Abstract
Many bacteria concentrate their chemoreceptors at the cell poles. Chemoreceptor location is important in Escherichia coli, since chemosensory responses are sensitive to receptor proximity. It is not known, however, whether chemotaxis in other bacteria is similarly regulated. To investigate the importance of receptor-receptor interactions in other bacterial species, we synthesized saccharide-bearing multivalent ligands that are designed to cluster relevant chemoreceptors. As has been shown with E. coli, we demonstrate that the behaviors of Bacillus subtilis, Spirochaete aurantia, and Vibrio furnissii are sensitive to the valence of the chemoattractant. Moreover, in B. subtilis, chemotactic responses to serine were increased by pretreatment with saccharide-bearing multivalent ligands. This result indicates that, as in E. coli, signaling information is transferred among chemoreceptors in B. subtilis. These results suggest that interreceptor communication may be a general mechanism for modulating chemotactic responses in bacteria.
Chemotaxis is a well-studied process that has been explored in diverse bacteria, including Escherichia coli, Salmonella enterica serovar Typhimurium, and Bacillus subtilis (11, 14, 18, 62, 76). Chemotaxis is mediated by a series of chemoreceptors that transform chemosensory information into a behavioral response through a two-component system. E. coli serves as the canonical model, and in this species, the two-component signaling system comprises the receptor-associated histidine kinase CheA and the cytoplasmic response regulator CheY (22, 65). Changes in chemoreceptor occupancy modulate the kinase activity of CheA, which in turn controls the concentrations of phosphorylated CheY and CheB. Phospho-CheY can interact with the flagellar motor protein FliM (74), thereby influencing the rotation of the flagella and the behavior of the cell (9). The cell is returned to its prestimulatus behavior by the methyltransferase CheR, an enzyme that transfers methyl groups from S-adenosylmethionine to glutamate residues on the cytoplasmic domains of the chemoreceptors (71, 75). Phospho-CheB regulates this adaptation through its methylesterase activity (78). Consequently, the chemoreceptors are termed methyl-accepting chemotaxis proteins (MCPs).
Self-association of the MCPs is important for their function. The structures of the MCPs from both E. coli and Salmonella serovar Typhimurium have been investigated by X-ray crystallography and directed cross-linking. The MCPs from both species are stable homodimers (12, 19, 40, 55, 56, 77). The receptor units are highly helical throughout their periplasmic and cytoplasmic domains. At their cytoplasmic ends, the homodimers associate in a four-helix bundle of two coiled-coils connected by hairpin turns. A recent crystallographic study (40) of the E. coli serine receptor demonstrates that MCPs can oligomerize through the association of their cytoplasmic domains to form a trimer of dimers. In addition, Ames et al. (6) recently provided genetic and biochemical evidence that suggests that the E. coli Tsr and Tar MCPs interact in “signaling teams” larger than dimers. Thus, data from diverse types of experiments suggest a role for higher-order oligomers in MCP function. In addition to these dimeric and oligomeric states, the chemoreceptors are concentrated at the cell poles in patches in E. coli (47), in other bacteria (5, 24, 32, 41), and in an archaeon (24). MCP localization is also observed in elongated cells of E. coli, Vibrio parahaemolyticus, and Proteus mirabilis, where, although the MCPs are not restricted to the poles, they remain concentrated in patches (24, 48). Thus, there is evidence that MCPs have three levels of self-association: helical-hairpin dimers (stable protein-protein contacts), trimers of dimers (higher-order assemblies), and localization in micrometer-sized patches or arrays.
One potential function for MCP-MCP interactions is modulation of responses to chemoeffectors. Models of oligomeric MCP signaling arrays have suggested such a function (13, 17, 67-69), and recent experiments provide evidence that higher-order oligomers play a role in signaling (6, 23, 44). E. coli senses small (1 to 10%) changes in the concentrations of stimuli over large dynamic ranges (as many as 6 orders of magnitude) (3, 35, 39, 53, 54, 66). Detection of low chemoeffector concentrations requires substantial signal amplification to generate a behavioral response equivalent to that induced by a high ligand concentration (35, 70, 72). Bray and others have suggested that interactions between MCPs could account for the necessary signal amplification (13, 17, 67-69). They propose that a functional “lattice” of chemoreceptors can transmit signals within a higher-order complex of chemoreceptors. Recently, experimental support in E. coli for this hypothesis has emerged (6, 23, 26, 46). A full complement of MCPs is required for maximum amplification of chemotactic responses, and the data indicate that clustering of the MCPs may be functionally important. It is not known, however, whether communication among MCPs functions is a general mechanism for chemotactic response amplification in other bacteria.
Although many aspects of the chemotactic machinery are conserved among bacteria, variations are common (8, 11). For example, the number of MCP family members varies among species; Vibrio cholerae has 45 MCP-like open reading frames (33), while E. coli has only five MCP-family receptors (Tsr, Tar, Trg, Tap, and Aer) (30). Other examples of differences between E. coli and other species include the importance of membrane depolarization in chemotactic signaling in Spirochaeta aurantia (28, 29), the mechanism of adaptation in B. subtilis, which is mediated by demethylation rather than methylation of the MCPs (11), and the presence of multiple types of flagellar structures, as seen in V. parahaemolyticus (52). Additionally, in some bacteria, signaling components not found in E. coli, such as the protein CheV (21, 51), are necessary for chemotaxis.
