Abstract
DNA arrays were used to investigate the global transcriptional profile of Bacillus subtilis grown in the presence of sulfate or methionine as the sole sulfur source. The expression of at least 56 genes differed significantly under the two growth conditions. The expression of several genes belonging to the S-box regulon was repressed in the presence of methionine probably in response to S-adenosylmethionine availability. The expression of genes encoding transporters (yhcL, ytmJKLMN, and yxeMO) was high when the sulfur source was methionine or taurine and reduced when it was sulfate.
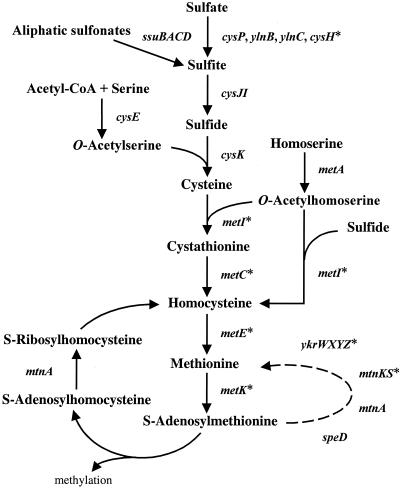
The pathways involved in the synthesis of sulfur-containing amino acids and the ways in which these pathways are regulated differ in various groups of organisms. The Bacillus subtilis pathways leading in the production of cysteine and methionine from inorganic sulfate were recently characterized (Fig. 1). The cysH operon encodes a sulfate permease (CysP) and enzymes catalyzing the conversion of sulfate into sulfite (12, 13). Sulfite is then incorporated into cysteine by sulfite reductase and cysteine synthase (27). Two alternative methionine biosynthesis pathways exist in B. subtilis (1). The first one requires the sequential action of cystathionine γ-synthase and cystathionine β-lyase with the intermediary formation of cystathionine. The second pathway bypasses cystathionine via direct sulfhydrylation of O-acetylhomoserine to homocysteine.
FIG. 1.
Cysteine and methionine biosynthesis pathways in B. subtilis. The different genes encoding proteins involved in sulfate assimilation, cysteine biosynthesis, and methionine biosynthesis are indicated. The functions of some genes are indicated in Table 1. ylnB, ATP sulfurylase; ylnC, adenosine phosphosulfate kinase; cysE, serine acetyltransferase; cysH, 3′-phosphoadenosine 5′-phosphosulfate sulfotransferase; metA, homoserine acetyltransferase; metI, cystathionine γ-synthase/O-acetylhomoserine sulfhydrylase; metC, cystathionine β-lyase; metK, S-adenosylmethionine synthetase; speD, S-adenosylmethionine decarboxylase; mtnK, methylthioribose kinase. The arrow between S-adenosylmethionine and methionine represents several enzymatic steps. Asterisks indicate genes that contain an S box in the leader region.
In Escherichia coli, regulation of the cysteine and methionine biosynthesis genes involves two LysR-type activators, CysB and MetR, and a repressor, MetJ. Full expression of the cysteine biosynthesis pathway requires the positive regulator CysB, the inducer N-acetylserine, and a limited amount of reduced sulfur (8, 17). Repression of methionine biosynthesis in the presence of methionine is mediated by the MetJ repressor. The MetJ-S-adenosylmethionine (AdoMet) complex binds to the Met box sequences present in the promoter regions of the met genes (20). The MetR activator is required for expression of both the metE and metH genes, which encode the two methionine synthases (5, 28). In B. subtilis, several genes involved in methionine metabolism are regulated by the S-box antitermination mechanism. Grundy and Henkin (6) proposed a model in which the 5′ portion of the leader forms an antiantiterminator structure that sequesters sequences required for the formation of an antiterminator, which, in turn, sequesters sequences required for the formation of the terminator. The only regulator known to be involved in the response to sulfur availability in B. subtilis is the LysR-like YtlI activator, which controls the expression of an operon containing an ABC transport system (3).
Complete genome sequences and expression profiling experiments provide a powerful tool for global transcriptional pattern analysis and gene function identification. DNA arrays have already been successfully used to study the B. subtilis responses to various growth conditions (15, 18, 29, 31).
Comparison of global gene expression profiles of B. subtilis grown with sulfate or methionine as the sole sulfur source.
