Abstract
Mutant strain 25-1 of the facultative chemoautotroph Ralstonia eutropha H16 had previously been shown to exhibit an obligately high-CO2-requiring (HCR) phenotype. Although the requirement varied with the carbon and energy sources utilized, none of these conditions allowed growth at the air concentration of CO2. In the present study, a gene designated can and encoding a β-carbonic anhydrase (CA) was identified as the site altered in strain 25-1. The mutation caused a replacement of the highly conserved glycine residue 98 by aspartate in Can. A can deletion introduced into wild-type strain H16 generated mutant HB1, which showed the same HCR phenotype as mutant 25-1. Overexpression of can in Escherichia coli and mass spectrometric determination of CA activity demonstrated that can encodes a functional CA. The enzyme is inhibited by ethoxyzolamide and requires 40 mM MgSO4 for maximal activity. Low but significant CA activities were detected in wild-type H16 but not in mutant HB1, strongly suggesting that the CA activity of Can is essential for growth of the wild type in the presence of low CO2 concentrations. The HCR phenotype of HB1 was overcome by complementation with heterologous CA genes, indicating that growth of the organism at low CO2 concentrations requires sufficient CA activity rather than the specific function of Can. The metabolic function(s) depending on CA activity remains to be identified.
Carbon dioxide and bicarbonate (dissolved inorganic carbon [DIC]) are essential growth factors for bacteria. The metabolic need for DIC is evident in autotrophs utilizing CO2 as the sole carbon source, but heterotrophs also fix significant amounts of both carbon species. Although sufficient CO2 is produced during catabolism, deprivation of atmospheric CO2 leads to growth inhibition or even death of heterotrophs (7, 14, 22). Pathogenic bacteria seem to be adapted to high DIC concentrations in their host environment, as they usually require 5 to 10% (vol/vol) CO2 for growth (9, 45, 60). Furthermore, elevated DIC was found to shorten the lag phase and accelerate growth of bacteria even though the organisms were not generally dependent on high DIC concentrations (46, 47, 60). This “sparking effect” is most pronounced when cultures are inoculated at low cell densities. The need for DIC is generally attributed to CO2 fixation in anaplerotic or other biosynthetic reactions. Consequently, the requirement is often satisfied by supplementation of the growth media with metabolites, particularly intermediates of the tricarboxylic acid cycle such as oxaloacetate and 2-oxoglutarate (28). Most high-CO2-requiring (HCR) mutants of Escherichia coli and other microorganisms regained the ability to grow at air concentrations of CO2 (0.035% [vol/vol]) upon provision with appropriate metabolites, but some depended strictly on high CO2 concentrations (5 to 10% [vol/vol]) (1, 10, 64). In contrast to their general DIC requirement, many microorganisms are inhibited by very high CO2 concentrations (ca. 20% [vol/vol] and above), an effect used in food preservation (15). However, the sensitivity towards CO2 varies widely among organisms and also depends on the nutritional and cultural conditions.
Although the uncatalyzed hydration-dehydration of CO2-HCO3− proceeds at significant rates, the metabolic reaction is catalyzed by carbonic anhydrase (CA) (EC 4.2.1.1) to support various physiological functions involving DIC. CAs are known to participate in transport and autotrophic fixation of CO2 in plants, algae, and cyanobacteria as well as in HCO3−- or H+-coupled ion transport, pH regulation, or carboxylation reactions in higher eukaryotes (8, 11, 17, 20, 59). Four phylogenetically unrelated families of CA (α, β, γ, and δ) are currently differentiated (26, 51, 63). However, while CAs are common in bacteria, with β-CA apparently as the dominant type, there is little information about the physiological significance of the enzyme in these organisms (36, 51).
Ralstonia eutropha (formerly Alcaligenes eutrophus) is a respiratory, facultatively chemoautotrophic bacterium. Organic acids such as pyruvate, lactate, or succinate are preferred organic substrates. Assimilation of CO2 during autotrophic growth with either hydrogen or formate as an energy source proceeds via the Calvin-Benson-Bassham cycle (5). An HCR mutant of R. eutropha H16, strain 25-1, that depended on increased CO2 concentrations for growth on all substrates tested was isolated previously, although the CO2 concentrations necessary for phenotypic restoration varied with the carbon source (1). Approximately 2.5% (vol/vol) CO2 was required for the mutant to regain wild-type growth rates on succinate, about 5% (vol/vol) was required on fructose, and even 10% (vol/vol) was not sufficient on lactate. Growth of the mutant on complex media also needed elevated CO2, as did lithoautotrophic growth. Supplementation with vitamins or various biosynthetic precursors did not alleviate the CO2 requirement of the mutant. Accumulation of the storage polyester poly-β-hydroxybutyrate was not affected, indicating that the main pathways involved in heterotrophic catabolism, autotrophic CO2 fixation, and synthesis of the storage compound were still functional.