In contrast to these differences, the localization of the MCPs is highly conserved. This suggested to us that, despite the differences among chemotactic systems in bacteria, the interreceptor communication observed in E. coli is a conserved mechanism by which chemotactic responses are amplified. This hypothesis is supported not only by the conserved localization of the MCPs (5, 24, 32, 41, 47) but also by the similar requirements among bacteria for sensitive sensory responses. Like E. coli, many types of bacteria, including B. subtilis (58, 60) and S. aurantia (31), respond to changes in chemoeffector concentration over ranges spanning 4 to 6 orders of magnitude. This sensitivity suggests that they also require a means of signal amplification. Understanding the general mechanisms by which chemotactic responses are amplified may lead to a better understanding of signaling in chemotaxis and other two-component systems.
Synthetic multivalent chemoattractants have provided insight into the role of MCP clustering in amplifying chemotactic responses in E. coli (23, 26). Multivalent ligands can be used to manipulate receptor proximity and elucidate the importance of this parameter in output responses (Fig. 1) (37). We report here the design, synthesis, and use of multivalent ligands to explore the mechanism of chemotactic response amplification in a range of bacteria. Using multivalent attractants, we demonstrate that bacteria other than E. coli, including those as divergent (15) as B. subtilis, amplify chemotactic responses through interreceptor communication.
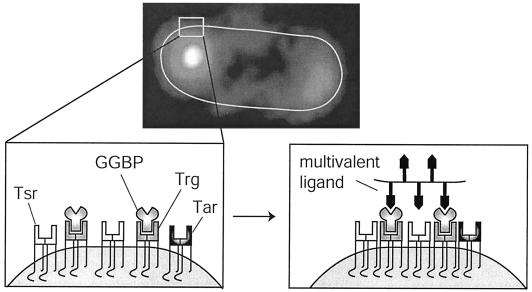
FIG. 1.
Reorganization of polar MCPs by synthetic multivalent ligands. (Top) Micrograph of an E. coli cell labeled with a fluorescent antibody against the MCPs. As has been observed previously (24, 47), the MCPs are concentrated at the poles. (Bottom left) Diagram of a collection of MCPs within the polar array. The MCPs are Tsr, the E. coli serine receptor, Tar, the aspartate receptor, and Trg, the receptor for glucose, galactose, and ribose. For simplicity the receptors are initially shown as isolated dimers, but a similar model would apply to oligomers. (Bottom right) Addition of a galactose-bearing multivalent ligand stabilizes the association between MCPs (23, 26). These multivalent ligands therefore serve as probes of processes that require receptor-receptor interactions. This figure depicts three levels of MCP self-association: dimers, oligomers, and polar arrays.
MATERIALS AND METHODS
Bacteria and growth conditions.
The bacterial strains used in these studies include E. coli AW405, B. subtilis OI1085, S. aurantia J1, and Vibrio furnissii SR1514. For E. coli, S. aurantia, and V. furnissii, motile cells were obtained by collecting cells from Luria-Bertani (LB) swim plates (0.25% agar) incubated at 30°C. For B. subtilis, motility was obtained on T broth plates (0.3% agar, 1% tryptone, 0.2 mM MgCl2, 0.5% NaCl, 0.01 mM MnCl2, 10 mM glucose, and 0.5% glycerol) grown at 37°C. Liquid cultures inoculated from these plates were supplemented with a 1 mM concentration of the monosaccharide relevant for the subsequent experiments.
Synthesis of chemoattractants.
Compounds 1 to 11 were synthesized as described previously (26, 36, 73). Briefly, monomers 1, 5, and 8 were generated by standard procedures utilized in ring-opening metathesis polymerization (ROMP) reactions to synthesize polymers 2 to 4, 6 and 7, and 9 to 11 (36). Termination of the polymerization reaction with a bifunctional capping agent (27, 61) provided the means to attach fluorophores. Compounds 12 to 14 were generated by this synthetic process. The valences (n) of compounds 2 to 4, 6 and 7, and 9 to 14 are reported here as the ratio between the monomer and the initiator used in the polymerization. This value can be used to predict the lengths of the resulting polymers (36, 50). Gel permeation chromatography (GPC) can be used to determine the degree of polymerization (DP), which is another estimation of the average number of monomer units incorporated into polymers. Reported DPs for these types of compounds are approximately 18 (n = 10) and 45 (n = 25) (73). The polydispersity indices (a measure of the heterogeneity of the polymer lengths produced) of polymers prepared by this method are on the order of 1.1 to 1.2 (a single compound has a polydispersity index of 1) (36). The lengths, and therefore the molecular masses, of these ligands were controlled to allow penetration of the outer membrane (in gram-negative bacteria). For example, compound 4 has an average molecular mass of 17 kDa. Globular proteins as large as 10 kDa have been shown to penetrate the outer membrane of E. coli (16).
Fluorescence microscopy.
Cells were prepared for fluorescence microscopy as described previously (24, 26). Compounds 12 to 14 were used at 500 μM. Concentrations are reported as the molar concentration of the saccharide (or saccharide residue), not the polymer concentration.