The genomic expression profiles of B. subtilis 168 grown in minimal medium in the presence of 1 mM sulfate or 1 mM methionine as the sole sulfur source were analyzed by using DNA macroarrays. The cells grew at similar rates on both sulfur sources. Exponentially growing cells were collected and broken by shaking in a Fastprep apparatus (Bio 101). Total RNA was then extracted by Trizol (Gibco-BRL) treatment. cDNAs, which were generated by using 1 μg of total RNA and B. subtilis CDS-specific primers, were hybridized to panorama B. subtilis gene arrays (Sigma-GenoSys Biotechnologies). The intensity of each dot was quantified with xdotsreader software (Cose). To account for unspecific variations, six experiments were carried out with five independent RNA preparations and two different sets of DNA arrays. Comparison of the intensities of the signals from duplicate or independent hybridizations (Fig. 2) showed that the procedures for RNA extraction, reverse transcription, and hybridization were reproducible. The nonparametric Wilcoxon statistical test (statview 5.0.1 package) allowed us to identify 101 genes, the expression levels of which were significantly different when cells were grown with sulfate or methionine (P ≤ 0.05). The genes whose expression levels differed by a factor ≥1.5 are listed in Table 1. Most of the genes identified during this work were further studied by using lacZ reporter fusions (Tables 1 and 2). The DNA array results were consistent with those of the lacZ fusion experiments, but the regulation factor was generally higher when lacZ fusions were used, suggesting that DNA arrays are less sensitive. We may therefore have missed some genes that are tightly regulated in response to sulfur availability.
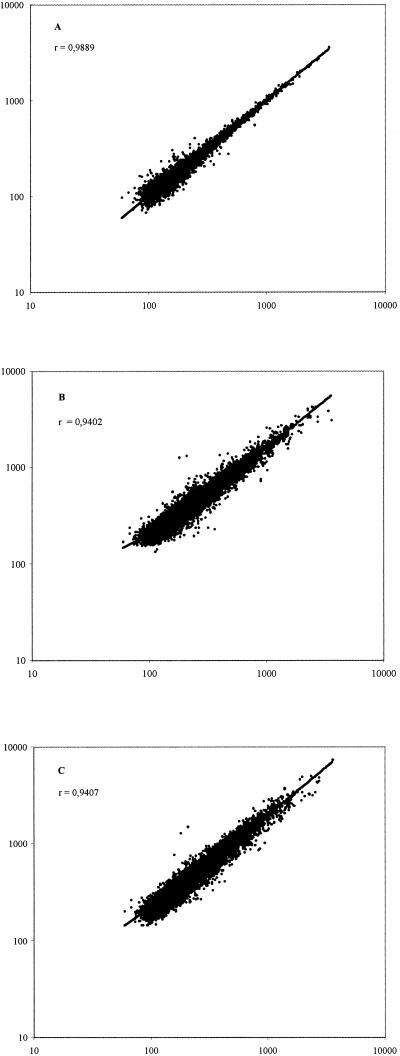
FIG. 2.
Comparison of the signal intensities of duplicate dots and independent hybridizations. The reproducibility of the DNA arrayresults obtained with 1 μg of total RNA extracted from B. subtilis 168 grown in minimal medium in the presence of sulfate as the sole sulfur source was assessed before subtraction of the background. In all experiments, the results showed a high degree of correlation (>0.94). (A) Comparison of signal intensities of pairs of dots corresponding to each gene. (B) Comparison of signal intensities of each gene in two independent hybridizations obtained from the same RNA sample but reverse transcribed and hybridized independently.(C) Comparison of signal intensities of each gene in two independent hybridizations of RNAs isolated from different cultures grown under the same conditions. Similar results were obtained with B. subtilis 168 grown in minimal medium in the presence of methionine as the sole sulfur source.
TABLE 1.
Genes differentially expressed in B. subtilis 168 grown in the presence of methionine or sulfate as the sole sulfur sourcea
| Gene | Function/similarity | Transcriptome analysis
|
β-Galactosidase activity (U mg of protein−1)b
|
|||
|---|---|---|---|---|---|---|
| Sulfate/methionine expression ratio | P value | Sulfate | Methionine | Sulfate/methionine expression ratio | ||
| Genes related to sulfur metabolism | ||||||
| cysJ | Sulfite reductase flavoprotein | 1.52 | 5 × 10−3 | |||
| cysI | Sulfite reductase hemoprotein | 1.94 | 1 × 10−2 | |||
| cysK | O-Acetylserine sulfhydrylase | 0.66 | 2 × 10−3 | |||
| mtnA | Methylthioadenosine nucleosidase | 0.66 | 3 × 10−2 | |||
| yrhA | Similar to O-acetylserine sulfhydrylase | 0.44 | 2 × 10−3 | 28 | 106 | 0.26 |
| yrhB | Similar to cystathionine γ-synthase | 0.26 | 1 × 10−2 | 51 | 206 | 0.25 |
| S-box family | ||||||
| metE | Probable cobalamin-independent methionine synthase | 3.54 | 5 × 10−3 | 110 | 13 | 8.5 |
| ykrW | Similar to ribulose bisphosphate carboxylase | 1.95 | 3 × 10−2 | |||
| ykrY | Similar to proteins of unknown function | 4.84 | 1 × 10−2 | |||
| ykrZ | Similar to proteins of unknown function | 1.50 | 2 × 10−3 | 151 | 58 | 2.6 |
| yoaD | Similar to phosphoglycerate dehydrogenase | 2.00 | 2 × 10−3 | 125 | 8 | 15.6 |
