Abstract
NAD(H)-dependent l-alanine dehydrogenase (EC 1.4.1.1) (Ald) catalyzes the oxidative deamination of l-alanine and the reductive amination of pyruvate. To assess the physiological role of Ald in Mycobacterium smegmatis, we cloned the ald gene, identified its promoter, determined the protein expression levels, and analyzed the combined effects of nutrient supplementation, oxygen availability, and growth stage on enzyme activity. High Ald activities were observed in cells grown in the presence of l- or d-alanine regardless of the oxygen availability and growth stage. In exponentially growing cells under aerobic conditions, supplementation with alanine resulted in a 25- to 50-fold increase in the enzyme activity. In the absence of alanine supplementation, 23-fold-higher Ald activities were observed in cells grown exponentially under anaerobic conditions. Furthermore, M. smegmatis ald null mutants were constructed by targeted disruption and were shown to lack any detectable Ald activity. In contrast, the glycine dehydrogenase (EC 1.4.1.10) (Gdh) activity in mutant cells remained at wild-type levels, indicating that another enzyme protein is responsible for the physiologically relevant reductive amination of glyoxylate. The ald mutants grew poorly in minimal medium with l-alanine as the sole nitrogen source, reaching a saturation density 100-fold less than that of the wild-type strain. Likewise, mutants grew to a saturation density 10-fold less than that of the wild-type strain under anaerobic conditions. In summary, the phenotypes displayed by the M. smegmatis ald mutants suggest that Ald plays an important role in both alanine utilization and anaerobic growth.
NAD(H)-dependent l-alanine dehydrogenase (EC 1.4.1.1) (Ald) catalyzes the oxidative deamination of l-alanine to pyruvate and ammonia (catabolic reaction) or, in the reverse direction, the reductive amination of pyruvate to l-alanine (biosynthetic reaction). Ald has been extensively studied in many microorganisms since it was first discovered in Bacillus subtilis (51). Ald plays a key role in the utilization of carbon and nitrogen sources in various microorganisms, thus providing a link between carbohydrate and amino acid metabolic pathways (6). In addition, Ald is required for spore formation in B. subtilis (37) and fruiting body development in Myxococcus xanthus (44). The specific functions of Ald are different in different bacterial species and include a catabolic role in B. subtilis for energy production during spore germination (37) and a biosynthetic role in Bradyrhizobium japonicum (38, 39), Rhizobium leguminosarum (1), and Pseudomonas sp. strain MA (4) for nitrogen assimilation. However, its primary functions in many other microorganisms remain unknown.
Ald activity was discovered in Mycobacterium tuberculosis H37Ra in 1959 (17). In the early 1990s, studies with monoclonal antibodies against M. tuberculosis led to identification of a 40-kDa antigen recognized by HBT-10. This protein was demonstrated to be a functional l-alanine dehydrogenase (2). Ald is one of the predominant antigens in M. tuberculosis culture filtrate, although it lacks a signal peptide, like several other M. tuberculosis extracellular proteins. It has been suggested that Ald may play a role in peptidoglycan biosynthesis since both l-alanine and d-alanine are important components of mycobacterial peptidoglycan. However, immunoblot analysis failed to detect Ald in Mycobacterium bovis BCG strains (2). Similar results were obtained by proteomic analysis (24). Thus, the physiological role of Ald in mycobacteria has not been elucidated.
Information about the regulation of Ald and its possible role in mycobacterial pathogenesis is limited. In the saprophytic organism Mycobacterium smegmatis, Ald is a cytosolic protein and is overproduced under anaerobic conditions which mimic the dormancy state of M. tuberculosis (14, 22, 32, 43). This result has led to the hypothesis that Ald contributes to maintenance of the NAD pool (i.e., recycling of NADH) (22). By microarray analysis, the M. tuberculosis ald transcript was recently shown to be overproduced under hypoxic conditions (36), under nutrient starvation conditions (5), or by sodium dodecyl sulfate (SDS) treatment in a sigE mutant (26). On the basis of biochemical evidence, Usha et al. (43) suggested that reductive amination of pyruvate and reductive amination of glyoxylate are catalyzed by the same protein in M. smegmatis, presumably Ald. Transcription induction of the ald gene has been observed upon infection of leopard frogs with Mycobacterium marinum (9). The role of Ald in the adaptation of pathogenic and saprophytic mycobacteria to physiological stresses needs to be investigated. In this study, we characterized the expression and regulation of the M. smegmatis ald gene. Increased Ald activity was observed in cells grown with alanine supplementation and under anaerobic conditions. Analysis of a null mutant demonstrated that Ald is necessary for proficient alanine utilization and sustained anaerobic growth.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani broth or agar. M. smegmatis strains were grown at 37°C with shaking (200 rpm; Innova 4300 incubator-shaker; New Brunswick Scientific, Edison, N.J.) in Middlebrook 7H9 broth (BBL Microbiology Systems, Cockeysville, Md.) supplemented with albumin-dextrose complex (ADC) and 0.05% Tween 80 (M-ADC-TW). When required, l-alanine was added at a concentration of 25 mM and d-alanine was added at a concentration of 45 mM as these concentrations support optimal growth of M. smegmatis (7). For analysis of nitrogen source utilization, M. smegmatis was grown in a minimal medium as previously described (8). Anaerobic conditions were achieved by using a GasPack system (BBL Microbiology Systems) with AnaeroPack (Mitsubishi Gas Chemical America, Inc., New York, N.Y.) as recommended by the manufacturer. To monitor the maintenance of anaerobic conditions in the system, indicator strips were placed in the jar and parallel cultures were grown in the presence of 1.5 μg of methylene blue (AMRESCO Inc., Solon, Ohio) per ml. The dye faded by 24 h and became colorless by 48 h, indicating that oxygen was gradually depleted. The system remained anaerobic during sample manipulation and thereafter. Transformations of E. coli and M. smegmatis were performed as described previously (15). The following antibiotics were added to the media when necessary: kanamycin (Sigma Chemical Co., St. Louis, Mo.) at a concentration of 50 μg ml−1, gentamicin (Sigma) at a concentration of 20 μg ml−1, or carbenicillin (Sigma) at a concentration of 50 μg ml−1 for E. coli strains; and kanamycin at a concentration of 10 μg ml−1, gentamicin at a concentration of 5 μg ml−1, or hygromycin B (Roche Molecular Biochemicals, Indianapolis, Ind.) at a concentration of 100 μg ml−1 for M. smegmatis strains.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference and/or source |
|---|---|---|
| Strains | ||