Capillary chemotaxis assay.
Capillary accumulation assays were performed as previously described (2, 10, 26, 31, 59). The buffer employed in experiments with E. coli was 10 mM potassium phosphate, pH 7.0, with 10 μM EDTA (2). For experiments with S. aurantia, a 10 mM potassium phosphate buffer, pH 7.0, with 0.2 mM cysteine was used (31). For V. furnissii experiments, a 10 mM phosphate buffer, pH 7.2, with 0.1 mM EDTA and 340 mM NaCl was employed (10, 64). For B. subtilis chemotaxis experiments, a 10 mM phosphate buffer, pH 7.0, with 10 μM EDTA, 0.05% glycerol, and 0.3 mM (NH4)2SO4 was used (59). Capillary assays were performed at 30°C for E. coli, 37°C for B. subtilis, and 25°C for S. aurantia and V. furnissii. Results are averages from at least three experiments performed in duplicate. The error was approximately 20%.
Motion analysis.
The behavior of B. subtilis was quantitated by analysis of bacterial motion, as described previously for E. coli (26, 63). The similar locomotion of these bacteria allowed the use of identical instrumentation. For experiments measuring the response of B. subtilis to compounds 5 to 7, cells were washed three times with chemotaxis buffer and resuspended to an optical density at 550 nm (OD550) of 0.01. A sample of this solution was placed under a coverslip and allowed to equilibrate for 1 to 2 min prior to addition of the attractant. After addition of the attractant, 2 min of bacterial behavior at 25°C was recorded. The paths of the cells were determined by using the ExpertVision system (Motion Analysis Corporation). Experiments measuring the response of B. subtilis to serine were performed similarly to those described above. After equilibration for 1 to 2 min, cells were treated with either a buffer or compounds 5 to 7 at a glucose residue concentration of 10 μM. Prior to treatment with serine, cells were incubated for 120 s, which allowed full adaptation to attractants 5 to 7. Serine was added to a final concentration of 10 μM, and motion was recorded for 60 s. The angular velocity was used as a measure of chemotactic response, but analysis of the linear velocity yielded similar results (data not shown). In contrast to experiments with E. coli (26), no EDTA treatment was required.
RESULTS
Design and synthesis of chemoattractants.
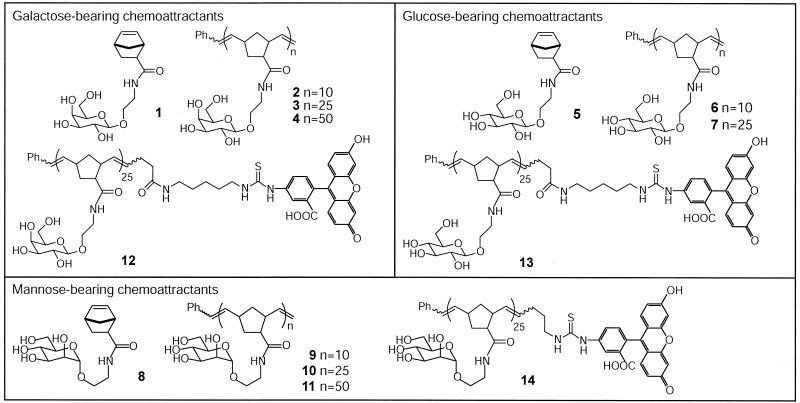
In our studies, we targeted E. coli, S. aurantia, V. furnissii, and B. subtilis to represent a range of gram-negative and gram-positive species (15). Each type of bacteria recognizes a distinct set of chemoattractants. For example, galactose is a potent chemoattractant for E. coli (3), but it is a poor attractant for B. subtilis (60). To investigate chemotactic responses in diverse bacteria, we sought to synthesize multivalent chemoattractants possessing a range of saccharides to provide access to reagents with broad utility. Toward this goal, the galactose-, glucose-, and mannose-bearing multivalent ligands 2 to 4, 6 and 7, and 9 to 11 were generated by ROMP (Fig. 2). Monomers 1, 5, and 8 were also tested as attractants for comparison. Such experiments were critical to verify that the modification of the attractant for incorporation into the polymer did not abolish its activity (25). In addition, fluorescent derivatives 12 to 14 were generated for colocalization studies with the MCPs (27, 61).
FIG. 2.
Chemical structures of saccharide-bearing compounds.
Fluorescent multivalent chemoattractants colocalize with the chemoreceptors.
To investigate whether synthetic multivalent chemoattractants bind specifically to the MCPs, we examined the colocalization of fluorescently labeled polymers 12 to 14 with fluorescent anti-MCP antibodies by microscopy. Our experiments are based on the observation by Hazelbauer and coworkers that antibodies raised against the E. coli chemoreceptors are cross-reactive with putative MCPs from other bacteria (4, 57). Examination by fluorescence microscopy of bacteria labeled with both fluorescent anti-MCP antibodies and compounds 12 to 14 revealed that the polymers colocalize with the anti-MCP antibodies in E. coli, B. subtilis, S. aurantia, and V. furnissii (Fig. 3). Importantly, when the saccharide moiety displayed from the ligand was not a chemoattractant for the species, no specific labeling was observed (data not shown). Additionally, high concentrations of unlabeled monosaccharide prevented binding of the fluorescent ligands (data not shown). These results suggest that synthetic saccharide-bearing polymers interact specifically with the MCPs. We therefore investigated the utility of the synthetic ligands as chemoattractants.