| yoaC | Similar to xylulokinase | 1.80 | 1 × 10−2 | |||
| yoaB | Similar to permease | 1.80 | 4 × 10−3 | |||
| yxjG | Similar to YxjH | 1.50 | 3 × 10−3 | 115 | 27 | 4.3 |
| yxjH | Similar to YxjG | 2.37 | 1 × 10−2 | 146 | 32 | 4.6 |
| Transporters and associated genes | ||||||
| ssuA | Aliphatic sulfonate ABC transporter (binding lipoprotein) | 0.47 | 2 × 10−3 | |||
| ssuC | Aliphatic sulfonate ABC transporter (permease) | 0.52 | 5 × 10−3 | |||
| ssuD | Aliphatic sulfonate monooxygenase | 0.04 | 2 × 10−3 | |||
| ygaN | Unknown | 0.08 | 2 × 10−3 | |||
| yhcL | Similar to sodium-glutamate symporter | 0.29 | 2 × 10−3 | 22 | 223 | 0.10 |
| ytmI | Similar to proteins of unknown function | 0.55 | 8 × 10−3 | |||
| ytmJ | Similar to amino acid ABC transporter (binding protein) | 0.19 | 5 × 10−3 | |||
| ytmK | Similar to amino acid ABC transporter (binding protein) | 0.17 | 5 × 10−3 | |||
| ytmL | Similar to amino acid ABC transporter (permease) | 0.12 | 2 × 10−3 | |||
| ytmM | Similar to amino acid ABC transporter (permease) | 0.55 | 2 × 10−3 | |||
| ytmN | Similar to amino acid ABC transporter (ATP-binding protein) | 0.31 | 2 × 10−3 | |||
| ytmO | Similar to proteins of unknown function | 0.16 | 2 × 10−3 | |||
| ytnI | Unknown | 0.35 | 6 × 10−3 | |||
| ribR | Riboflavin kinase | 0.29 | 3 × 10−3 | |||
| ytnL | Similar to aminohydrolase | 0.51 | 3 × 10−3 | |||
| ytnM | Unknown | 0.16 | 2 × 10−3 | |||
| yxeK | Similar to monooxygenase | 0.40 | 3 × 10−2 | 55 | 292 | 0.19 |
| yxeL | Similar to proteins of unknown function | 0.66 | 1 × 10−2 | |||
| yxeM | Similar to amino acid ABC transporter (binding protein) | 0.50 | 2 × 10−2 | 33 | 353 | 0.09 |
| yxeO | Similar to amino acid ABC transporter (ATP-binding protein) | 0.66 | 1 × 10−2 | 15 | 150 | 0.10 |
| yxeQ | Unknown | 0.37 | 2 × 10−3 | 20 | 311 | 0.06 |
| Regulator, ytlI | Similar to transcriptional regulator (LysR family) | 0.66 | 4 × 10−2 | 12 | 216 | 0.05 |
| Genes with other functions | ||||||
| accA | Acetyl-CoA carboxylase (alpha subunit) | 2.15 | 2 × 10−2 | |||
| appB | Oligopeptide ABC transporter (permease) | 0.52 | 5 × 10−2 | |||
| asnB | Asparagine synthetase | 1.55 | 1 × 10−2 | |||
| atpI | ATP synthase (subunit i) | 1.80 | 4 × 10−3 | |||
| folD | Probable methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase | 1.90 | 1 × 10−2 | |||
| glnR | Transcriptional repressor of the glutamine synthetase gene | 1.90 | 3 × 10−2 | |||
| hisB | Imidazoleglycerol-phosphate dehydratase | 1.80 | 1 × 10−2 | |||
| hisD | Histidinol dehydrogenase | 1.76 | 8 × 10−3 | |||
| hisI | Phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP pyrophosphohydrolase | 2.23 | 2 × 10−3 | |||
| katA | Vegetative catalase 1 | 0.33 | 4 × 10−3 | 77 | 216 | 0.35 |
| nadA | Probable quinolinate synthetase | 1.95 | 3 × 10−3 | |||
| nadC | Probable nicotinate-nucleotide pyrophosphorylase | 1.40 | 2 × 10−3 | |||
| nadB | l-Aspartate oxidase | 1.69 | 3 × 10−3 | |||
| nrgA | Ammonium transporter | 1.70 | 1 × 10−2 | |||
| pyrAA | Carbamoyl-phosphate synthetase (glutaminase subunit) | 2.12 | 8 × 10−3 | |||
| pyrE | Orotate phosphoribosyltransferase | 2.93 | 6 × 10−3 | |||
| yciA | Similar to proteins of unknown function | 3.09 | 1 × 10−2 | |||
| yciB | Similar to proteins of unknown function | 1.80 | 6 × 10−3 | 3 | 0.8 | 3.70 |
| yciC | Involved in low-affinity zinc transport system | 2.44 | 1 × 10−2 | 161 | 55 | 3.00 |
| ydbM | Similar to butyryl-CoA dehydrogenase | 0.30 | 3 × 10−3 | 7 | 32 | 0.21 |
The results obtained are representative of at least six hybridizations from five independent RNA extractions. The expression intensities of 3,830 genes were above the background level. Only genes with a P value of ≤0.05 in a Wilcoxon test and whose expression differed by a factor of ≥1.5 between the two growth conditions are listed. Genes are grouped according to function. Column two indicates protein function according to the SubtiList database (http://genolist.pasteur.fr/SubtiList/).