In the present study we identified the can gene as the site of mutation in mutant 25-1 of R. eutropha H16. The gene encodes a β-CA whose function is essential for growth at air concentrations of CO2. The HCR phenotype of mutant 25-1 was confirmed by the generation of a can deletion strain. High CA activities detected in E. coli after heterologous expression of can provided evidence that the gene encodes a functional CA. Phenotypic complementation of the mutants with several heterologous CA genes showed that there is a requirement for sufficient CA activity in R. eutropha but not for the specific function of the can-encoded CA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains of R. eutropha were propagated in nutrient broth or mineral salts medium at 30°C as described previously (65). The mineral medium was supplemented with organic substrates at a final concentration of 0.2% (wt/vol). Lithoautotrophic cultures were incubated under an atmosphere consisting of H2, CO2, and O2 (8:1:1, vol/vol/vol). For growth of R. eutropha mutants 25-1 and HB1, small liquid cultures (10 or 50 ml) or agar plates were incubated in desiccators under air plus 10% (vol/vol) CO2. Larger cultures (1 liter) used in growth experiments were continuously supplied with this gas mixture at a rate of 2.5 liters/min and stirred magnetically at 600 rpm. CO2-limited cultures of HB1 were first gassed with air plus 10% (vol/vol) CO2 until an optical density at 436 nm (OD436) of 0.3 to 0.4 was reached. The cultures were then bubbled with ambient air for several hours.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| R. eutropha | ||
| H16 | Cfx Hox Fox; wild type | ATCC 17699, DSM 428 |
| 25-1 | Cfx Hox Fox HCR; can; mutant of H16 | 1 |
| HB1 | Cfx Hox Fox HCR; Δcan; mutant of H16 | This study |
| E. coli | ||
| S17-1 | Smr Mod+ Res−; thi pro recA; integrated RP4 (Tc::Mu-Km::Tn7) | 48 |
| JW1 | ara Δ(lac-proAB) rpsL thi φ80(lacZΔM15) F′(lacIqlacZΔM15 proA+B+) | 34 |
| BH1214 | Hfr; pyrE41 metB1 tonA22 relA1 spoT PA02 | 24 |
| BUM012 | HfrR5; ΔcynT lacY+ | 24 |
| Plasmidsb | ||
| pUC19 | Apr; lacPOZ′ | 66 |
| pBluescript KS | Apr; lacPOZ′ | Stratagene |
| pLAFR1 | Tcr Mob+ Tra−; λ-cos; broad-host-range cosmid | 19 |
| pMP92 | Tcr Mob+ Tra−; broad host range | 54 |
| pNHG1 | Kmr Tcr Mob+; sacB RP4-oriT ColE1-ori | 29 |
| pBH2240 | pUC19::224-bp BglII/HinfI | 39 |
| pMP2240 | pMP92(PstI-XbaI)::224-bp BglII/HinfI, recloned from pBH2240 as PstI-XbaI fragment | This study |
| pKR1 | pLAFR 1::20+3.1+1.7-kb EcoRI | This study |
| pKR100 | pMP92::3,067-bp EcoRI from pKR1 | This study |
| pKR200 | pMP92::821-bp KpnI-EcoRI from pKR100 | This study |
| pCAN3000/3001 | pBluescript KS::3067-bp EcoRI; different orientations of insert | This study |
| pCAN1701 | pBluescript KS::1736-bp BglII-EcoRI from pCAN3001 | This study |
| pCAN1701Δ | pCAN1701 carrying a 423-bp HincII in-frame deletion in can | This study |
| pNHG1701Δ | pNHG1(XbaI-PmeI)::1,313-bp BglII-EcoRI from pCAN1701Δ, recloned as XbaI-EcoRV | This study |
| pCAN8210 | pUC19::821-bp KpnI-EcoRI from pCAN3000 | This study |
| pAL12 | pUC118::1.3-kb EcoRI-HindIII carrying cynT from E. coli | 24 |
| pHCAII | pICB1::0.8-kb XhoI-BglII carrying the human HCAII gene | 16 |
| pMP-cynT | pMP2240(BamHI-BglII)::0.7-kb HincII-BglII from pAL12 | This study |
| pMP-HCAII | pMP2240(EcoRI-BglII)::0.8-kb XhoI-BglII from pHCAII | This study |
Apr, ampicillin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant; Cfx, ability for autotrophic CO2 fixation; Hox, ability for H2 oxidation; Fox, ability for formate oxidation; HCR, high CO2 requirement.
The last digit of the pCAN plasmid designations indicates the relative orientation of the cloned genes relative to lacZ′ of the vector: 0, can colinear to lacZ′; 1, can divergent to lacZ′.