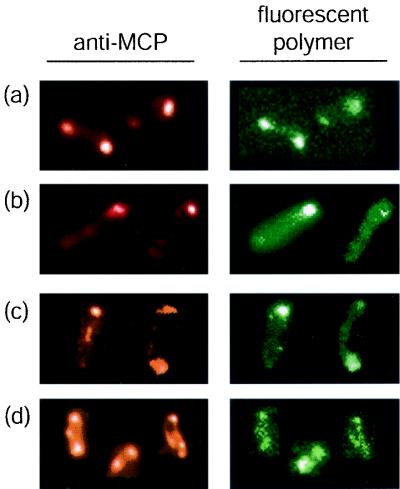
FIG. 3.
Binding of fluorescent multivalent ligands to the MCPs. (a) E. coli treated with an anti-MCP antibody and fluorescent galactose-bearing polymer 12. (b) B. subtilis treated with an anti-MCP antibody and fluorescent glucose-bearing polymer 13. (c) S. aurantia treated with an anti-MCP antibody and fluorescent glucose-bearing polymer 13. (d) V. furnissii treated with an anti-MCP antibody and mannose-bearing polymer 14.
Chemoattractant valence influences chemotactic response.
It has been shown previously that multivalent chemoattractants of sufficient length are capable of clustering the MCPs of E. coli (23, 26). Additionally, the concentration of a multivalent ligand required for chemotactic responses decreases with increasing ligand valence (26). To extend these findings to evolutionarily diverse bacteria, we conducted capillary assays on cells of B. subtilis, S. aurantia, and V. furnissii. We used the ligands at concentrations normalized to the amount of saccharide derivative to allow comparisons between attractants of various lengths. Addition of compounds 1 to 11 to these cells revealed that chemotactic responses were dependent on the valence of the attractant (Table 1). As the valence of the ligand increased, its chemotactic potency also increased. These data suggest that chemotaxis in these bacteria may be influenced by MCP proximity. Consistent with our fluorescence microscopy results, only those ligands displaying saccharides that serve as chemoattractants for the individual bacterial species were effective in this assay.
TABLE 1.
Effect of chemoattractant valence on chemotactic responses
| Strain and saccharide | Chemotactic response (relative potency)a to:
|
|||
|---|---|---|---|---|
| Monomer | 10-mer | 25-mer | 50-mer | |
| E. coli | ||||
| Galactoseb | 1 (1) | 1 (1) | 0.25 (4) | 0.1 (10) |
| Glucose | 10 (1) | 1 (10) | 0.5 (20) | ND |
| B. subtilis | ||||
| Galactose | Nonchemotacticc | Nonchemotactic | Nonchemotactic | Nonchemotactic |
| Glucoseb | 1 (1) | 0.1 (10) | 0.01 (100) | ND |
| V. furnissii, mannose | 20 (1) | 10 (2) | 5 (4) | 0.5 (40) |
| S. aurantia | ||||
| Galactose | Nonchemotacticc | Nonchemotactic | Nonchemotactic | Nonchemotactic |
| Glucose | 0.01 (1) | 0.1 (10) | 0.0001 (100) | ND |
Chemotactic response is reported as the millimolar saccharide concentration of maximum bacterial accumulation in the capillary. Relative potency is the potency of the chemoattractant relative to the monomer. The result for the most potent ligand for each bacterium is boldfaced. ND, not determined. Maximum accumulation ranges for multivalent ligands and unmodified saccharides, respectively are as follows: E. coli, 4,000 to 5,000 and 150,000; B. subtilis, 2,000 to 3,000 and 2,000; V. furnissii, 15,000 to 25,000 and not determined; S. aurantia, 200,000 to 400,000 and 40,000.
See reference 26.
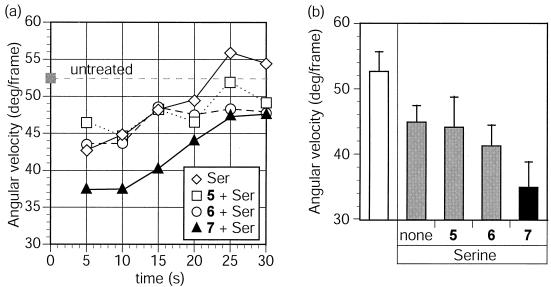
The results of capillary assays can sometimes be difficult to interpret. Complicating factors include chemoeffector diffusion, accessibility, and turnover (1, 63). To examine chemotactic activity using a different assay, B. subtilis chemotactic responses were investigated by motion analysis (26, 63). In this assay, an increase in attractant concentration is expected to reduce directional changes (angular velocity) and enhance swimming speed (translational velocity). In agreement with the results obtained from capillary assays, the behavior of B. subtilis was dependent on the valence of the chemoattractant (Fig. 4). The highest-valence glucose-bearing ligand 7 generated an angular velocity that was decreased by 11% relative to that caused by the monovalent ligand 5 at an identical glucose concentration. This result indicates that the multivalent ligand 7 is a more potent chemoattractant than its monovalent counterpart. This assay also provided information about the adaptation of B. subtilis to compounds 5 to 7. Unlike that in E. coli (23), adaptation in B. subtilis was not significantly affected by ligand valence; times for adaptation to compounds 5 to 7 were all approximately 110 to 130 s.