Cells were grown in minimal medium (6 mM K2HPO4, 4.4 mM KH2PO4, 0.3 mM trisodium citrate, 5 mM MgCl2, 0.5% glucose, 50 mg of l-tryptophan liter−1, 22 mg of ferric ammonium citrate liter−1, 0.1% l-glutamine) containing 1 mM sulfate or 1 mM methionine as the sulfur source. β-Galactosidase activities were determined in extracts prepared from exponentially growing cells. The values shown are averages from at least three independent experiments.
TABLE 2.
Bacterial strains used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| BSIP1142 | trpC2 amyE::pmetI′-lacZ cat | 1 |
| BSIP1159 | trpC2 amyE::pmetK′-lacZ cat | 21 |
| BSIP1214 | trpC2 ytlI::aphA3 | 3 |
| BSIP1215 | trpC2 amyE::pytlI′-lacZ cat | pDIA5575→168 |
| BSIP1306 | trpC2 amyE::pmetE′-lacZ cat | pDIA5626→168 |
| BSIP1307 | trpC2 amyE::p(G−273→A) metE′-lacZ cat | pDIA5627→168 |
| BSIP1324 | trpC2 amyE::pkatA′-lacZ cat | pDIA5624→168 |
| BSIP1379 | trpC2 metK1 sacB::φPveg-metK+amyE::pmetI′-lacZ cat | pDIA5510→SA29 |
| BSIP1382 | trpC2 metK1 sacB::φPveg-metK+amyE::pmetE′-lacZ cat | pDIA5626→SA29 |
| BSIP1385 | trpC2 metK1 sacB::φPveg-metK+ amyE::p(G−273→A) metE′-lacZ cat | pDIA5627→SA29 |
| BSIP1386 | trpC2 yxjH′::lacZ cat | pDIA5628→168 |
| BSIP1387 | trpC2 metK1 sacB::φPveg-metK+yxjH′::lacZ cat | pDIA5628→SA29 |
| BFS1605b | trpC2 yhcL′::lacZ erm | S. Bron |
| BFS1850b | trpC2 ykrT′::lacZ erm | K. M. Devine |
| BFS1853b | trpC2 ykrZ′::lacZ erm | K. M. Devine |
| BFS2048b | trpC2 yoaD′::lacZ erm | W. Schumann |
| BFS2062b | trpC2 yrhB′::lacZ erm | W. Schumann |
| BFS2063b | trpC2 yrhA′::lacZ erm | W. Schumann |
| BFS3022b | trpC2 yitJ′::lacZ erm | S. J. Seror |
| BFS4035c | trpC2 yxjG′::lacZ erm | Y. Fujita |
| BFS4069c | trpC2 yxeQ′::lacZ erm | Y. Fujita |
| BFS4071c | trpC2 yxeO′::lacZ erm | Y. Fujita |
| BFS4073c | trpC2 yxeM′::lacZ erm | Y. Fujita |
| BFS4075c | trpC2 yxeK′::lacZ erm | Y. Fujita |
| BFS4363c | trpC2 yciB′::lacZ erm | K. Yamane |
| BFS4364c | trpC2 yciC′::lacZ erm | K. Yamane |
| BFS4451c | trpC2 ydbM′::lacZ erm | F. Kawamura |
| SA29 | 62378 with sacB::φPveg-metK+ (formerly metE+) | 30 |
| 1A607d | trpC2 metC85::Tn917 | BGSCe |
Arrows indicate construction by transformation. cat is the pC194 chloramphenicol acetyl-transferase gene, aphA3 is the Enterococcus faecalis kanamycin resistance gene, and erm is an erythromycin resistance gene.
Strain constructed as part of the EC project for the functional characterization of the B. subtilis genome (http://locus.jouy.inra.fr/cgi-bin/genmic/madbase/progs/madbase.operl).
Strain constructed as part of the Japanese project for functional characterization of the B. subtilis genome (http://bacillus.genome.ad.jp).
In the metC85::Tn917 mutant, the transposon is inserted in the metE (formerly metC) gene.
BGSC, Bacillus Genetic Stock Center.
Interestingly, most of the genes that were upregulated in the presence of methionine encode proteins containing no cysteine residues or one cysteine residue. This suggests that the growth of B. subtilis in the presence of methionine is associated to sulfur limitation conditions.
Links with different metabolic pathways.