Air-grown cells were obtained by initially growing the culture under air plus 10% (vol/vol) CO2 to an OD436 of 0.3 to 0.4 followed by gassing with ambient air. To achieve better control of DIC (bicarbonate) concentration during growth experiments, the pH of the mineral medium was kept at 6.5. Anaerobic, denitrifying growth was tested under an atmosphere of N2 or N2 plus CO2 (9:1, vol/vol) on fructose mineral agar containing 0.2% (wt/vol) KNO3. The utilization of cyanate as a nitrogen source was checked on fructose mineral agar, in which the NH4Cl of the standard medium was replaced by 0.05% (wt/vol) KCNO. Succinate mineral medium supplemented with various metabolites at a final concentration of 1 mM was used to test if the metabolites supported growth of R. eutropha HB1 at ambient CO2.
E. coli was routinely grown in Luria-Bertani medium at 37°C. Growth in cyanate-containing Luria-Bertani medium was done as detailed earlier (24). Required antibiotics were added to media at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml for E. coli, 450 μg/ml for R. eutropha in mineral medium, and 120 μg/ml for R. eutropha in nutrient broth; and tetracycline, 15 μg/ml for E. coli and 20 μg/ml for R. eutropha.
Manipulation of DNA.
Standard protocols were employed for DNA isolation and cloning (3). Restriction and DNA-modifying enzymes were used as recommended by the manufacturers. Oligodeoxynucleotide primers were purchased from MWG-Biotech (Ebersberg, Germany). PCRs were performed with Taq (Qiagen, Hilden, Germany) or Pfu (Promega, Mannheim, Germany) DNA polymerase. For Southern hybridizations, digested DNA was separated by agarose gel electrophoresis and vacuum blotted onto a Biodyne B nylon membrane (Pall, Dreieich, Germany). The DNA probes were labeled nonradioactively with digoxigenin, and DNA-DNA hybrids on the blots were detected in a staining reaction involving nitroblue tetrazolium, 5-bromo-4-chloro-3-indolyl phosphate, and alkaline phosphatase conjugated to antidigoxigenin Fab fragments (Roche, Mannheim, Germany).
DNA sequencing and analysis.
DNA sequences were determined by the dideoxy chain termination method, using cycle sequencing reactions (SequiTherm cycle sequencing kit; Biozym, Hessisch Oldendorf, Germany) with α-35S-dATP (ICN Biomedicals, Eschwege, Germany) as the labeled nucleotide. The can gene of R. eutropha was amplified by PCR with primers CA-PCR1-2 (20-mer) and CA-PCR2 (24-mer), which annealed to positions 2244 to 2263 and 3032 to 3009, respectively, of the 3,067-bp EcoRI fragment. Amplified fragments from two independent PCRs were sequenced to localize the site of mutation in strain 25-1. Basic analyses of nucleotide and deduced protein sequences were performed with the Genetics Computer Group (Madison, Wis.) program package, version 10.0. Similarity searches within various sequence databases were done using the BLAST programs (2). Sequence alignments were generated with the ClustalX program, version 1.8 (61).
Phenotypic complementations.
Broad-host-range cosmid (pLAFR1) clones of a genomic library of R. eutropha H16 (18) were conjugally transferred from E. coli S17-1 into HCR can mutant 25-1. Transconjugants were selected under air plus 10% (vol/vol) CO2 on fructose mineral agar containing tetracycline and subsequently checked for growth on the same agar under low (air) CO2 concentrations. For verification, the transferred plasmids were isolated from apparently complemented transconjugants, retransformed into E. coli S17-1, and finally retransferred into mutant 25-1. Cosmids complementing the mutant were subjected to restriction analysis. Subclones were generated and used for complementation of mutants 25-1 and HB1 to delimit the size of the complementing DNA fragment. Mutant HB1 was also complemented with heterologous CA genes cloned in pMP2240. Transconjugants were tested for growth under air or air plus 10% (vol/vol) CO2 on mineral agar containing various carbon and energy sources.
Construction of plasmids.