| E. coli XL10-GOLD | Tetr Δ(mcrA)183183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZ ΔM15 Tn10 (Tetr) Amy Camr] | Stratagene |
| M. smegmatis mc2155 | High-transformation mutant of M. smegmatis mc26 | 40 |
| M. smegmatis GPM258 | mc2155(pBUN246); overproducing Ald | This study |
| M. smegmatis GPM267 | mc2155Δald::aph; ald null mutant | This study |
| M. smegmatis GPM268 | mc2155Δald::aph; ald null mutant | This study |
| M. smegmatis GPM269 | mc2155Δald::aph; ald null mutant | This study |
| M. smegmatis GPM270 | mc2155Δald::aph; ald null mutant | This study |
| M. smegmatis GPM276 | GPM267(pBUN279); M. smegmatis ald mutant complemented with the wild-type ald gene integrated at the mycobacteriophage L5 attB site | This study |
| M. smegmatis GPM282 | mc2155(pBUN286a); wild-type M. smegmatis carrying an additional copy of the ald promoter at the mycobacteriophage L5 attB site | This study |
| Plasmids | ||
| pBluescript II KS+ | Standard E. coli cloning vector | Stratagene |
| pCR2.1 | TA cloning vector | Invitrogen |
| pUC4K | Plasmid containing an excisable aminoglycoside 3′-phosphotransferase gene from Tn903 | Amersham Pharmacia (28) |
| pMV262 | E. coli-Mycobacterium shuttle plasmid | MedImmune (41) |
| pPR27 | E. coli-Mycobacterium shuttle plasmid with sacB and the temperature-sensitive mycobacterial ori | 31 |
| pYUB178 | E. coli-Mycobacterium integration-proficient vector; integrates at the mycobacteriophage L5 attB site | 29 |
| pYUB412 | E. coli-Mycobacterium integration-proficient vector; integrates at the mycobacteriophage L5 attB site | 30 |
| pBUN246 | Recombinant plasmid isolated from an M. smegmatis genomic library which hybridized with the M. tuberculosis ald gene; 8.0-kb insert | This study |
| pBUN253 | pCR2.1 with 1.5-kb PCR fragment containing complete M. smegmatis ald gene amplified with SMALDCF1 and SMALDCR | This study |
| pBUN261 | pCR2.1 with 0.4-kb PCR fragment containing the M. smegmatis ald promoter amplified with SMALDCF1 and SMALDPE | This study |
| pBUN268 | pBluescript II KS+ with 1.5-kb EcoRI fragment from pBUN253 in the EcoRI site | This study |
| pBUN270a | pBUN268 carrying the 1.2-kb blunted EcoRI fragment of pUC4K containing aph gene for the 0.7-kb NruI fragment internal of the ald gene | This study |
| pBUN272 | pPR27 with 2.2-kb EcoRV/XbaI fragment from pBUN270a in the BamHI (polished)/XbaI site | This study |
| pBUN279 | pYUB412 with 1.5-kb EcoRI fragment from pBUN253 in the EcoRI site | This study |
| pBUN286a | pYUB178 with 0.4-kb EcoRI fragment from pBUN261 in the EcoRI site | This study |
Oligonucleotide primers, PCR, and primer extension analysis.
The oligonucleotide primers (Integrated DNA Technologies, Inc., Coralville, Iowa) used for PCR amplification of the complete ald gene from M. tuberculosis were TBALDF (5′-TAT CCC GAC AGT GTC CGC TAA C-3′) and TBALDR (5′-ACA TCA TCG CTT CCC TTA CTC C-3′). The following primer pairs were used for amplification of the M. smegmatis ald gene: SMALDF (5′-CGA TCA TGC TCG TCG GAA T-3′) and SMALDR (5′-CGA TCA CGG TTC GGT TGT TA-3′), which amplify a 1.1-kb fragment used in Southern blot analysis; and SMALDCF1 (5′-CGG ATC ACA CCG ATC TCC-3′) and SMALDCR (5′-CCT CAT CGG GGA ACT CAC-3′), which amplify a 1.5-kb fragment used for construction of mutant and complemented strains. Primer SMALDPE (5′-GTC AGC TCT GCC ACT CCA G-3′) was used in primer extension analysis. PCR amplifications were performed with a Perkin-Elmer GeneAmp 9600 thermal cycler (Roche Molecular Systems, Branchburg, N.J.) by using the Expand High Fidelity PCR system (Roche) as recommended by the manufacturer. For Southern blot analysis, restriction digestion, ligation, and agarose gel electrophoresis, standard procedures were followed as previously described (34). Total RNA from M. smegmatis strains was isolated by using RNAWIZ (Ambion, Inc., Austin, Tex.) with minor modifications, as previously described (3). Primer extension analysis of the ald mRNA was carried out as described previously (12).
M. smegmatis genomic library construction and cloning of the M. smegmatis ald gene.
Chromosomal DNA from M. smegmatis mc2155 was prepared (50) and partially digested with Sau3AI, and 3.0- to 4.0-kb fragments were ligated into the E. coli-Mycobacterium shuttle plasmid pMV262 (41). The ligation mixture was transformed into E. coli XL10-GOLD (Stratagene, La Jolla, Calif.), and approximately 6,000 recombinants were obtained for a theoretical representation of P > 99% of the M. smegmatis genome. Since only the M. tuberculosis genomic database was available at the start of these experiments, the M. smegmatis ald gene was isolated from a genomic library by using a probe from M. tuberculosis. The M. tuberculosis ald gene was amplified from H37Rv genomic DNA by using primers TBALDF and TBALDR and was radiolabeled by using the Rediprime II labeling system (Amersham Pharmacia Biotech, Piscataway, N.J.). After three rounds of library screening, recombinant plasmid pBUN246 was obtained and confirmed to contain the complete coding region of the ald gene. Based on the partial sequence obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org), the full-length sequence of the ald gene in pBUN246 was obtained by primer walking at the DNA Sequencing & Synthesis Facility of Iowa State University (Ames, Iowa).
Two-dimensional gel electrophoresis and protein N-terminal sequencing.
Cell crude extract from M. smegmatis strains was prepared as previously described (8). Two-dimensional gel electrophoresis was performed by using a PROTEAN IEF cell (Bio-Rad) with immobilized pH gradient strips that were 7 cm long and had a pH range of 4 to 7 (Bio-Rad). Crude extracts containing approximately 600 μg of protein were precipitated with 10% trichloroacetic acid and resuspended in rehydration buffer containing 8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 10 mM dithiothreitol, and 0.2% Bio-Lytes 3/10. The strips were actively rehydrated at 50 V for 12 h and focused at 4,000 V for a total of 60,000 V · h. Subsequently, the strips were equilibrated in a solution containing 6 M urea, 275 mM Tris-HCl (pH 6.8), 2% SDS, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and 20% glycerol and placed on the second dimension (10 to 20% linear gradient of polyacrylamide; Bio-Rad). The gel was electrophoresed at 125 V in 0.3% SDS-Tris-glycine buffer (pH 8.3), and polypeptide spots were visualized by Coomassie blue staining. Proteins from a parallel preparation were electroblotted for 2 h at 45 V onto polyvinylidene difluoride membranes in electrotransfer buffer (10% methanol, Tris-glycine; pH 8.3) by using a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). Protein spots on the membranes were visualized by amido black staining, and the proteins of interest were excised from the membranes for automated Edman sequencing. Protein sequencing was performed with an ABI-Procise 494 sequencer (Applied Biosystems, Foster City, Calif.) by using manufacturer-recommended protocols.