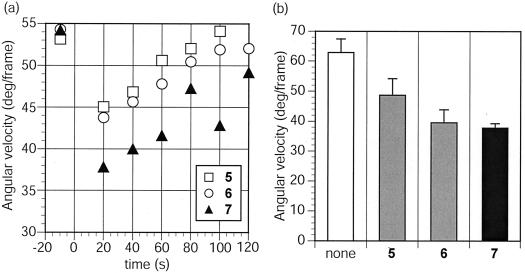
FIG. 4.
Effects of synthetic ligands on chemotactic response in B. subtilis. (a) Time course for monitoring of the angular velocities of B. subtilis after treatment at time zero with compounds 5 to 7 at 10 μM. Ligand concentrations are calculated based on the glucose residue concentration. The angular velocity of the cells prior to treatment is shown. Data points are averages of 10-s intervals. Approximately 2 s were lost during movement of the microscope objective prior to and after addition of ligand. Data are from a single experiment that is representative of three replicates. (b) The average angular velocities of B. subtilis during the first 60 s after treatment with compounds 5 to 7 at 10 μM. Data are averages from three independent experiments performed in triplicate. Error bars, single standard deviations.
Clustering of one chemoreceptor type influences responses mediated through another.
In previous experiments using E. coli (23, 26), we found that high-valence galactose-bearing ligands were capable of stabilizing clusters of MCPs. These clusters contained copies of the MCPs that mediate responses to both galactose (Trg) and serine (Tsr). Responses to serine were potentiated in the presence of a multivalent ligand that could stabilize these clusters. The potentiation of serine responses within clusters strongly supports the involvement of interreceptor interactions in signaling. To examine the same issue in other species, we added attractants 5 to 7 to B. subtilis and then allowed the cells to adapt to the synthetic ligands for various times. The subsequent response to serine was monitored by motion analysis (Fig. 5). The highest-valence ligand, compound 7, caused a 20% increase in the response to serine. No potentiation was observed upon pretreatment with the monovalent ligand 5, and only a minor effect was observed with the shorter oligovalent ligand 6. These results suggest that stabilizing inter-MCP interactions enhances the amplification of chemotactic responses mediated by receptors within the clusters.
FIG. 5.
Behavioral response of B. subtilis to serine after pretreatment with compounds 5 to 7. (a) Cells were treated either with a buffer or with compounds 5 to 7 at 10 μM. After 120 s, serine was added to a final concentration of 10 μM. Data points are the average angular velocity of 5-s intervals. Results are representative of four independent experiments performed in triplicate. (b) First 15 s of the response of B. subtilis to serine after pretreatment with a buffer or compounds 5 to 7. Results are averages from four independent experiments performed in triplicate. Error bars, standard deviations.
DISCUSSION
In E. coli, there is evidence that interreceptor communication within a polar lattice is important in the amplification of chemotactic responses (6, 13, 23). We have investigated whether this mechanism is conserved in other bacteria. Using synthetic multivalent ligands, we found that, as in E. coli, chemotactic responses of B. subtilis, V. furnissii, and S. aurantia are sensitive to ligand valence. Our results focus on chemotactic behavior, and this behavior is directly related to signaling. Thus, the data suggest that changes in MCP localization influence the phosphorylation and methylation of the relevant components of the chemotactic signaling system. These results suggest that communication between MCPs is evolutionarily conserved, as is the importance of receptor-receptor interactions in the amplification of chemotactic responses. Our data provide a functional role for MCP localization in the many bacteria that localize these proteins.
In addition to their similarities, there are differences between the chemotactic responses and signaling mechanisms of various bacteria. Our results indicate that variation within the chemotactic machinery of a particular species can be illuminated with multivalent ligands. Specifically, it had been found previously that increasing the valence of a chemoattractant increased the adaptation time of E. coli approximately twofold (23). In our studies with B. subtilis, however, we observed no effect of ligand valence on adaptation times (Fig. 4). This result is consistent with the different mechanisms for adaptation in E. coli and B. subtilis (11). Additionally, the sensitivity of S. aurantia to ligand valence was far greater than that of E. coli. In S. aurantia, a 100-fold increase in potency was observed for the multivalent ligands, and by comparison, a maximum 10-fold increase was observed in experiments with E. coli (Table 1). This variation could be related to the differences between the chemotactic systems of these organisms. Chemotaxis in S. aurantia involves membrane depolarization (28, 29), but in E. coli it does not. Alternatively, differences in sensitivity could be related to structural differences between the MCPs in these organisms. For instance, multivalent ligands might cluster the chemoreceptors more effectively in one species than in another. Conversely, changes in chemoreceptor clustering might have different effects on kinase or methyltransferase activities in different species. Thus, these synthetic reagents may serve as sensitive probes of differences in chemotactic machinery.