The 3.1-kb EcoRI fragment of pKR1 was recloned into pBluescript KS to yield pCAN3000 and pCAN3001. Cloning of the same fragment into broad-host-range vector pMP92 generated pKR100. Digestion of pCAN3001 with BamHI and BglII removed a 0.7-kb BamHI-BglII and a 0.6-kb BglII fragment prior to religation, producing pCAN1701. To delete a can-internal 423-bp HincII fragment (Fig. 1) from the 1.74-kb insert of pCAN1701, the plasmid was cleaved with HincII and the resulting large vector-insert and 341-bp HincII fragments were religated in their original orientations, yielding pCAN1701Δ. The 1.31-kb insert of pCAN1701Δ was recloned as an XbaI-EcoRV fragment into pNHG1, producing pNHG1701Δ. Plasmid pCAN8210 was constructed by cloning the 821-bp KpnI-EcoRI fragment of pCAN3000 into pUC19. Recloning of this fragment in pMP92 gave pKR200. Expression vector pMP2240 was constructed by inserting the 0.23-kb XbaI-PstI fragment of pBH2240, containing the chromosomal cbb operon promoter of R. eutropha, into pMP92. The human HCAII gene was recloned from pHCAII as a 0.8-kb XhoI-BglII fragment into pMP2240 to generate pMP-HCAII. For the construction of pMP-cynT the 0.7-kb HincII-BglII fragment of pAL12, containing the E. coli cynT gene, was ligated into pMP2240.
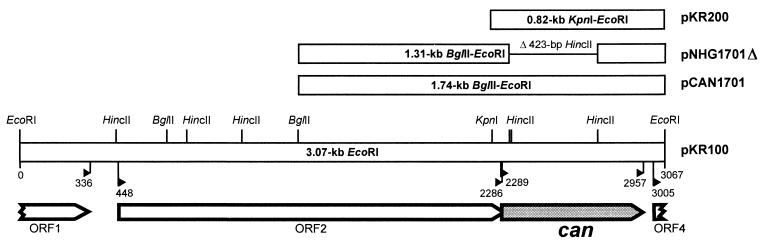
FIG. 1.
Genetic organization within the 3,067-bp EcoRI fragment of R. eutropha H16. The orientation and sizes of the identified ORFs (ORF1, ORF2, can, and ORF4) are indicated by arrows. Fragments cloned in the respective plasmids are shown by bars together with the cleavage sites of relevant restriction endonucleases. Flagged marks indicate the positions (in base pairs) of the start and stop codons of the ORFs.
Construction of can deletion strain HB1.
A can deletion strain of R. eutropha H16 was constructed by gene replacement mutagenesis. For this purpose plasmid pNHG1701Δ carrying a 423-bp in-frame deletion within can (Fig. 1) was conjugally transferred from E. coli S17-1 into R. eutropha H16. Subsequent recombinations and selections of hetero- and homogenotes were done as described previously (29). Since the can mutant was expected to exhibit an HCR phenotype, recombinants were grown under air plus 10% (vol/vol) CO2. The mutant genotype was verified by Southern hybridization and PCR.
Overexpression of can and preparation of cell extracts.
The can gene in pCAN8210 was expressed in E. coli JW1 after induction by isopropyl-β-d-thiogalactopyranoside as described before (38). Proteins in lysates obtained from the induced cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Crude cell extracts of E. coli and R. eutropha were prepared from cells suspended in CA buffer (50 mM Bicine-NaOH [pH 8.0] containing 40 mM MgSO4, 5 mM dithioerythritol, and 1 mM phenylmethylsulfonyl fluoride) and disrupted by sonication. The extracts were obtained after centrifugation at 14,000 × g for 20 min to remove unbroken cells and cell debris. Up to 100 μl of the extracts was assayed for CA activity.
Determination of CA activities.
CA activities were determined by a mass spectrometric method based on the loss of 18O from doubly labeled 13C18O2 to water (58). The 18O decline was monitored with a quadrupole mass spectrometer (MSD 5970; Hewlett-Packard, Waldbronn, Germany) coupled to a 10-ml thermostatted (30°C) reaction cuvette via a membrane inlet system. Changes in mass signals m/z = 45 (13C16O2), m/z = 47 (13C18O16O), and m/z = 49 (13C18O2) were recorded and used to calculate the 18O fraction (as percent enrichment) in doubly labeled CO2 as follows: log (% enrichment) = log (13C18O2/13CO2) = log [100 − 49(45 + 47 + 49)].
H13C18O3− was added (1 mM) to the assay buffer (50 mM Bicine-NaOH [pH 8.0] plus 40 mM MgSO4), and the cuvette was closed. Isotopic changes in CO2 were recorded until isotopic equilibrium (blank) was reached. The CA-catalyzed reaction was then initiated by injecting cell extract. CA activity was calculated from the linear decrease in log (% enrichment) before and after addition of the sample as [log (% enrichment)sample − log (% enrichment)blank]/log (% enrichment)blank and normalized on a protein basis (58).
For in vivo CA assays, cells were harvested from freshly grown cultures (OD436 of 2 to 3) and resuspended in mineral salts medium at an OD436 of 50. Up to 400 μl of the cell suspension, corresponding to about 2.5 mg of total cell protein, was used per assay. Addition of cells caused a biphasic decline of log (% enrichment) resulting in an initial and a final slope. The apparent internal CA activity was expressed as the difference in loss of enrichment by extrapolating the final slope back to the time point of cell addition (43, 57). Control assays were run with mineral medium to evaluate the effects of pH change and dilution.