Enzyme assays.
l-Alanine dehydrogenase activity was measured by using a spectrophotometric method in the direction of oxidative deamination coupled to the production of NADH as described previously (23). All samples were measured in triplicate. Specific activities were expressed in micromoles of l-alanine (substrate) consumed per milligram per minute. No net changes in absorbance were detected when l-alanine was omitted from the reaction mixtures containing active extracts. The reductive amination reaction could not be measured in crude cell extracts due to interference by the lactate dehydrogenase enzyme that uses pyruvate and NADH as substrates. Glycine dehydrogenase activity was determined based on optical measurement of the rate of oxidation of NADH coupled with the reductive amination of glyoxylate as described previously (18). All samples were measured in triplicate. Specific activities were expressed in micromoles of sodium glyoxylate (substrate) consumed per milligram per minute. No net changes in absorbance were detected when sodium glyoxylate was omitted from the reaction mixtures containing active extracts. Consistent with previous observations (18, 43, 48), the oxidative deamination activity could not be detected.
Construction of M. smegmatis ald null mutants.
To construct M. smegmatis ald null mutants, the internal 0.7-kb NruI fragment of the ald gene in pBUN268 was replaced by the 1.3-kb kanamycin-resistant determinant from pUC4K (28), yielding pBUN270a. The inactivated allele was cloned into pPR27, a plasmid carrying a gentamicin resistance marker, a temperature-sensitive mycobacterial origin of replication, and a sacB gene as a counterselectable marker (31). The resulting plasmid, pBUN272, was electroporated into M. smegmatis mc2155, and recombinants were selected at 32°C on medium containing 10 μg of kanamycin per ml. One of these recombinants was grown in M-ADC-TW to saturation at 32°C, and approximately 109 cells were plated onto Middlebrook 7H9-ADC agar containing kanamycin and 10% sucrose and incubated at 39°C. Five days later, kanamycin- and sucrose-resistant (Kanr Sucr) colonies appeared, and they were screened individually for gentamicin sensitivity (Gens), as the Kanr Sucr Gens phenotype was expected if there was allelic exchange and the plasmid vector was lost. Of the 476 Kanr Sucr colonies obtained, 4 were confirmed to be Gens. The remaining colonies were Kanr Sucr Genr and probably resulted from inactivation of the sacB gene and/or the leaky temperature-sensitive phenotype of the plasmid. The four Kanr Sucr Gens colonies were chosen as putative mutants for further study and were designated GPM267, GPM268, GPM269, and GPM270.
Nucleotide sequence accession number.
The sequence of the M. smegmatis l-alanine dehydrogenase gene cloned in pBUN246 has been deposited in the GenBank database under accession number AF304867.
RESULTS
Cloning and sequence analysis of the M. smegmatis ald gene.
A genomic library of mc2155 was constructed in the E. coli-Mycobacterium replicating vector pMV262, and E. coli recombinant clones were screened by colony hybridization. This screening analysis yielded a positive clone carrying a recombinant plasmid with an 8.0-kb insert, which was designated pBUN246. This multicopy plasmid was introduced into wild-type strain mc2155 to generate recombinant strain GPM258. To confirm that pBUN246 carries a functional full-length ald gene, a cell extract was prepared and assayed for enzymatic activity. As expected, GPM258 displayed a higher level of Ald activity (approximately 90-fold higher) than the wild-type strain (data not shown).
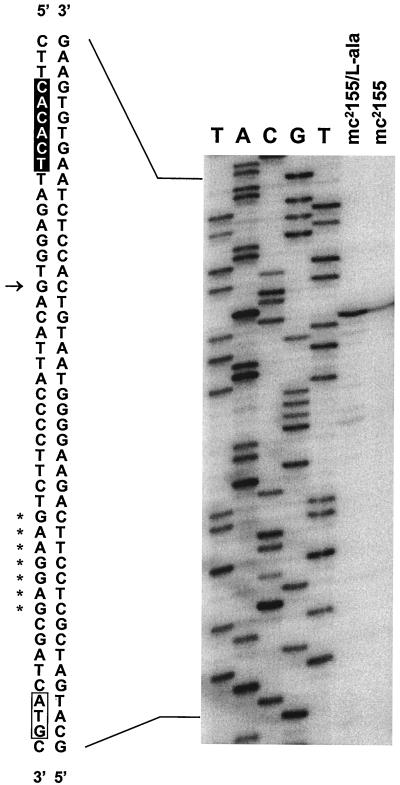
Sequence analysis of the insert in pBUN246 revealed four open reading frames coding for a putative AsnC family transcriptional regulator homologous to the M. tuberculosis Rv2779c, alanine dehydrogenase (Ald), a putative β-lactamase homologous to Mycobacterium fortuitum BlaF (42), and a putative peptidase homologous to M. tuberculosis PepR (Rv2782c). The ald gene appears to comprise a monocistronic message since the flanking open reading frames are encoded on the strand opposite the strand containing ald. Consistent with this hypothesis, primer extension analysis of RNA isolated from cells grown in Middlebrook medium under aerobic conditions revealed the presence of a promoter immediately upstream from the ald gene (Fig. 1). A transcriptional start site was identified at a guanosine residue, 27 nucleotides upstream from the start codon (ATG) followed by a putative ribosome binding site (GAAGGAG) five nucleotides upstream. No alternative transcriptional start site was observed when primer extension was performed with RNA isolated from cells grown in minimal medium and shifted to anaerobic conditions (data not shown). The coding region of the M. smegmatis ald gene contains 1,116 nucleotides, and it encodes a 39.0-kDa protein consisting of 371 amino acids. A putative rho-independent transcriptional terminator (ATCGCCCGGCCGGAA and TTCCGGCCGGGCGAT) was located 13 nucleotides downstream of the stop codon (TAA).
FIG. 1.
Primer extension analysis of the ald transcript from M. smegmatis. Total RNA (50 μg) from mc2155 cells, grown in either M-ADC-TW or M-ADC-TW supplemented with l-alanine, was annealed with primer SMALDPE and extended as described in Materials and Methods. Lanes A, C, G, and T show a dideoxy sequencing ladder of the ald gene generated with the same primer. The transcriptional start site is indicated by an arrow. The putative −10 box deduced from the consensus of mycobacterial promoter sequences is highlighted (19). The start codon (ATG) is enclosed in a box, and the putative ribosome binding site (GAAGGAG) is indicated by asterisks.
Alignment of the amino acid sequences of several bacterial l-alanine dehydrogenases showed that the M. smegmatis Ald polypeptide exhibits 79, 80, and 76% amino acid identity to the homologous polypeptides of Mycobacterium leprae (11), M. tuberculosis (10), and Mycobacterium avium (TIGR), respectively. It also exhibits approximately 60% identity to isozymes of Streptomyces coelicolor (33) and B. subtilis (37). The multiple alignment revealed two highly conserved regions with the consensus sequences K73VKEP77 and v173iGgGtaGyNAAriA-GmGa-VTvlDvn200, which correspond to the enzyme active site for pyruvate binding (13) and the NAD cofactor binding region (2), respectively.