There are several mechanisms by which the multivalent ligands might potentiate chemotactic responses. The synthetic attractants are designed to stabilize interreceptor interactions (Fig. 1), and they have been shown to be capable of this function (26). An alternative mode of action for these materials, however, is potentiation of responses via increased avidity. Multivalent ligands often have higher avidity for their target than monovalent derivatives (37, 38, 43, 49). Although a multivalent chemoattractant can exhibit an increase in binding through avidity (25), this mechanism does not account for all of the data. If the enhanced chemotactic response were due to avidity alone, saccharide-bearing ligands should not influence B. subtilis responses to serine (Fig. 5). The potentiation of a response through the serine-sensing receptor, which does not directly bind the synthetic multivalent chemoattractants, provides evidence of communication between the glucose- and serine-sensing receptors. Therefore, our data support a mechanism that involves MCP clustering.
Changes in MCP clustering could influence the structure of the signaling complex and/or the stoichiometry of signaling components within it. Specifically, clustering may favor direct protein-protein contacts between glucose- and serine-sensing MCPs, similar to those observed in vitro in dimers and oligomers (40, 44). Thus, changes in conformation induced by chemoeffector binding would be relayed to other MCP members of the complex and ultimately to the signaling components. Moreover, MCP clustering may serve to concentrate or exclude cytoplasmic components, such as CheA or CheB (70). Additional experiments to further elucidate the molecular mechanisms of signal amplification are ongoing.
Two-component systems mediate many prokaryotic and some eukaryotic responses to stimuli (7, 34, 42, 45). Additionally, two-component systems, including those governing chemotaxis (20), have been implicated in virulence and pathogenicity. We suggest that synthetic molecules designed to control interreceptor proximity may serve to uncover relationships between receptor location and signaling in a range of physiologically and medically important two-component systems.
Acknowledgments
Bacterial strains were provided by J. Adler (E. coli AW405), G. Ordal (B. subtilis OI1085), E. P. Greenberg (S. aurantia JI), and L. McCarter (V. furnissii SR1514). For supplying antibodies we thank G. Hazelbauer (anti-Trg), J. S. Parkinson (anti-Tsr), and G. Ordal (anti-McpB). S. Bednarek supplied access to fluorescence microscopy equipment. We thank F. Boehm for providing synthetic intermediates.
This research was supported in part by a grant from the NIH (GM 55984). A.C.L. acknowledges the NSF predoctoral fellowship program for support. J.E.G. thanks the NIH Biotechnology Training Program for a predoctoral fellowship (GM 08349). L.E.S. was supported by an NIH predoctoral fellowship (GM 18750). R.M.O. was supported by a Pharmacia-Upjohn fellowship and a fellowship from Eastman Chemical Company.
REFERENCES
- 1.Adler, J. 1969. Chemoreceptors in bacteria. Science 166:1588-1597. [DOI] [PubMed] [Google Scholar]
- 2.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 3.Adler, J., G. L. Hazelbauer, and M. M. Dahl. 1973. Chemotaxis towards sugars in Escherichia coli. J. Bacteriol. 115:824-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam, M., and G. L. Hazelbauer. 1991. Structural features of methyl-accepting taxis proteins conserved between Archaebacteria and Eubacteria revealed by antigenic cross-reaction. J. Bacteriol. 173:5837-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alley, M. R. K., J. R. Maddock, and L. Shapiro. 1992. Polar localization of a bacterial chemoreceptor. Genes Dev. 6:825-836. [DOI] [PubMed] [Google Scholar]
- 6.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appleby, J. L., J. S. Parkinson, and R. B. Bourret. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86:845-848. [DOI] [PubMed] [Google Scholar]
- 8.Armitage, J. P., and R. Schmitt. 1997. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology 143:3671-3682. [DOI] [PubMed] [Google Scholar]
- 9.Barak, R., and M. Eisenbach. 1992. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry 31:1822-1826. [DOI] [PubMed] [Google Scholar]
- 10.Bassler, B. L., P. J. Gibbons, C. Yu, and S. Roseman. 1991. Chitin utilization by marine bacteria. J. Biol. Chem. 266:24268-24275. [PubMed] [Google Scholar]
- 11.Bischoff, D. S., and G. W. Ordal. 1992. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol. Microbiol. 6:23-28. [DOI] [PubMed] [Google Scholar]
- 12.Boyd, A., K. Kendall, and M. I. Simon. 1983. Structure of the serine chemoreceptor in Escherichia coli. Nature 301:623-626. [DOI] [PubMed] [Google Scholar]
- 13.Bray, D., M. D. Levin, and C. J. Morton-Firth. 1998. Receptor clustering as a cellular mechanism to control sensitivity. Nature 393:85-88. [DOI] [PubMed] [Google Scholar]
- 14.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal perception. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown, J. R., and W. F. Doolittle. 1997. Archaea and the prokaryotic-to-eukaryotic transition. Microbiol. Mol. Biol. Rev. 61:456-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, G., A. Hayhurst, J. G. Tomas, B. R. Harvey, B. L. Iverson, and G. Georgiou. 2001. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS). Nat. Biotechnol. 19:537-542. [DOI] [PubMed] [Google Scholar]