Nucleotide sequence accession number.
The nucleotide sequence of the 3,067-bp EcoRI fragment was deposited in the EMBL/GenBank/DDBJ databases under accession number AJ310671.
RESULTS AND DISCUSSION
The defect in HCR mutant 25-1.
Phenotypic complementation of mutant 25-1 by hybrid cosmids from a genomic library of wild-type strain H16 resulted in the isolation of a transconjugant harboring pKR1. Growth of the transconjugant at ambient CO2 concentrations was restored. Subcloning identified a 3.07-kb EcoRI fragment (pKR100) that complemented the mutation. A 0.82-kb KpnI-EcoRI subfragment (pKR200) (Fig. 1) was eventually shown to be sufficient for complementation, indicating that the mutation resided within this region.
Sequencing of the EcoRI fragment revealed 3,067 bp comprising two incomplete and two complete open reading frames (ORFs) (Fig. 1). The deduced product of the first incomplete ORF (ORF1) shares highest sequence similarity (74% amino acid identity) with a putative isovaleryl coenzyme A dehydrogenase from Pseudomonas aeruginosa (55). ORF2, located 111 bp downstream of ORF1, is complete and resembles aceK, encoding the dual-function isocitrate dehydrogenase kinase/phosphatase in E. coli (33). The amino acid sequence of AceK is 42% identical to the deduced amino acid sequence of ORF2 (613 residues).
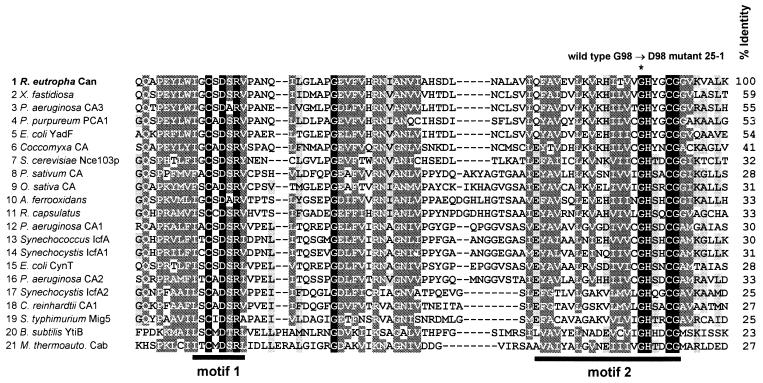
The second complete ORF, designated can, encodes a polypeptide of 223 amino acid residues that has a calculated molecular mass of 24,909 Da and represents a presumptive β-CA. Sequencing of can amplified by PCR from 25-1 DNA identified a G→A transition at position 293 of the gene that converted a highly conserved glycine residue of the Can protein into an aspartate (G98D) (Fig. 2). To verify the phenotypic effect of the mutation in strain 25-1, a 423-bp in-frame deletion was introduced into the can gene of wild-type H16, generating mutant HB1. Like mutant 25-1, HB1 was unable to grow at air concentrations of CO2 regardless of the substrate provided, confirming an essential physiological role of Can. In addition, the HCR phenotype of the deletion mutant was relieved by providing the can-containing 0.82-kb KpnI-EcoRI fragment in trans (pKR200). Growth of the wild-type strain at ambient CO2 concentrations therefore requires a functional can gene.
FIG. 2.
Partial sequence alignment of various (putative) β-CAs. The selected segment corresponds to amino acid residues 33 through 110 of Can from R. eutropha and covers the two most conserved motifs (motifs 1 and 2, indicated by bars) involved in binding of Zn2+ within the active center of the enzyme. The alignment was performed by means of the program ClustalX (version 1.8) with a blosum62 matrix (gap-opening penalty, 11; gap extension penalty, 1). Different shadings indicate the relative similarity of amino acid residues (dark shading, 100% identity; medium shading, 100 to 75% conservation; light shading, 74 to 50% conservation) based on the following groupings: (D, E, H, K, R), (N, Q, S, T), and (L, I, V, M, F, Y, W, A, G). The asterisk marks the highly conserved glycine residue found to be the site of mutation in R. eutropha HCR mutant 25-1. Overall identities of the CA sequences with that of R. eutropha Can are given at the right. Origins of sequences (accession number or source): 1, R. eutropha (AJ310671); 2, X. fastidiosa (AAF83690); 2, P. aeruginosa (AAG08063); 4, P. purpureum PCA1 (D86050); 5, E. coli YadF (AE000122); 6, Coccomyxa sp. (U49976); 7, S. cerevisiae (U52369); 8, Pisum sativum (X52558); 9, Oryza sativa (AB016283); 10, Acidithiobacillus ferrooxidans (The Institute for Genomic Research); 11; Rhodobacter capsulatus (University of Chicago and Institute of Molecular Genetics, Prague); 12, P. aeruginosa (AAG03492); 13, Synechococcus sp. strain PCC 7942 IcfA (M77095); 14, Synechocystis sp. strain PCC 6803 IcfA1 (U45962); 15, E. coli CynT (AE000141); 16, P. aeruginosa (AAG05441); 17, Synechocystis sp. strain PCC 6803 IcfA2 (D64001); 18, Chlamydomonas reinhardtii CA1 (CRU41189); 19, Salmonella enterica serovar Typhimurium Mig5 (AF020806); 20, Bacillus subtilis YtiB (Z991119); and 21, Methanobacterium thermoautotrophicum Cab (AE000918). The sequences of the putative CAs from A. ferrooxidans and R. capsulatus were derived from unfinished genome sequences and may thus contain errors.