Ald activity is regulated by nutrient supplementation, oxygen availability, and growth stage.
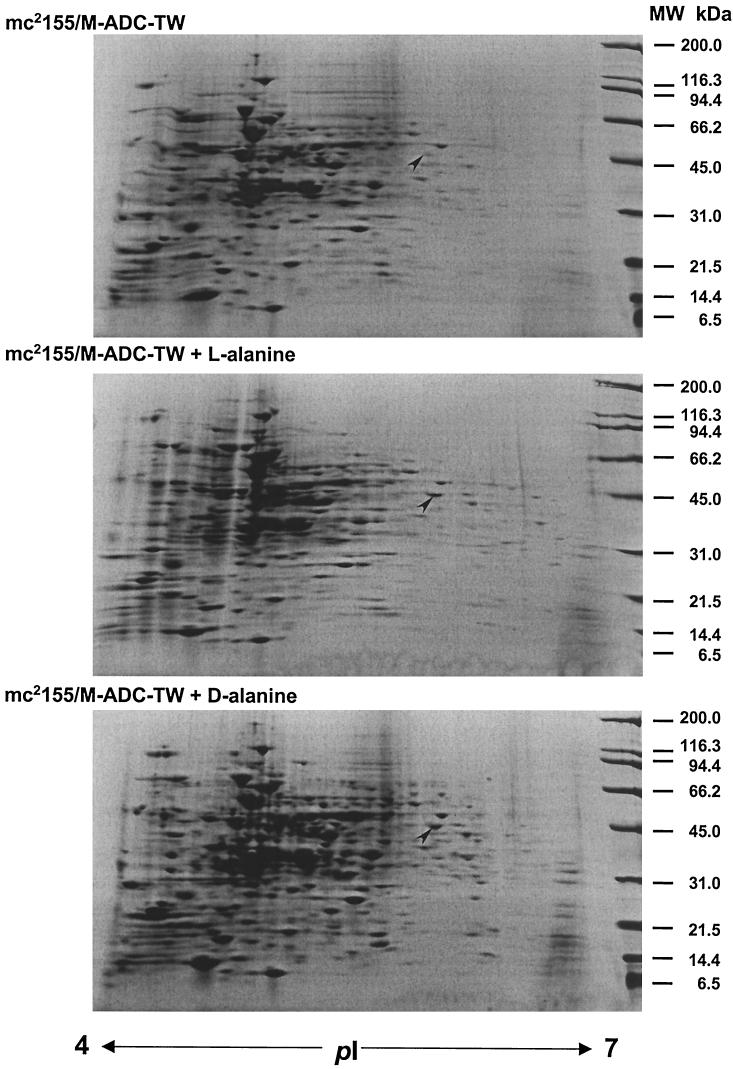
Regulation of Ald has been studied in various microorganisms. In Bacillus spp. Ald is produced at high levels upon induction by alanine, during sporulation, and at the onset of the stationary phase (16, 27, 37). In M. smegmatis and M. tuberculosis, expression of the ald gene has been shown to be up-regulated under limiting oxygen conditions (22, 36, 43). These findings suggest that multiple factors are involved in the regulation of Ald. However, previous studies of mycobacterial Ald have not investigated the combined effects of nutrient supplementation, oxygen availability, and growth stage. To gain further insight into the function and regulation of Ald, we analyzed the expression of the M. smegmatis ald gene in exponentially growing or stationary cells in the presence of l- or d-alanine under aerobic or anaerobic conditions. We first analyzed the effect of alanine on the expression of the ald gene. A dramatic increase in the transcription of the ald gene was observed when cells were grown in the presence of l-alanine (Fig. 1). The increase in the mRNA level corresponded to a similar level of overproduction of a polypeptide spot (molecular weight, 42,000; pI 5.8) induced by l- or d-alanine, as observed in two-dimensional gels (Fig. 2). We confirmed that this differentially expressed spot corresponded to Ald since N-terminal sequencing analysis yielded a sequence (MLVGIPTEIK) identical to that deduced from translation of the M. smegmatis ald gene.
FIG. 2.
Two-dimensional gel electrophoresis analysis of M. smegmatis mc2155 cellular proteins. Cells were grown in M-ADC-TW or M-ADC-TW supplemented with l- or d-alanine as indicated. Cell extracts were prepared as described in Materials and Methods. Approximately 600 μg of total proteins was resolved by two-dimensional gel electrophoresis and stained with Coomassie blue. Isoelectric points (pI) are indicated at the bottom, and the positions of molecular mass size markers are indicated on the right. The location of the Ald enzyme (42 kDa, pI 5.8) in each gel is indicated by an arrowhead.
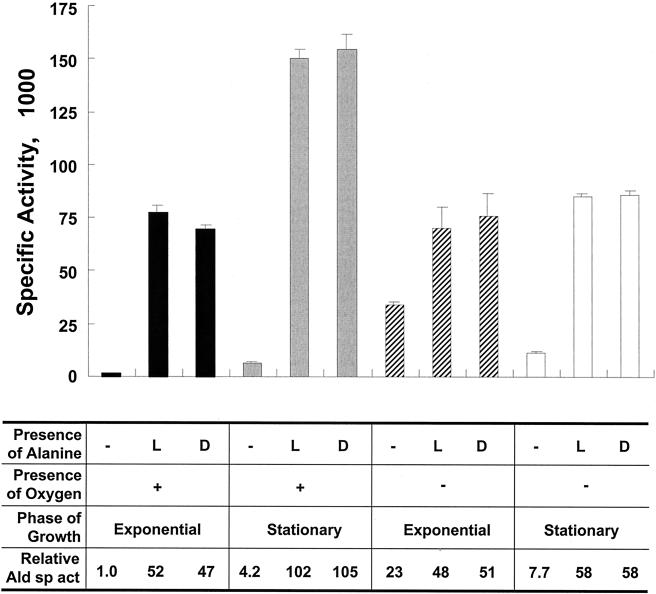
To quantify the increase in Ald enzyme activity in cells grown with l- or d-alanine under aerobic conditions, crude cell extracts were prepared and assayed (Fig. 3). In exponentially growing cells, addition of either l- or d-alanine to the growth medium resulted in an approximately 50-fold increase in enzyme activity. In stationary cells, Ald was produced at a level that was 25-fold greater than the basal level in cells grown without alanine. The basal level of Ald activity in stationary cells was approximately fourfold higher than the level in exponentially growing cells. Under anaerobic conditions, the basal level of Ald activity in exponentially growing cells was 23-fold higher than the level in cells grown under aerobic conditions. Addition of l- or d-alanine increased Ald activity by twofold, and the activity reached approximately the same level as the level in exponentially growing cells under aerobic conditions. When anaerobically grown cells reached the stationary phase, the basal level of Ald activity decreased by threefold, while in the presence of alanine the Ald activity increased to a level similar to that in exponentially growing cells. In summary, high levels of Ald activity were observed in cells grown in the presence of l- or d-alanine regardless of the oxygen availability and growth stage. However, in the absence of alanine, high basal levels of Ald activity were observed in cells growing exponentially under anaerobic conditions.