- 17.Duke, T. A. J., and D. Bray. 1999. Heightened sensitivity of a lattice of membrane receptors. Proc. Natl. Acad. Sci. USA 96:10104-10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13:457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falke, J. J., and D. E. Koshland, Jr. 1987. Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science 237:1596-1600. [DOI] [PubMed] [Google Scholar]
- 20.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possess two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredrick, K. L., and J. D. Helmann. 1994. Dual chemotaxis signaling pathways in Bacillus subtilis: a σD-dependent gene encodes a novel protein with both CheW and CheY homologous domains. J. Bacteriol. 176:2727-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 23.Gestwicki, J. E., and L. L. Kiessling. 2002. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415:81-84. [DOI] [PubMed] [Google Scholar]
- 24.Gestwicki, J. E., A. C. Lamanna, R. M. Harshey, L. L. McCarter, L. L. Kiessling, and J. Adler. 2000. Evolutionary conservation of methyl-accepting chemotaxis protein location in Bacteria and Archaea. J. Bacteriol. 182:6499-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gestwicki, J. E., L. E. Strong, S. L. Borchardt, C. W. Cairo, A. M. Schnoes, and L. L. Kiessling. 2001. Designed potent multivalent chemoattractants for Escherichia coli. Bioorg. Med. Chem. 9:2387-2393. [DOI] [PubMed] [Google Scholar]
- 26.Gestwicki, J. E., L. E. Strong, and L. L. Kiessling. 2000. Tuning chemotactic responses using synthetic multivalent ligands. Chem. Biol. 7:583-591. [DOI] [PubMed] [Google Scholar]
- 27.Gordon, E. J., J. E. Gestwicki, L. E. Strong, and L. L. Kiessling. 2000. Synthesis of end-labeled multivalent ligands for exploring cell-surface-receptor-ligand interactions. Chem. Biol. 7:9-16. [DOI] [PubMed] [Google Scholar]
- 28.Goulbourne, E. A. J., and E. P. Greenberg. 1981. Chemotaxis of Spirochaeta aurantia: involvement of membrane potential in chemosensory signal transduction. J. Bacteriol. 148:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulbourne, E. A. J., and E. P. Greenberg. 1983. A voltage clamp inhibits chemotaxis of Spirochaeta aurantia. J. Bacteriol. 153:916-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grebe, T. W., and J. Stock. 1998. Bacterial chemotaxis: the five sensors of a bacterium. Curr. Biol. 8:R154-R157. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg, E. P., and E. Canale-Parola. 1977. Chemotaxis in Spirochaeta aurantia. J. Bacteriol. 130:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison, D., J. Skidmore, J. Armitage, and J. Maddock. 1999. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol. Microbiol. 31:885-892. [DOI] [PubMed] [Google Scholar]
- 33.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoch, J. A., and K. I. Varughese. 2001. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 183:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jasuja, R., Y. Lin, D. R. Trentham, and S. Khan. 1999. Response tuning in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 96:11346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanai, M., K. H. Mortell, and L. L. Kiessling. 1997. Varying the size of multivalent ligands: the dependence of concanavalin A binding on neoglycopolymer length. J. Am. Chem. Soc. 119:9931-9932. [Google Scholar]
- 37.Kiessling, L. L., J. E. Gestwicki, and L. E. Strong. 2000. Synthetic multivalent ligands in the exploration of cell surface interactions. Curr. Opin. Chem. Biol. 4:696-703. [DOI] [PubMed] [Google Scholar]
- 38.Kiessling, L. L., and N. L. Pohl. 1996. Strength in numbers: non-natural polyvalent carbohydrate derivatives. Chem. Biol. 3:71-77. [DOI] [PubMed] [Google Scholar]
- 39.Kim, C., M. Jackson, R. Lux, and S. Khan. 2001. Determinants of chemotactic signal amplification in Escherichia coli. J. Mol. Biol. 307:119-135. [DOI] [PubMed] [Google Scholar]
- 40.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 41.Kirby, J. R., T. B. Niewold, S. Maloy, and G. W. Ordal. 2000. CheB is required for behavioral responses to negative stimuli during chemotaxis in Bacillus subtilis. Mol. Microbiol. 35:44-57. [DOI] [PubMed] [Google Scholar]
- 42.Koretke, K. K., A. N. Lupas, P. V. Warren, M. Rosenberg, and J. R. Brown. 2001. Evolution of two-component signal transduction. Mol. Biol. Evol. 17:1956-1970. [DOI] [PubMed] [Google Scholar]
- 43.Lee, Y. C., and R. T. Lee. 1995. Carbohydrate-protein interactions: basis of glycobiology. Acc. Chem. Res. 28:321-327. [Google Scholar]
- 44.Liu, Y., M. Levit, R. Lurz, M. G. Surette, and J. B. Stock. 1997. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 16:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loomis, W. F., G. Shaulsky, and N. Wang. 1997. Histidine kinases in signal transduction pathways of eukaryotes. J. Cell Sci. 110:1141-1145. [DOI] [PubMed] [Google Scholar]