The 5′-terminal region of the second incomplete ORF (ORF4) was identified 46 bp downstream of can. Its potential product comprises 21 amino acid residues and shows high similarity (68% identity within 19 residues) to the phaA-encoded β-ketothiolase (EC 2.3.1.16) of P. aeruginosa (55). ORF4 may represent the gene of a third β-ketothiolase in R. eutropha, which has been postulated before (50). Conspicuous promoter structures were not detected on the entire fragment. The putative operon-like organization did not provide a hint at a common metabolic function of all genes.
can encodes a β-CA.
The amino acid sequence of the Can protein is more than 50% identical to those of putative β-CAs identified in Xylella fastidiosa (49) and P. aeruginosa (55), the almost-identical PCA1 and PCA2 of the red alga Porphyridium purpureum (41), and the yadF-encoded CA of E. coli (13). However, β-CAs are highly diverse, with overall sequence identities ranging to below 25%, limiting the similarities almost to the Zn2+-binding motifs within the catalytic centers of the enzymes (Fig 2). The mutated glycine 98 in Can of R. eutropha mutant 25-1 is located next to a histidine residue that is present in all known β-CAs and that has been shown to participate in binding of Zn2+ (13, 32, 42, 56). Thus, the mutant Can in 25-1 is either nonfunctional or less active than the wild-type enzyme.
In bacteria, β-CA seems to be the most frequent type (25, 51). These enzymes are also found in some archaea, lower eukaryotes such as yeast and fungi, and generally in algae and higher plants. However, the number and type of CAs in bacteria vary widely, and many organisms have multiple genes encoding CAs that belong to the same or a different family (25, 51). In P. aeruginosa three putative β-CA (Fig. 2) and γ-CA genes were recognized (55). Two β-CAs, CynT and YadF, are known to function in E. coli (Fig. 2), while a third protein, CaiE, represents a presumptive γ-CA (51). In Neisseria gonorrhoeae and Helicobacter pylori, putative β- and γ-CA genes were identified (51, 62). The α-CAs of these organisms have been characterized recently (12, 27). The cyanobacterium Synechococcus sp. strain PCC 7942 produces an α-CA (EcaA) (53) and a β-CA (IcfA [CcaA]) (21), whereas another cyanobacterium, Synechocystis sp. strain PCC 6803, has two β-CAs (CcaA and EcaB) (30, 52). Although there is preliminary evidence for additional CA genes in R. eutropha H16, the potential CA activities of their products are apparently not sufficient to support growth of the organism under air in the absence of Can.
Heterologous expression of can.
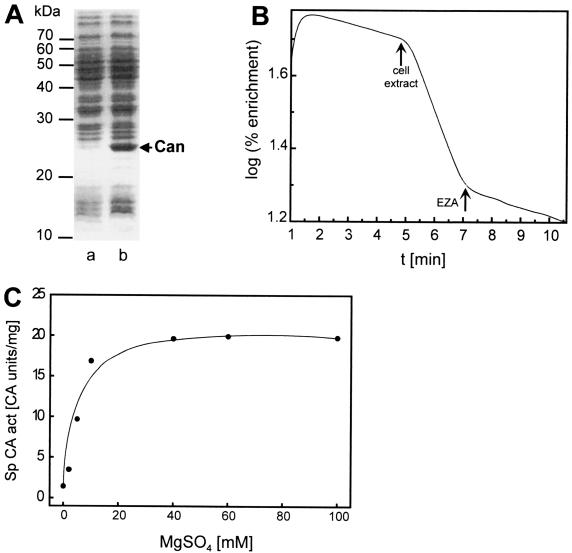
The can gene cloned in pCAN8210 was expressed in E. coli, resulting in an overproduced protein with a molecular mass of about 25 kDa (Fig. 3A), which is in close agreement with the value calculated from the deduced amino acid sequence of Can. High specific CA activity (20.5 CA U/mg) was detected in cell extracts of transformant E. coli(pCAN8210), whereas the reference strain E. coli(pUC19) showed only very low activity (0.2 CA U/mg), strongly suggesting that can encodes a functional CA. The general CA inhibitor ethoxyzolamide (EZA) (40) almost completely inhibited this activity at a concentration of 0.5 mM (Fig. 3B). Maximal CA activity was observed in the presence of 40 mM MgSO4 (Fig. 3C), similar to the case for IcfA of Synechococcus sp. strain PCC 7942, which requires 20 mM MgSO4 (67). MgCl2 only slightly stimulated Can. This effect was also seen with IcfA and might be attributed to inhibition exerted by the chloride anions counteracting the Mg2+ stimulation of Can. However, in contrast to the case for IcfA, the reducing agent dithioerythritol showed no significant influence on the CA activity of Can (data not shown).