FIG. 3.
Analysis of M. smegmatis l-alanine dehydrogenase activities. Cells were grown under either aerobic or anaerobic conditions to the exponential or stationary phase in M-ADC-TW or M-ADC-TW supplemented with l- or d-alanine as indicated. The relative Ald specific activities were calculated by assigning a value of 1.0 to the specific activity of the extract prepared from exponentially growing cells incubated under aerobic conditions without alanine. The specific activities are expressed in micromoles of l-alanine per milligram per minute. The values are means ± standard deviations of triplicate measurements.
Isolation and characterization of M. smegmatis ald null mutants.
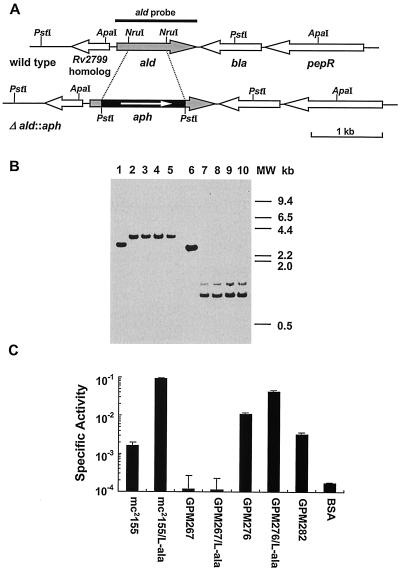
To further analyze the physiological role of Ald, M. smegmatis ald null mutants were generated by allelic exchange (Fig. 4A) as described in Materials and Methods. Four putative mutants (GPM267 to GPM270) were analyzed further. To determine whether the mutant strains carry the inactivated ald allele, genomic DNA was isolated and amplified by PCR with primers SMALDCF1 and SMALDR. As expected for inactivation of the ald gene, genomic DNA from all four mutant strains yielded a single 2.2-kb product distinct from the 1.5-kb fragment from mc2155 DNA (data not shown). In addition, Southern blot analysis was used to verify the occurrence of homologous recombination events in these putative ald mutants. Genomic DNA was isolated, digested with either ApaI or PstI, transferred to a membrane, and hybridized with the wild-type ald gene fragment as a probe. After digestion with ApaI, wild-type strain mc2155 gave a 3.0-kb band (Fig. 4B, lane 1) and all of the mutants yielded a 3.6-kb band (Fig. 4B, lanes 2 to 5); after digestion with PstI, mc2155 gave a 2.7-kb band (Fig. 4B, lane 6) and all of the mutants yielded two bands at approximately 0.9 and 1.1 kb (Fig. 4B, lanes 7 to 10). These patterns were expected for the predicted recombinational events, validating the construction of the ald null mutants. No differences in colony morphology were observed between mc2155 and the ald null mutants when the organisms were plated on Middlebrook 7H9-ADC agar. Likewise, no differences in growth rate and saturation density were observed in M-ADC-TW broth (data not shown). Therefore, the ald gene of M. smegmatis can be inactivated and is nonessential.
FIG. 4.
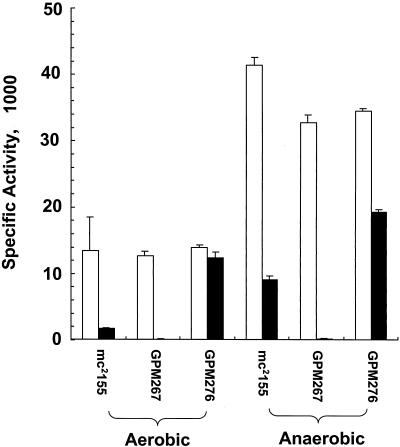
Construction and characterization of M. smegmatis ald null mutants. (A) Open reading frame organization at the ald locus in both strain mc2155 (wild type) and the ald null mutant strain (Δald::aph). Relevant restriction enzyme sites and the PCR fragment used as the ald probe are indicated. (B) Southern blot analysis of genomic DNA from wild-type and ald null mutant strains. Genomic DNA was digested with ApaI (lanes 1 to 5) or PstI (lanes 6 to 10). The strains examined are wild-type strain mc2155 (lanes 1 and 6) and ald mutants GPM267 (lanes 2 and 7), GPM268 (lanes 3 and 8), GPM269 (lanes 4 and 9), and GPM270 (lanes 5 and 10). Blots were hybridized with the radiolabeled 1.1-kb PCR fragment containing the ald gene and washed under stringent conditions as described in Materials and Methods. MW, molecular weight. (C) Analysis of l-alanine dehydrogenase activities in M. smegmatis wild-type, mutant, and recombinant strains. Extracts from organisms grown in both M-ADC-TW and M-ADC-TW supplemented with l-alanine were prepared and analyzed. Ald specific activities are expressed in micromoles of l-alanine per milligram per minute. The values are means ± standard deviations of triplicate measurements. A mock assay was also carried out with bovine serum albumin (BSA) in place of equivalent amounts of the cell extracts.
M. smegmatis ald mutants have no detectable l-alanine dehydrogenase activity.
To determine whether M. smegmatis ald mutants are or are not devoid of Ald activity, crude cell extracts from wild-type strain mc2155, ald mutant GPM267, and the complemented strain GPM276 were prepared and assayed for Ald activity (Fig. 4C). Crude extracts prepared from GPM267 cells grown in the presence or absence of l-alanine lacked any detectable Ald activity. These extracts yielded background levels of Ald activity that were not significantly different from the levels obtained by substituting bovine serum albumin for GPM267 extracts in the reaction mixture. In complemented strain GPM276, which resulted from integration of a single copy of the wild-type ald gene into the GPM267 chromosome at the mycobacteriophage L5 attB site, the Ald activity was restored. When the organisms were grown in M-ADC-TW supplemented with l-alanine, the Ald activity of complemented strain GPM276 was slightly lower than that of the wild-type strain. This activity was about sixfold higher than that of the wild-type strain when the organisms were grown without l-alanine. The Ald activity of recombinant strain GPM282, which carried an additional copy of the ald promoter at the mycobacteriophage L5 attB site, was increased by approximately twofold. This suggests that the presence of two copies of the ald promoter and positional effects within the chromosome may moderately alter expression of the gene. We concluded that inactivation of the ald gene results in no detectable Ald activity, indicating that Ald is the only protein responsible for this activity in M. smegmatis.
Inactivation of the ald gene does not affect glycine dehydrogenase activity.