- 46.Lybarger, S. R., and J. R. Maddock. 2000. Differences in the polar clustering of the high- and low-abundance chemoreceptors of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8057-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 48.Maki, N., J. E. Gestwicki, E. M. Lake, L. L. Kiessling, and J. Adler. 2000. Motility and chemotaxis in filamentous cells of Escherichia coli. J. Bacteriol. 182:4337-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mammen, M., S.-K. Choi, and G. M. Whitesides. 1998. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 37:2755-2794. [DOI] [PubMed] [Google Scholar]
- 50.Mann, D. A., M. Kanai, D. J. Maly, and L. L. Kiessling. 1998. Probing low affinity and multivalent interactions with surface plasmon resonance: ligands for concanavalin A. J. Am. Chem. Soc. 120:10575-10582. [Google Scholar]
- 51.Martin, A. C., G. H. Wadhams, and J. P. Armitage. 2001. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol. Microbiol. 40:1261-1272. [DOI] [PubMed] [Google Scholar]
- 52.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mesibov, R., and J. Adler. 1972. Chemotaxis toward amino acids in Escherichia coli. J. Bacteriol. 112:315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mesibov, R., G. W. Ordal, and J. Adler. 1973. The range of attractant concentrations for bacterial chemotaxis and the threshold and size over this range. J. Gen. Physiol. 62:203-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milburn, M. V., G. G. Prive, D. L. Milligan, W. G. Scott, J. Yeh, J. Jancarik, D. E. Koshland, Jr., and S.-H. Kim. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342-1347. [DOI] [PubMed] [Google Scholar]
- 56.Milligan, D. L., and D. E. Koshland, Jr. 1988. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 263:6268-6275. [PubMed] [Google Scholar]
- 57.Morgan, D. G., J. W. Baumgartner, and G. L. Hazelbauer. 1993. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J. Bacteriol. 175:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ordal, G. W., and K. J. Gibson. 1977. Chemotaxis toward amino acids by Bacillus subtilis. J. Bacteriol. 129:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ordal, G. W., and D. J. Goldman. 1975. Chemotaxis away from uncouplers of oxidative phosphorylation in Bacillus subtilis. Science 189:802-805. [DOI] [PubMed] [Google Scholar]
- 60.Ordal, G. W., D. P. Villani, and M. S. Rosendahl. 1979. Chemotaxis towards sugars by Bacillus subtilis. J. Gen. Microbiol. 115:167-172. [DOI] [PubMed] [Google Scholar]
- 61.Owen, R. M., J. E. Gestwicki, T. Young, and L. L. Kiessling. 2002. Synthesis and applications of end-labeled neoglycopolymers. Org. Lett. 4:2293-2296. [DOI] [PubMed] [Google Scholar]
- 62.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 63.Sager, B. M., J. J. Sekelsky, P. Matsumura, and J. Adler. 1988. Use of a computer to assay motility in bacteria. Anal. Biochem. 173:271-277. [DOI] [PubMed] [Google Scholar]
- 64.Sar, N., L. McCarter, M. Simon, and M. Silverman. 1990. Chemotactic control of the two flagellar systems of Vibrio parahaemolyticus. J. Bacteriol. 172:334-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuster, S. C., R. V. Swanson, L. A. Alex, R. B. Bourret, and M. I. Simon. 1993. Assembly and function of a quaternary signal transduction complex by surface plasmon resonance. Nature 365:343-347. [DOI] [PubMed] [Google Scholar]
- 66.Segall, J. E., S. M. Block, and H. C. Berg. 1986. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 83:8987-8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi, Y. 2001. Effects of thermal fluctuation and the receptor-receptor interaction in bacterial chemotactic signaling and adaptation. Phys. Rev. E 64:1910.. [DOI] [PubMed] [Google Scholar]
- 68.Shi, Y., and T. Duke. 1998. Cooperative model of bacterial sensing. Phys. Rev. E 58:6399-6406. [Google Scholar]
- 69.Shimizu, T. S., N. Le Novére, M. D. Levin, A. J. Beavil, B. J. Sutton, and D. Bray. 2000. Molecular model of a lattice of signaling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2:792-796. [DOI] [PubMed] [Google Scholar]
- 70.Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 99:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Springer, W. R., and D. E. Koshland, Jr. 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. USA 74:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stock, A. M. 1999. A nonlinear stimulus-response relation in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 96:10945-10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strong, L. E., and L. L. Kiessling. 1999. A general synthetic route to defined, biologically active multivalent arrays. J. Am. Chem. Soc. 121:6193-6196. [Google Scholar]
- 74.Welch, M., K. Oosawa, S.-I. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch. Proc. Natl. Acad. Sci. USA 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu, J., J. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 76.Wylie, D., A. Stock, C.-Y. Wong, and J. Stock. 1988. Sensory transduction in bacterial chemotaxis involves phosphotransfer between Che proteins. Biochem. Biophys. Res. Commun. 151:891-896. [DOI] [PubMed] [Google Scholar]
- 77.Yeh, J. I., H.-P. Biemann, J. Pandit, D. E. Koshland, and S.-H. Kim. 1993. The three-dimensional structure of the ligand-binding domain of a wild-type chemotaxis receptor. J. Biol. Chem. 268:9787-9792. [PubMed] [Google Scholar]
- 78.Yonekawa, H., H. Hayashi, and J. S. Parkinson. 1983. Requirement of the cheB function for sensory adaptation in Escherichia coli. J. Bacteriol. 156:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]