FIG. 3.
Heterologous overexpression of the can gene from R. eutropha H16 in E. coli (A) and mass spectrometric assays of CA activities in a crude cell extract of transformant E. coli JW1(pCAN8210) (B and C). (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (14% [wt/vol] acrylamide) of cell lysates of transformants harboring pUC19 (lane a) or pCAN8210 (lane b). The arrow indicates the Can protein overproduced in E. coli JW1(pCAN8210). Molecular masses of reference proteins are indicated. (B) Representation of a CA activity assay in a cell extract of E. coli JW1(pCAN8210). The initial slope represents the uncatalyzed exchange reaction between 13C18O2 and unlabeled CO2 after addition of NaH13C18O3. Arrows mark the time points at which cell extract or EZA (0.5 mM) was added to the assay mixture. (C) Dependence of the CA activity on the presence of MgSO4.
CA activities in R. eutropha.
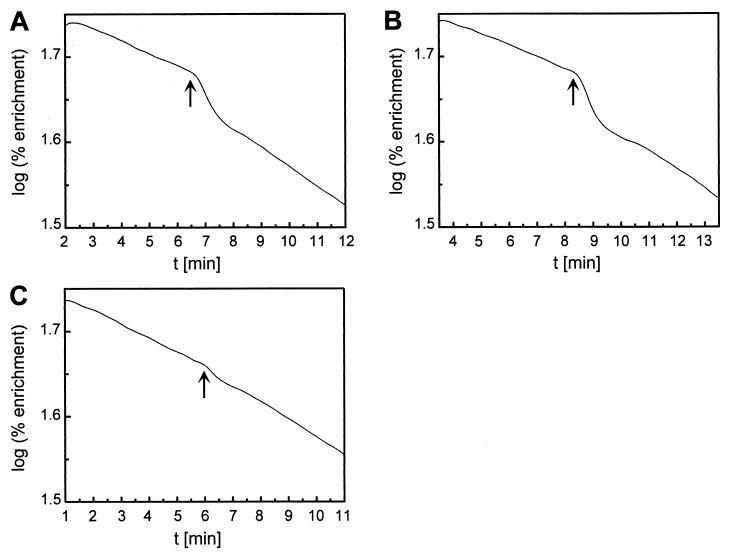
Very low but significant CA activities were found in cell extracts (Table 2) as well as whole cells (Fig. 4) of wild-type strain H16, as determined by the mass spectrometric assay. The conventional assay measuring the drop in pH as result of CO2 hydration was not sufficiently sensitive to detect these activities. EZA partially inhibited the CA activity detected in the cell extract that was apparently absent in the can mutant HB1. These findings strongly correlate the activity with Can and suggest that Can is the most prominent CA present in the cells under the growth conditions tested. The biphasic reaction kinetics of the whole-cell assays (Fig. 4) were indicative of a nonhomogeneous distribution of Can in the assay mixture and suggested an intracellular localization of the enzyme (43). Although the measured CA activity was low, it is apparently essential for growth of strain H16 at air concentrations of CO2. Similarly low activities were also found for the β-CAs CynT of E. coli (24) and IcfA of Synechococcus sp. strain PCC 7942 (4, 67). Moreover, inactivation of the corresponding genes led to HCR phenotypes of the resulting mutants (21, 24). Aeration of the R. eutropha cultures by low (air) or high (10% [vol/vol]) CO2 concentrations as well as shifts from high to low CO2 concentrations did not significantly alter the CA activities (Table 2). The can gene of R. eutropha might thus be expressed constitutively at low levels, a conclusion supported by preliminary transcriptional studies (data not shown).
TABLE 2.
