of whole-cell lysates and secreted-protein fractions were prepared as described in Materials and Methods. Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and subjected to immunodetection using the specified antibodies.
SpiC mutants do not have a generalized TTSS secretion defect.
Several SPI2 effector and translocon components have recently been demonstrated to be secreted into the culture supernatant under certain in vitro growth conditions (12). One of the more abundant of the translocated effectors secreted into the cultured supernatant is SseJ, which in this work was demonstrated to require SpiC for translocation into the eukaryotic cell cytoplasm after bacterial infection of macrophages and epithelial cells. Therefore, to test if spiC mutants had a generalized type III secretion defect, an allele of sseJ with the coding sequence for a carboxy-terminal HA epitope tag was constructed so that secretion of SseJ into the culture supernatant could be assayed. As in the allele of spiC fused to the M45 coding sequence, expression of SseJ-HA was driven by the native sseJ promoter. The plasmid-encoded HA epitope-tagged SseJ (pJAF111) was introduced into wild-type and mutant Salmonella serovar Typhimurium. As shown in Fig. 7, SseJ-HA is secreted into the culture supernatant by wild-type but not ssaT mutant bacteria. Surprisingly, the spiC deletion mutant secreted SseJ at levels equivalent to those seen in wild-type bacteria. This result indicated that SpiC is required for the specific secretion of SseB and SseC by the SPI2 TTSS but not for the secretion of all substrate proteins. Therefore, SpiC appears to function as a cytoplasmic protein that is required for type III secretion of SseB and SseC and subsequent assembly of the translocon but not the secretion of translocated effectors. This indicates that SpiC is not required for the assembly of the type III cytoplasmic machinery or the needle complex.
DISCUSSION
This work provides further elucidation and definition of the role of SPI2 TTSS protein SpiC. A nonpolar deletion spiC mutant was constructed and was demonstrated to produce a mouse virulence phenotype equivalent to that produced by mutation of the SPI2 TTSS apparatus. This result was explained by the observation that SpiC is a cytoplasmic protein required for translocation of SPI2 effector proteins. Furthermore, SpiC was demonstrated to be required for the surface localization of translocon proteins SseB and SseC. SpiC was also shown to not be required for type III secretion of effector protein SseJ into the culture supernatant in vitro. Therefore, SpiC is essential for assembly of the translocon and delivery of effector proteins, indicating that the spiC virulence phenotypes are most likely the result of an inability to translocate all SPI2 effector proteins.
The lack of translocation of SPI2 effectors displayed by the ΔspiC mutant was not explained by an inability to replicate and survive within infected macrophages. Translocation of cyclase fusions to SPI2 effectors was assayed before an intracellular replication defect could be measured, as equal numbers of wild-type and SPI2 mutant bacteria were recovered at the time that translocation was assayed (data not shown). In contrast, a sifA mutant which is defective in intracellular replication could translocate a variety of effectors. The inability of the ΔspiC strain to translocate SPI2 effectors was also not due to polar effects on downstream genes, for this mutant was later shown to be able to secrete SseJ-HA into the culture supernatant, a function requiring genes downstream of spiC. These results indicate that, similar to other TTSS, the SPI2 translocon is not required for the in vitro secretion of other TTSS substrate proteins. Therefore, secretion of SseJ into the culture supernatant was observed in the absence of SpiC although, within macrophages, the spiC mutants were unable to translocate SseJ. Furthermore, the translocation defect was not limited to the strain with spiC deleted or to the cyclase assay, for the strain with the spiC::aphT allele was also defective for translocation of M45 epitope-tagged SseJ as determined by indirect immunofluorescence. This indicates that SpiC is not required for type III secretion and serves a more specialized function related to the translocon.
Despite its requirement for translocation of SPI2 effectors, an M45 epitope-tagged SpiC was not observed to be translocated to the cytoplasm of infected macrophages. Indeed, SpiC appears to be limited to the bacterial cytoplasm and is not secreted or localized to the cell surface when bacteria are grown in vitro under conditions that lead to the surface localization of translocon proteins SseB and SseC and the secretion of effector protein SseJ. These results are in conflict with those reported by Uchiya et al., who identified SpiC as a translocated effector protein that functions in bacterial virulence to inhibit phagolysosomal fusion. This result was further buttressed by the fact that purified SpiC could inhibit vesicle fusion in vitro. Uchiya et al. initially demonstrated SpiC translocation by indirect immunofluorescence (38). Their results using polyclonal antibodies raised against SpiC are in direct conflict with the results here using monoclonal antibodies against M45 epitope-tagged SpiC. The lack of translocation or secretion of M45-tagged SpiC was unlikely to be the result of an inability of this protein to fold or function correctly, for the spiC-M45 allele was able to complement a spiC mutant and restore the secretion and surface localization of SseB. Though it seems unlikely, the possibility exists that the M45 epitope is processed from SpiC and that, once processed, SpiC is free to be secreted and translocated.
The immunofluorescence studies in this work utilized detergents that do not permeabilize the bacteria. Protein retained within the bacteria was thus not detected in this analysis. Therefore, in the images presented here, only proteins present on the bacterial cell surface or delivered into eukaryotic cells would be detected. By this procedure, SseJ-M45 was detected within the macrophage cytoplasm upon translocation from intracellular bacteria, while no SpiC-M45 was detected, indicating that this protein was not translocated. In contrast, Uchiya et al. performed their immunofluorescence experiments using detergents that permeabilize the bacterial cell, such that bacterial cytoplasmic protein was stained (38). A review of their images indicated that most of the SpiC staining is associated with the bacteria, representing untranslocated protein. Thus, it is possible that the extrabacterial SpiC staining reflects leakage of SpiC from the permeabilized bacteria.