Increased glycine dehydrogenase (EC 1.4.1.10) (Gdh) activity has been found in M. tuberculosis and M. bovis BCG during anaerobic growth, suggesting that Gdh contributes to maintenance of the NAD pool (25, 45, 47). Based on biochemical evidence, two recent reports suggested that reductive amination of pyruvate and reductive amination of glyoxylate are catalyzed by the same protein in both M. smegmatis (43) and M. tuberculosis (V. Usha, B. C. Elias, R. Jayaram, S. Ravishankar, and K. S. Das, Abstr. Tuberculosis Past, Present, Future, abstr. 151, 2000). If the physiological Gdh activity is carried by Ald, the M. smegmatis ald null mutant cells would be expected to lack Gdh activity. To test this hypothesis, we assayed Ald and Gdh activities in crude extracts of the wild-type, ald mutant, and complemented strains by using cells grown either under aerobic conditions or following an anaerobic shift (Fig. 5). Ald activity was significantly higher under anaerobic conditions for the wild-type and complemented stains, while it was undetectable for the mutant strain either under aerobic conditions or following an anaerobic shift. However, all of the strains displayed similar Gdh activities under either aerobic or anaerobic conditions, with an approximately fourfold increase following an anaerobic shift, as previously observed (43). Since the Gdh activity in the ald null mutant was not affected, we concluded that another enzyme protein is responsible for the physiological Gdh activity in M. smegmatis.
FIG. 5.
Analysis of l-alanine dehydrogenase and glycine dehydrogenase activities in wild-type, ald null mutant, and complemented strains. M. smegmatis was grown in M-ADC-TW to an optical density at 600 nm of approximately 0.8 under aerobic conditions. Approximately one-half of each culture was harvested, and the remaining cells were shifted to anaerobic growth for 24 h. Crude cell extracts were prepared and assayed for Ald (solid bars) and Gdh (open bars) activities as described in Materials and Methods. Specific activities are expressed in micromoles of substrate per milligram per minute. The values are means ± standard deviations of triplicate measurements.
M. smegmatis ald mutants are impaired in the utilization of alanine as a sole nitrogen source.
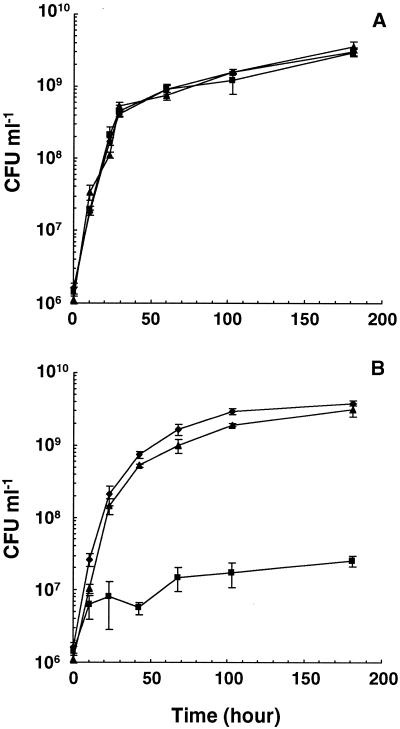
To further characterize the role of the Ald enzyme in nitrogen metabolism, the wild-type, ald null mutant, and complemented strains were tested for the ability to utilize various nitrogen sources. When ammonium chloride or l-asparagine was used as the sole nitrogen source, the ald null mutant utilized either source as proficiently as wild-type strain mc2155 and the complemented strain (Fig. 6A and data not shown). However, the mutant grew poorly on l-alanine as the sole nitrogen source, while the wild-type and complemented strains displayed similar growth rates and saturation densities on both l-alanine and ammonium chloride (Fig. 6B). With l-alanine as the sole nitrogen source, the mutant reached a saturation cell density of 2.5 × 107 CFU ml−1, which was approximately 10-fold greater than the density of the initial inoculum but 100-fold less than the densities of the wild-type and complemented strains. Similar results were obtained when d-alanine was used as the sole nitrogen source in broth, as well as when cells were grown on solid agar with either l-alanine or d-alanine as the sole nitrogen source (data not shown). Therefore, M. smegmatis Ald is required for proficient utilization of alanine as a nitrogen source.
FIG. 6.
Utilization of nitrogen sources by M. smegmatis wild-type, ald null mutant, and complemented strains. Exponentially growing cells of mc2155 (♦), GPM267 (▴), and GPM276 (▪) were harvested, washed, and inoculated into minimal medium containing ammonium chloride (A) or l-alanine (B) as the sole nitrogen source at an initial density of approximately 2.0 × 106 CFU ml−1. At various times, the number of viable cells was determined.
M. smegmatis ald mutants are impaired for anaerobic growth.
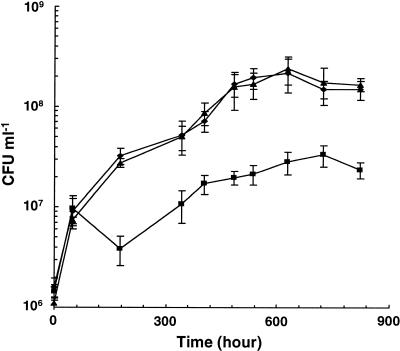
Since mycobacterial Ald is overproduced during growth under low-oxygen conditions (22, 36, 43; also see above), we tested the effect of inactivation of the M. smegmatis ald gene on anaerobic growth. The growth of the wild-type strain was substantial during the first 7 days and then continued at a lower rate, and the cell density reached approximately 2 × 108 CFU ml−1 at 20 days postinoculation. The ald null mutant was able to reach a cell density similar to that of the wild-type strain 2 days after inoculation. However, at later time points, its growth was minimal, and the saturation density was 10-fold lower than that of the wild-type strain (Fig. 7). No differences were observed between the complemented and wild-type strains. We concluded that the M. smegmatis Ald is necessary for optimal growth under anaerobic conditions.
FIG. 7.
Growth of M. smegmatis wild-type, ald null mutant, and complemented strains under anaerobic conditions. Exponentially growing cells of mc2155 (♦), GPM267 (▴), and GPM276 (▪) were inoculated into M-ADC-TW at an initial density of approximately 2.0 × 106 CFU ml−1 in a GasPack system as described in Materials and Methods. At various times, the number of viable cells was determined.
DISCUSSION
Regulation of l-alanine dehydrogenase is determined by multiple factors. In Bacillus spp., l-alanine dehydrogenase is repressed by glucose and is induced by alanine and other amino acids (16), entry into the stationary phase, or spore germination (27, 37). In mycobacterial species, Ald is overproduced during anaerobic growth of M. smegmatis (22, 43). Likewise, the M. tuberculosis ald gene is up-regulated 10-fold under hypoxic conditions (36). Ald from Mycobacterium strain HE5 is stimulated by l-alanine and the xenobiotic compound morpholine (35). In our studies of M. smegmatis Ald, we found that the presence of either stereoisoform of alanine led to a 50-fold increase in Ald activity for exponentially growing cells under aerobic conditions and a 25-fold increase for stationary cells, reflecting the catabolic role of Ald when alanine is abundant. The fourfold induction of the basal Ald activity in the absence of alanine by entry into the stationary phase may be due to protein turnover with the concomitant increase in alanine in the intracellular amino acid pool. The elevated Ald activity observed when wild-type cells are supplemented with alanine under aerobic conditions is consistent with the impaired utilization of alanine as a sole nitrogen source in the ald mutant. These data suggest that oxidative deamination by Ald is the main route by which M. smegmatis utilizes alanine.