The expression of several genes not directly related to sulfur metabolism was modified, depending on the sulfur source. Some genes involved in histidine biosynthesis (hisB, hisD, and hisI), pyrimidine biosynthesis (pyrAA and pyrE), one-carbon metabolism (folD), nitrogen metabolism (glnR and nrgA), NAD biosynthesis (nadA and nadB), and energy production (atpI) were upregulated in the presence of sulfate. The synthesis of proteins implicated in the metabolism of one-carbon units and in the biosynthesis of nucleotides has been shown to be repressed under sulfur-limiting conditions (3). This is consistent with our transcriptome data. As one-carbon units are used in the synthesis of methionine, these two metabolic pathways seem to be linked in B. subtilis, as observed in E. coli (14). Two genes, accA and ydbM, are related to lipid metabolism (Table 1). The accA transcript level was higher in the presence of sulfate, while the expression of the ydbM gene, which encodes a polypeptide with similarities to acyl coenzyme A (acyl-CoA) dehydrogenases, was increased in the presence of methionine (Table 1). The expression of yciA, yciB, and yciC, which are adjacent on the chromosome, was increased in the presence of sulfate when both transcriptome and lacZ fusions were used (Table 1). The YciC protein is an integral membrane protein that may be part of a low-affinity zinc transport system (4).
Interestingly, the expression of the katA gene, which encodes the major catalase in growing cells (10), increased in the presence of methionine. A transcriptional fusion of the katA promoter region (from position −261 to position +6 relative to the translational start site) and the promoterless lacZ gene was inserted at the amyE locus (Table 2). During the exponential growth phase, β-galactosidase activity was 216 U mg of protein−1 in methionine-grown cells, 77 U mg of protein−1 in sulfate-grown cells, 76 U mg of protein−1 in cysteine-grown cells, 82 U mg of protein−1 in taurine-grown cells, and 12 U mg of protein−1 in methionine-plus-sulfate-grown cells. Therefore, katA expression was regulated in response to sulfur availability. We tested the resistance of the wild-type strain to oxidative stress on minimal-medium plates by using a disk impregnated with 2 μl of a 30% hydrogen peroxide solution. The area of growth inhibition was 50% smaller in the presence of methionine than in the presence of sulfate. More catalase activity was also observed in cells grown in the presence of methionine than in cells grown in the presence of sulfate (data not shown). Thus, there is a correlation between (i) the sulfur source used for growth and (ii) catalase activity, which is consistent with the regulation of katA gene expression.
Genes of the S-box family.
Of the 11 transcriptional units containing an S box in the leader region (6), 5 showed increased transcript levels in the presence of sulfate: metE, yxjH, yxjG, yoaDCB, and ykrWXYZ (Table 1). The yoaDCB operon encodes proteins similar to a phosphoglycerate dehydrogenase, a xylulokinase, and a transporter, respectively. The ykrWXYZ operon is involved in the scavenging of methylthioribose to generate methionine (16, 23). The amino acid sequences of YxjG and YxjH are 70% identical to each other. These polypeptides have moderate similarities to a C-terminal portion of the MetE protein (7), which is highly similar to the E. coli cobalamin-independent methionine synthase. A mutation in a methionine auxotrophic mutant (strain 1A607; Table 2) mapped to the same region as the metE gene. We determined that the transposon is inserted 102 bp downstream of the translational start site of the metE gene in this mutant. The metE mutant was unable to grow in the presence of sulfate, cysteine, cystathionine, or homocysteine as the sole sulfur source, but it grew similarly to the wild-type strain in the presence of methionine (data not shown). Thus, the metE gene appears to encode the unique methionine synthase in B. subtilis.
Surprisingly, six genes or operons containing an S-box motif were not downregulated in the presence of methionine in our transcriptome experiments. We further compared the data obtained with the DNA arrays with those obtained with lacZ fusions constructed either during the B. subtilis functional-analysis project or during this work (Tables 1 and 2). A fragment corresponding to the metE promoter region (nucleotides −369 to +56 relative to the translational start site) was inserted into pAC6 (25). This metE′-lacZ fusion was then integrated at the amyE locus of the wild-type strain. An internal fragment of the yxjH gene (nucleotides +7 to +359 relative to the translational start site) was cloned into the integrative plasmid pDIA5307 (2). The yxjH′-lacZ fusion was integrated into the yxjH locus by a Campbell-type mechanism (Table 2).
The expression levels of the lacZ fusions with several representative genes from the S-box family were determined after growth in minimal medium plus sulfate or methionine (Table 1 and data not shown). All of the met genes implicated in methionine metabolism are members of the S-box family, with the exception of the metA gene, which is hardly regulated, depending on the sulfur source (S. Auger, unpublished results). The expression of the metE gene was 8.5-fold higher in the presence of sulfate than in the presence of methionine, whereas the expression of the metI and metK genes was only three- and twofold higher in the presence of sulfate, respectively (1, 30; this work). Thus, the focal point of transcriptional regulation of the methionine biosynthesis pathway corresponds to the metE gene.
All members of the S-box family are not tightly regulated by methionine under the growth conditions used. Indeed, the regulation factors observed with transcriptional fusions range from ≤2 for mtnK (ykrT), yitJ, and cysH to 15.5 for yoaD (11) (Table 1 and data not shown). This is in contrast to the data presented by Grundy and Henkin (6), who found a much higher factor of repression by methionine for the yitJ and mtnK genes under more drastic methionine starvation conditions. It is possible that different genes from the S-box regulon respond to different levels of methionine starvation (6) and/or are controlled by other regulatory signals (11, 24). A comparison of the ΔG° values of the terminator and antiterminator of all of the S-box structures did not establish a clear correlation between the stability of the terminator or antiterminator and the efficiency of methionine-dependent repression.