Regardless of whether SpiC is delivered to the cytosol of eukaryotic cells, its virulence phenotype is clearly a result of an inability to assemble a functional translocon that contains SseB and SseC. Thus, what is the role of SpiC in TTSS function? Since SpiC is required for translocon assembly, it is tempting to speculate that SpiC may itself be a component of the translocon, functioning perhaps to either anchor other translocon components to the cell surface or escort these proteins to this location. However, such proteins are usually secreted and are not usually required for the secretion of other translocon components such as SseB and SseC. Though SpiC was only detected in the bacterial cytoplasm, it is possible that a limited quantity is secreted or that it is secreted under conditions different than those used here. If SpiC is a component of the translocon, it is possible that small amounts may be directed into eukaryotic cells, for translocation of some translocon proteins into eukaryotic cells has been observed (10). The results presented here, however, suggest that SpiC is limited to the bacterial cytoplasm and is not directed either to the bacterial cell surface or into eukaryotic cells. Thus, acting within the cytoplasm, SpiC may function as a chaperone for, or regulator of, SseB and SseC secretion. In support of this hypothesis, SpiC is a small acidic protein (pI 5.4) that is required for the secretion of only a subset of proteins. Encoded immediately upstream of SseC, however, is SscA, a protein that has significant homology with Yersinia type III secretion chaperone LcrH. Since LcrH is required for the secretion of the SseC homolog, YopB, SscA may serve as the chaperone for SseC (41). Therefore, it is impossible at this time to easily classify SpiC. The experiments described here indicate that SpiC is a cytoplasmic protein that promotes SseB and SseC secretion and that all virulence phenotypes resulting from the loss of spiC are not due to the loss of a single effector but rather are due to the loss of all SPI2 effector proteins.
Acknowledgments
This work was supported by RO1 AI48683 from the National Institutes of Health, a grant of the Deutsche Forschungsgemeinschaft (HE1964/2-4), and a predoctoral fellowship of the Ludwig-Maximilians-Universität of C.R.
We thank E. A. Groisman for providing strain EG10128 and plasmid pEG9127 and C. A. Scherer for construction of plasmid pCAS61. We are indebted to J. Heesemann for generous support of this work and stimulating discussion. We thank Gabrielle Deckers-Hebestreit (Universität Osnabrück) for antisera against E. coli ATP-synthase subunit b.
REFERENCES
- 1.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brumell, J. H., S. L. Marcus, and B. B. Finlay. 2000. N-terminal conservation of putative type III secreted effectors of Salmonella typhimurium. Mol. Microbiol. 36:773-774. [DOI] [PubMed] [Google Scholar]
- 4.Brumell, J. H., C. M. Rosenberger, G. T. Gotto, S. L. Marcus, and B. B. Finlay. 2001. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell. Microbiol. 3:75-84. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., M. R. Smith, K. Thirumalai, and A. Zychlinsky. 1996. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 15:3853-3860. [PMC free article] [PubMed] [Google Scholar]
- 6.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 7.Daniell, S. J., N. Takahashi, R. Wilson, D. Friedberg, I. Rosenshine, F. P. Booy, R. K. Shaw, S. Knutton, G. Frankel, and S. Aizawa. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865-871. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deiwick, J., T. Nikolaus, J. E. Shea, C. Gleeson, D. W. Holden, and M. Hensel. 1998. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J. Bacteriol. 180:4775-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis, M. S., and H. Wolf-Watz. 1998. YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol. Microbiol. 29:799-813. [DOI] [PubMed] [Google Scholar]
- 11.Galyov, E. E., M. W. Wood, R. Rosqvist, P. B. Mullan, P. R. Watson, S. Hedges, and T. S. Wallis. 1997. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 25:903-912. [DOI] [PubMed] [Google Scholar]
- 12.Hansen-Wester, I., B. Stecher, and M. Hensel. 2002. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect. Immun. 70:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardt, W. D., and J. E. Galan. 1997. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA 94:9887-9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual, p. 457-469. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Hayward, R. D., and V. Koronakis. 1999. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 16.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, M. A., M. W. Wood, P. B. Mullan, P. R. Watson, T. S. Wallis, and E. E. Galyov. 1998. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect. Immun. 66:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaniga, K., D. Trollinger, and J. E. Galan. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaniga, K., J. Uralil, J. B. Bliska, and J. E. Galan. 1996. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol. Microbiol. 21:633-641. [DOI] [PubMed] [Google Scholar]
- 21.Klein, J. R., and B. D. Jones. 2001. Salmonella pathogenicity island 2-encoded proteins SseC and SseD are essential for virulence and are substrates of the type III secretion system. Infect. Immun. 69:737-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 25.Miller, S. I., E. L. Hohmann, and D. A. Pegues. 1995. Salmonella (including Salmonella typhi), vol. 2. Churchill Livingston Inc., New York, N.Y.
- 26.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schroder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obert, S., R. J. O'Connor, S. Schmid, and P. Hearing. 1994. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14:1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 31.Scherer, C. A., E. Cooper, and S. I. Miller. 2000. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 37:1133-1145. [DOI] [PubMed] [Google Scholar]
- 32.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shea, J. E., C. R. Beuzon, C. Gleeson, R. Mundy, and D. W. Holden. 1999. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect. Immun. 67:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 36.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 37.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 38.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 40.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 41.Wattiau, P., B. Bernier, P. Deslee, T. Michiels, and G. R. Cornelis. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. USA 91:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]