Under anaerobic conditions, the Ald activity was elevated 23-fold under basal conditions, and addition of alanine resulted in only a twofold increase. This indicates that Ald may play an important role in growth under anaerobic conditions since this increase is observed in exponentially growing cells. This hypothesis is consistent with our observation that there was a threefold decrease in the basal Ald activity upon entry into the stationary phase under anaerobic conditions. At physiological pH, M. tuberculosis Ald seems to play a biosynthetic role in catalyzing the reductive amination of pyruvate, which is the thermodynamically favored conversion (Keq = 5.5 × 10−11) (17, 23). In addition to generating alanine for protein and peptidoglycan biosynthesis, this reaction may also contribute to the recycling of NADH, which is necessary for growth under anaerobic conditions when oxygen as the terminal electron acceptor becomes depleted. Consistent with this biosynthetic role of Ald under anaerobic conditions, we found that the M. smegmatis ald null mutants displayed a defective phenotype during anaerobic growth. However, the mutant multiplied to a cell density approximately 10 times greater than that of the initial inoculum, suggesting that another enzyme(s) may also participate in the recycling of NADH under anaerobic conditions. In the Wayne dormancy culture model, the 10-fold increase in M. tuberculosis Gdh activity has been proposed to contribute to maintenance of the NAD pool by reductive amination of glyoxylate (47, 48). However, the corresponding glycine dehydrogenase gene has not yet been identified. In the M. tuberculosis genome, one gene (gcvB) codes for a putative glycine dehydrogenase, but its product probably catalyzes the decarboxylation of glycine instead of the reductive amination of glyoxylate (10, 48). Purified Ald from either Bacillus megaterium or Mycobacterium strain HE5 has been reported to catalyze the reductive amination of glyoxylate (20, 35). Two recent reports have also suggested that the same enzyme protein, presumably Ald, from either M. smegmatis or M. tuberculosis is responsible for the physiological Ald and Gdh activities (43; also see above). In contrast, our data showed wild-type levels of Gdh activity in the ald mutant under either aerobic or anaerobic conditions. Consistently, M. bovis BCG lacks a functional Ald enzyme (2, 24) as a consequence of a frameshift mutation in the ald gene (21), while increased Gdh activity was observed under anaerobic conditions (25). Therefore, the identities of the Gdh enzyme and the corresponding gene in mycobacteria remain unknown.
In summary, the phenotypes displayed by the M. smegmatis ald mutants suggest that Ald plays an important role in both alanine utilization and anaerobic growth. In strains of the M. tuberculosis complex, the roles of Ald and other enzymes involved in adaptation to anaerobiosis are less clear. M. bovis BCG (Ald−) (2, 24) and M. tuberculosis (Ald+) (2) behave in similar ways in the Wayne model of adaptation to anaerobiosis (25, 45). In M. tuberculosis, increased nitrate reduction by nitrate reductase encoded by the narGHIJ cluster was observed during shift-down from aerobic to anaerobic growth (46). Although homologous genes have been identified in the M. smegmatis genome (TIGR), it was observed that M. smegmatis mc2155 does not display this enzyme activity even under anaerobic conditions (49). The recently constructed M. tuberculosis ald mutant should be a useful tool to further assess the role of Ald in pathogenesis (S. C. Woolwine, C. Ko, S. Tyagi, and W. R. Bishai, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. U-53, 2002). Comparative analysis of M. smegmatis and M. tuberculosis ald mutants may reveal whether there are important differences in the adaptations of saprophytic and pathogenic mycobacteria to anaerobic growth conditions.
Acknowledgments
This research was supported by funds from the University of Nebraska Department of Veterinary and Biomedical Sciences, by a Nebraska Agriculture Experiment Station interdisciplinary research award, by USDA Cooperative State Research Service project NEB 14-108, and by the University of Nebraska-Lincoln Tobacco Settlement Biomedical Research Enhancement Fund. N.E.C. and G.S. are supported by the Center for Biotechnology, University of Nebraska-Lincoln, funded through the Nebraska Research Initiative. Z.F. is a recipient of a Maude Hammond Fling-Bukey Memorial Fund fellowship from the University of Nebraska-Lincoln Graduate Studies. Sequencing of M. smegmatis mc2155 and M. avium MAC104 was accomplished with support from NIAID.
We thank A. K. Benson, O. Chacon, J. D. Cirillo, N. B. Harris, and D. K. Zinniel for critical reviews of the manuscript and helpful discussions. We thank S. Hinkley for translation and C. L. Joor for technical assistance with preliminary experiments. Preliminary sequence data were obtained from the TIGR website (http://www.tigr.org).
Footnotes
A contribution of the University of Nebraska Agricultural Research Division, Lincoln. Journal Series no. 13651.
REFERENCES
- 1.Allaway, D., E. M. Lodwig, L. A. Crompton, M. Wood, R. Parsons, T. R. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36:508-515. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, Å. B., P. Andersen, and L. Ljungqvist. 1992. Structure and function of a 40,000-molecular-weight protein antigen of Mycobacterium tuberculosis. Infect. Immun. 60:2317-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashyam, M. D., and A. Tyagi. 1994. An efficient and high-yielding method for isolation of RNA from mycobacteria. BioTechniques 17:834-836. [PubMed] [Google Scholar]
- 4.Bellion, E., and F. Tan. 1987. An NAD+-dependent alanine dehydrogenase from a methylotrophic bacterium. Biochem. J. 244:565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 6.Brunhuber, N. M., and J. S. Blanchard. 1994. The biochemistry and enzymology of amino acid dehydrogenases. Crit. Rev. Biochem. Mol. Biol. 29:415-467. [DOI] [PubMed] [Google Scholar]
- 7.Cáceres, N. E. 1999. Ph.D. thesis. University of Nebraska, Lincoln.
- 8.Chacon, O., Z. Feng, N. B. Harris, N. E. Cáceres, L. G. Adams, and R. G. Barletta. 2002. Mycobacterium smegmatis d-alanine racemase mutants are not dependent on d-alanine for growth. Antimicrob. Agents Chemother. 46:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, K., T. Knaak, L. Satkamp, O. Humbert, S. Falkow, and L. Ramakrishnan. 2002. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc. Natl. Acad. Sci. USA 99:3920-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, R. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 12.Davis, L. G., W. M. Kuehl, and J. F. Battey. 1994. Basic methods in molecular biology, 2nd ed. Appleton & Lange, Norwalk, Conn.