Role of AdoMet in the regulation of the genes controlled by the S box.
To determine the mechanisms that are involved in the regulation of the S-box regulon, the role of methionine and AdoMet in this regulation has been further examined. Overexpression of AdoMet synthetase leads to methionine auxotrophy in B. subtilis, suggesting that AdoMet is a corepressor of methionine biosynthesis in this organism (30). The expression of the metE′-lacZ, metI′-lacZ, and yxjH′-lacZ transcriptional fusions in response to methionine limitation was then tested in the wild-type strain and the SA29 strain, which overproduces AdoMet synthetase. The metE′-lacZ, metI′-lacZ, and yxjH′-lacZ fusions were 3- to 20-fold less strongly expressed in the SA29 strain than in the wild type, both in the presence and in the absence of methionine (Table 3). The downregulation of metE and metI gene expression in strain SA29 can explain its methionine auxotrophy. The sixfold increase in AdoMet synthetase activity observed in this strain (30) probably decreases the cellular content of methionine and increases the cellular concentration of AdoMet. Methionine depletion was expected to derepress the transcription of the S-box regulon (1, 6). In contrast, we demonstrated that overproduction of AdoMet synthetase led to lower levels of transcription of yxjH and of some met genes, strongly suggesting that this effect is due to an increase in the concentration of AdoMet.
TABLE 3.
Effect of AdoMet synthetase overproduction on expression of metE′-lacZ, metI′-lacZ, and yxjH′::lacZ transcriptional fusionsa
| Strain | Relevant genotype | β-Galactosidase activity (U mg of protein−1)
|
||||
|---|---|---|---|---|---|---|
| t0 | t1 + methionine | t1 − methionine | t2 + methionine | t2 − methionine | ||
| BSIP1306 | amyE::pmetE′-lacZ | 10 | 9.5 | 121 | 9 | 172 |
| BSIP1382 | metK1 sacB::φPveg-metK+amyE::pmetE′-lacZ | 2 | 2.5 | 11 | 2 | 32 |
| BSIP1307 | amyE::p(G−273→A) metE′-lacZ | 710 | 721 | 634 | 1,185 | 905 |
| BFS1385 | metK1 sacB::φPveg-metK+amyE::p(G−273→A) metE′-lacZ | 661 | 734 | 633 | 961 | 993 |
| BSIP1142 | amyE::pmetI′-lacZ | 91 | 89 | 452 | 107 | 510 |
| BSIP1379 | metK1 sacB::φPveg-metK+amyE::pmetI′-lacZ | 22 | 15 | 23 | 16 | 56 |
| BSIP1386 | yxjH′::lacZ | 30 | 26 | 139 | 30 | 149 |
| BSIP1387 | metK1 sacB::φPveg-metK+yxjH′::lacZ | 10 | 10.5 | 35 | 6 | 29 |
Cells were grown in minimal medium (6 mM K2HPO4, 4.4 mM KH2PO4, 0.3 mM trisodium citrate, 5 mM MgCl2, 0.5% glucose, 50 mg of l-tryptophan liter−1, 22 mg of ferric ammonium citrate liter−1, 0.1% l-glutamine) supplemented with 1 mM methionine. The cells were collected by centrifugation and resuspended in minimal medium in the presence or absence of methionine. Samples were taken before resuspension (t0), 1 h after resuspension (t1), and 2 h after resuspension (t2). To overproduce AdoMet synthetase in the SA29 strain, the metK gene was placed under the control of a strong constitutive promoter, Pveg (30).
In the wild-type background, the metE′-lacZ and metI′-lacZ fusions were induced 19- and 5-fold 2 h after the removal of methionine (Table 3). In strain SA29, these fusions were induced 16- and 3.5-fold 2 h after methionine was removed, respectively. Overproduction of AdoMet synthetase may not be sufficient to completely repress the expression of the met gene in the absence of methionine. Under conditions of drastic methionine starvation, the AdoMet concentration of the cell probably remains low even when AdoMet synthetase is overproduced.
We obtained a metE′-lacZ fusion containing a substitution in the S box. During the cloning of the metE promoter fragment into pAC6, we isolated a plasmid that conferred high-level constitutive expression of the metE′-lacZ fusion on B. subtilis 168 (Table 3). Sequencing revealed the presence of a G→A substitution at position −273 relative to the translational start site of metE. This mutation, located in the region between 5′-half helix 1 and 5′-half helix 2 of the S box, alters a highly conserved residue but does not disrupt the structural features of the S-box region (6). Thus, it may lead to loss of binding of a negative regulatory factor rather than destabilization of the RNA structure. The AdoMet synthetase overproduction in strain SA29 did not modify the expression of the constitutive p(G−273→A)metE′-lacZ fusion, while the expression of the wild-type metE′-lacZ fusion was decreased 4- to 10-fold in this strain (Table 3). Thus, it appears that expression of the metE gene in response to AdoMet synthetase overproduction is mediated via the S box. We therefore propose that AdoMet could act as a corepressor to modulate the binding of a negative regulatory factor to the S-box region. However, we cannot exclude the possibility that AdoMet has an indirect effect. Further information is needed about the nature of the repressor and its ability to interact with the regulatory S-box region to elucidate how AdoMet functions in this system.
The YtlI regulator.
In addition to the S-box transcription antitermination system, the YtlI activator was recently shown to be involved in the control of transcription in response to sulfur availability. The ytlI gene is transcribed divergently from the ytmI operon. The YtlI protein controls the expression of this operon (3). Interestingly, DNA arrays showed that the expression of the ytlI gene was 1.5-fold higher in methionine than in sulfate (Table 1). To confirm this result, a transcriptional ytlI′-lacZ fusion was constructed by inserting a 213-bp DNA fragment containing the promoter region of the ytlI gene (nucleotides −209 to +4 relative to the translational start site) into pAC6 (25). The resulting ytlI′-lacZ fusion was integrated at the amyE locus of a wild-type strain (Table 2). β-Galactosidase activity was tested after growth in the presence of various sulfur sources. The level of expression of the ytlI gene was high in the presence of methionine (215 U mg of protein−1) or taurine (80 U mg of protein−1) and 8- to 30-fold lower in the presence of sulfate (12 U mg of protein−1) or cysteine (7 U mg of protein−1). This suggests that a regulatory cascade controls the expression of the ytmI operon.
Regulation of genes involved in the uptake and metabolism of sulfur compounds.
Other genes involved in the metabolism of sulfur-containing amino acids were differently expressed in DNA array experiments (Table 1). The expression of the genes ensuring the last two steps of cysteine biosynthesis (Fig. 1) was regulated in the opposite way. Whereas the level of expression of the cysJI operon was higher in the presence of sulfate than in the presence of methionine, the expression of the cysK gene was downregulated in sulfate-grown cells (Table 1). The B. subtilis genome contains two other putative cysteine synthases encoded by the yrhA and ytkP genes (9). As observed for cysK, the amount of yrhA mRNA was lower in the presence of sulfate whereas no difference in expression was detected for the ytkP gene. Two other genes whose expression was increased in the presence of methionine were yrhB and mtnA, which encode a putative cystathionine γ-synthase and a methylthioadenosine nucleosidase (22), respectively. The role of YrhA, YrhB, and YtkP polypeptides remains to be determined, but YrhA and YrhB are probably involved in the recycling of methionine to cysteine (7) (Auger, unpublished).
Several sulfur compounds including methionine, homocysteine, cysteine, sulfate, sulfite, and thiosulfate are taken up by B. subtilis. The expression of the ssu operon, which encodes an ABC permease system for aliphatic sulfonates and an oxygenase, is derepressed in the presence of methionine (Table 1) (3, 26). Several genes encoding transporters (yhcL, ytmJ, ytmK, ytmL, ytmM, ytmN, yxeM, and yxeO) are also expressed more strongly in the presence of methionine than in the presence of sulfate (Table 1). The YtmJKLMN, YxeMNO, and YhcL polypeptides are good candidates for the transport of sulfur-containing compounds. The yhcL gene encodes a membrane protein that shows homology with the sodium-dicarboxylate symporter family. The YtmJKLMN and YxeMNO proteins exhibit substantial sequence similarities to ABC transporters specific for polar amino acids (19). The YtmJKLMN permease belongs to the large ytmIJKLMNO-ytnIJ-ribR-ytnLM operon (3). The yxeMNO genes are located close to the yxeK, yxeL, and yxeQ genes, which were also found to be upregulated in the presence of methionine (Table 1). The transcriptional fusions between lacZ and all of the genes from yxeK to yxeR were expressed more strongly in the presence of methionine than in the presence of sulfate. These results strongly suggest the existence of a large operon stretching from yxeK to yxeR.
The transcription level of the ytmI gene is high in the presence of methionine, taurine, or glutathione and very low in the presence of sulfate, thiosulfate, and cysteine (3). To determine whether the yhcL gene and the yxeK operon are regulated together with the ytmI operon, the expression of yhcL′-lacZ and yxeK′-lacZ transcriptional fusions was measured after growth in the presence of taurine, methionine, or sulfate. The β-galactosidase activities of these fusions were 10- and 8-fold higher in the presence of methionine than in the presence of sulfate, respectively (Table 1). These results confirm the data obtained by transcriptome analysis. The expression of these fusions was also high in the presence of taurine (data not shown). The expression of the yhcL gene and that of the yxeK and ytmI operons appear, therefore, to be coordinately regulated. The ssu operon is expressed under the same conditions (26). However, the expression level of the ytmI and ssu operons is about 200- to 1,000-fold higher in the presence of methionine than in the presence of sulfate while the yxeK and yhcL genes are only 5- to 10-fold more strongly expressed in the presence of methionine (3, 26) (this study). It is noteworthy that the ssu operon is regulated at two different levels (initiation and termination of transcription) (26), whereas a cascade of regulation modulates the expression of the ytmI operon. Whether these genes are all controlled by one or several regulatory systems remains to be determined. However, we failed to identify a target sequence common to the promoter regions of ssuB, ytmI, ytlI, yxeK, and yhcL. The identification of other regulators and the systematic characterization of cis-acting targets in the promoter regions of these genes will help to define the sulfur regulatory network in B. subtilis.