- 13.Delforge, D., B. Devreese, M. Dieu, E. Delaive, J. Van Beeumen, and J. Remacle. 1997. Identification of lysine 74 in the pyruvate binding site of alanine dehydrogenase from Bacillus subtilis. Chemical modification with 2,4,6-trinitrobenzenesulfonic acid, n-succinimidyl 3-(2-pyridyldithio)propionate, and 5′-(p-(fluorosulfonyl)benzoyl)adenosine. J. Biol. Chem. 272:2276-2284. [DOI] [PubMed] [Google Scholar]
- 14.Dick, T., B. H. Lee, and B. Murugasu-Oei. 1998. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 163:159-164. [DOI] [PubMed] [Google Scholar]
- 15.Foley-Thomas, E. M., D. L. Whipple, L. E. Bermudez, and R. G. Barletta. 1995. Phage infection, transfection and transformation of Mycobacterium avium complex and Mycobacterium paratuberculosis. Microbiology 141:1173-1181. [DOI] [PubMed] [Google Scholar]
- 16.Freese, E., and J. Oosterwyk. 1963. The induction of alanine dehydrogenase. Biochemistry 2:1212-1216. [DOI] [PubMed] [Google Scholar]
- 17.Goldman, D. S. 1959. Enzyme system in the mycobacteria. VII. Purification, properties and mechanism of action of the alanine dehydrogenase. Biochim. Biophys. Acta 34:527-539. [DOI] [PubMed] [Google Scholar]
- 18.Goldman, D. S., and M. J. Wagner. 1962. Enzyme systems in the mycobacteria. XIII. Glycine dehydrogenase and the glyoxylic acid cycle. Biochim. Biophys. Acta 65:297-306. [DOI] [PubMed] [Google Scholar]
- 19.Gomez, M., and I. Smith. 2000. Determinants of mycobacterial gene expression, p. 111-130. In G. F. Hatfull, and W. R. J. Jacobs (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 20.Honorat, A., F. Monot, and D. Ballerini. 1990. Synthesis of l-alanine and l-valine by enzyme systems from Bacillus megaterium. Enzyme Microb. Technol. 12:515-520. [Google Scholar]
- 21.Hutter, B. 1996. Ph.D. thesis. University of Braunschweig, Braunschweig, Germany.
- 22.Hutter, B., and T. Dick.1998. Increased alanine dehydrogenase activity during dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 167:7-11. [DOI] [PubMed] [Google Scholar]
- 23.Hutter, B., and M. Singh. 1999. Properties of the 40 kDa antigen of Mycobacterium tuberculosis, a functional l-alanine dehydrogenase. Biochem. J. 343:669-672. [PMC free article] [PubMed] [Google Scholar]
- 24.Jungblut, P. R., U. E. Schaible, H. J. Mollenkopf, U. Zimny-Arndt, B. Raupach, J. Mattow, P. Halada, S. Lamer, K. Hagens, and S. H. Kaufmann. 1999. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 33:1103-1117. [DOI] [PubMed] [Google Scholar]
- 25.Lim, A., M. Eleuterio, B. Hutter, B. Murugasu-Oei, and T. Dick. 1999. Oxygen depletion-induced dormancy in Mycobacterium bovis BCG. J. Bacteriol. 181:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 27.McCowen, S. M., and P. V. Phibbs, Jr. 1974. Regulation of alanine dehydrogenase in Bacillus licheniformis. J. Bacteriol. 118:590-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 29.Pascopella, L., F. M. Collins, J. M. Martin, M. H. Lee, G. F. Hatfull, C. K. Stover, B. R. Bloom, and W. R. Jacobs, Jr. 1994. Use of in vivo complementation in Mycobacterium tuberculosis to identify a genomic fragment associated with virulence. Infect. Immun. 62:1313-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavelka, M. S., Jr., and W. R. Jacobs, Jr. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H 37Rv by allelic exchange. J. Bacteriol. 181:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raynaud, C., G. Etienne, P. Peyron, M. A. Laneelle, and M. Daffé. 1998. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 144:577-587. [DOI] [PubMed] [Google Scholar]
- 33.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schuffenhauer, G., T. Schrader, and J. R. Andreesen. 1999. Morpholine-induced formation of l-alanine dehydrogenase activity in Mycobacterium strain HE5. Arch. Microbiol. 171:417-423. [DOI] [PubMed] [Google Scholar]
- 36.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siranosian, K. J., K. Ireton, and A. D. Grossman. 1993. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J. Bacteriol. 175:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, M. T., and D. W. Emerich. 1993. Alanine dehydrogenase from soybean nodule bacteroids. Kinetic mechanism and pH studies. J. Biol. Chem. 268:10746-10753. [PubMed] [Google Scholar]
- 39.Smith, M. T., and D. W. Emerich. 1993. Alanine dehydrogenase from soybean nodule bacteroids: purification and properties. Arch. Biochem. Biophys. 304:379-385. [DOI] [PubMed] [Google Scholar]
- 40.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 41.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. J. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 42.Timm, J., M. G. Perilli, C. Duez, J. Trias, G. Orefici, L. Fattorini, G. Amicosante, A. Oratore, B. Joris, J. M. Frere, A. P. Pugsley, and B. Gicquel. 1994. Transcription and expression analysis, using lacZ and phoA gene fusions, of Mycobacterium fortuitum beta-lactamase genes cloned from a natural isolate and a high-level beta-lactamase producer. Mol. Microbiol. 12:491-504. [DOI] [PubMed] [Google Scholar]
- 43.Usha, V., R. Jayaraman, J. C. Toro, S. E. Hoffner, and K. S. Das. 2002. Glycine and alanine dehydrogenase activities are catalyzed by the same protein in Mycobacterium smegmatis: upregulation of both activities under microaerophilic adaptation. Can. J. Microbiol. 48:7-13. [DOI] [PubMed] [Google Scholar]
- 44.Ward, M. J., H. Lew, and D. R. Zusman. 2000. Disruption of aldA influences the developmental process in Myxococcus xanthus. J. Bacteriol. 182:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wayne, L. G., and L. G. Hayes. 1998. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber. Lung Dis. 79:127-132. [DOI] [PubMed] [Google Scholar]
- 47.Wayne, L. G., and K. Y. Lin. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect. Immun. 37:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 49.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F. C. Bange. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017-1025. [DOI] [PubMed] [Google Scholar]
- 50.Whipple, D. L., R. B. Le Febvre, R. E. Andrews, Jr., and A. B. Thiermann. 1987. Isolation and analysis of restriction endonuclease digestive patterns of chromosomal DNA from Mycobacterium paratuberculosis and other Mycobacterium species. J. Clin. Microbiol. 25:1511-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiame, J. M., and A. Pierard. 1955. Occurrence of an l(+)-alanine-dehydrogenase in Bacillus subtilis. Nature 176:1073-1075.13272750 [Google Scholar]