Acknowledgments
We thank J. Pero and R. Yocum for the gift of the SA29 strain and I. Guillouard, M. F. Hullo, A. Sekowska, E. Krin, F. Hommais, I. Moszer, and S. Moreira for helpful discussions. We are also grateful to our European and Japanese colleagues for the construction of the BFS mutants.
This research was supported by grants from the Ministère de l'Education Nationale de la Recherche et de la Technologie, the Centre National de la Recherche Scientifique (URA 2171), the Institut Pasteur, the Université Paris 7, and the European Biotech Program (contract QLG2 CT9901455).
REFERENCES
- 1.Auger, S., W. H. Huen, A. Danchin, and I. Martin-Verstraete. 2002. The metIC operon involved in methionine biosynthesis in Bacillus subtilis is controlled by transcription antitermination. Microbiology 148:507-518. [DOI] [PubMed] [Google Scholar]
- 2.Calogero, S., R. Gardan, P. Glaser, J. Schweitzer, G. Rapoport, and M. Débarbouillé. 1994. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 176:1234-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppée, J. Y., S. Auger, E. Turlin, A. Sekowska, J. P. Le Caer, V. Labas, V. Vagner, A. Danchin, and I. Martin-Verstraete. 2001. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 147:1631-1640. [DOI] [PubMed] [Google Scholar]
- 4.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and J. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 6.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 7.Grundy, F. J., and T. M. Henkin. 2001. Synthesis of serine, glycine, cysteine, and methionine, p.245-254. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, D.C.
- 8.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and J. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 9.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 10.Loewen, P. C., and J. Switala. 1987. Genetic mapping of katA, a locus that affects catalase 1 level in Bacillus subtilis. J. Bacteriol. 169:5848-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansilla, M. C., D. Albanesi, and D. de Mendoza. 2000. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in L-cysteine biosynthesis in Bacillus subtilis. J. Bacteriol. 182:5885-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansilla, M. C., and D. de Mendoza. 2000. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology 146:815-821. [DOI] [PubMed] [Google Scholar]
- 13.Mansilla, M. C., and D. de Mendoza. 1997. L-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J. Bacteriol. 179:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews, R. G. 1996. One-carbon metabolism, p. 600-611. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and J. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 15.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 16.Murphy, B. A., F. J. Grundy, and, T. M. Henkin. 2002. Prediction of gene function in methylthioadenosine recycling from regulatory signals. J. Bacteriol. 184:2314-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostrowski, J., G. Jagura-Burdzy, and N. M. Kredich. 1987. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 262:5999-6005. [PubMed] [Google Scholar]
- 18.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 20.Saint-Girons, I., N. Duchange, G. N. Cohen, and M. M. Zakin. 1984. Structure and autoregulation of the metJ regulatory gene in Escherichia coli. J. Biol. Chem. 259:14282-14285. [PubMed] [Google Scholar]
- 21.Sekowska, A., J. Y. Coppee, J. P. Le Caer, I. Martin-Verstraete, and A. Danchin. 2000. S-Adenosylmethionine decarboxylase of Bacillus subtilis is closely related to archaebacterial counterparts. Mol. Microbiol. 36:1135-1147. [DOI] [PubMed] [Google Scholar]
- 22.Sekowska, A., and A. Danchin. 1999. Identification of yrrU as the methylthioadenosine nucleosidase gene in Bacillus subtilis. DNA Res. 6:255-264. [DOI] [PubMed] [Google Scholar]
- 23.Sekowska, A., and A. Danchin. 2002. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekowska, A., L. Mulard, S. Krogh, J. K. Tse, and A. Danchin. 2001. MtnK, methylthioribose kinase, is a starvation-induced protein in Bacillus subtilis. BMC Microbiol. 1:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 26.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Expression of the Bacillus subtilis sulphonate-sulphur utilization genes is regulated at the levels of transcription initiation and termination. Mol. Microbiol. 39:1356-1365. [PubMed] [Google Scholar]
- 27.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol. Lett. 201:29-35. [DOI] [PubMed] [Google Scholar]
- 28.Weissbach, H., and N. Brot. 1991. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 5:1593-1597. [DOI] [PubMed] [Google Scholar]
- 29.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yocum, R. R., J. B. Perkins, C. L. Howitt, and J. Pero. 1996. Cloning and characterization of the metE gene encoding S-adenosylmethionine synthetase from Bacillus subtilis. J. Bacteriol. 178:4604-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]