CA activities in cell extracts of R. eutropha wild-type strain H16 and can mutant HB1 grown in pyruvate- mineral medium under different aerations
| Strain | Aerationa | Sp act of CA (CA U/mg of protein)b
|
|
|---|---|---|---|
| Without EZA | With EZA | ||
| Wild type H16 | Air | 0.06 | 0.03 |
| Air + CO2 | 0.08 | 0.04 | |
| Shift | 0.07 | 0.05 | |
| Mutant HB1 | Air + CO2 | <0.01 | NDc |
| Shift | <0.01 | ND | |
Aeration of the cultures with air or air plus 10% (vol/vol) CO2. For shifting from high to low CO2, cells were initially grown under air plus 10% (vol/vol) CO2 before the CO2 supply was turned off when the culture reached an OD436 of about 0.3. Cells were harvested at an OD436 of about 1 or 4 h after the shift.
CA activities were determined by the mass spectrometric assay in the absence or presence (0.5 mM) of the CA inhibitor EZA.
ND, not determined.
FIG. 4.
Mass spectrometric assay of CA activity in whole cells of R. eutropha wild-type strain H16 and can mutant HB1. Cells were grown in pyruvate-mineral medium under different aerations. (A) H16 grown under air. (B) H16 grown under air plus 10% (vol/vol) CO2. (C) HB1 grown under air plus 10% (vol/vol) CO2. Arrows mark the time points at which cells were added to the assay mixture.
Growth characteristics of HCR mutant HB1.
Like the original HCR mutant 25-1, can deletion mutant HB1 failed to grow both heterotrophically and autotrophically regardless of the carbon and energy sources supplied when incubated on agar plates under air or air containing ≤0.1% (vol/vol) CO2. Supplementation of the media with metabolites (malonate, 2-oxoglutarate, proline, histidine, arginine, hypoxanthine, adenine, thymine, uracil, or oleate) known to replace high CO2 requirements of mutants of other microorganisms (6, 10, 28, 64) was not effective for HB1. Even growth on complex nutrient broth medium was dependent on high CO2 concentrations. Wild-type growth of HB1 was restored in the presence of highly elevated CO2 (3 to 10% [vol/vol]) in air, depending on the substrate utilized. Similar obligate HCR phenotypes have been described previously for mutants of E. coli (10) and Streptomyces coelicolor (64) without assigning them to a specific metabolic/genetic defect.
The obligate HCR phenotype of mutant HB1 excludes a specific involvement of Can in autotrophic metabolism of R. eutropha. In contrast, IcfA appears to play a specific role in the autotrophic CO2 assimilation of Synechococcus sp. strain PCC 7942. IcfA is a β-CA located in the carboxysomes of the organism that functions as a component of the CO2-concentrating mechanism required for photosynthetic growth at low CO2 concentrations (31). Inactivation of the icfA gene caused an HCR phenotype of Synechococcus (21, 44, 67). Because the cyanobacterium is obligately autotrophic, it is not easily possible to differentiate between the specific CA function of IcfA in CO2 assimilation and a potential additional role in the general CO2 metabolism of the organism. An increased sensitivity towards oxygen as reported for the nce103 mutant strain of the yeast Saccharomyces cerevisiae, which is defective in a putative β-CA (23), was not observed with R. eutropha mutant HB1. The HCR phenotype of HB1 was also evident when it was grown anaerobically under denitrifying conditions (data not shown). In contrast, yeast mutant nce103 was found to grow well aerobically at elevated CO2 concentrations (D. Sültemeyer, unpublished data).
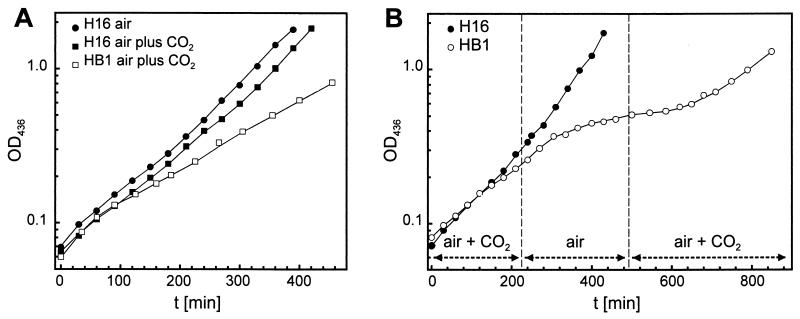
Growth of wild-type H16 and mutant HB1 was studied in more detail in liquid culture with mineral medium containing fructose or pyruvate as a substrate. As anticipated, the mutant failed to grow when gassed with air, whereas air plus 10% (vol/vol) CO2 restored wild-type growth rates on fructose (data not shown). In contrast, growth of the mutant on pyruvate remained significantly slower than that of H16 even under air plus 10% (vol/vol) CO2 (doubling time of 2.5 versus 1.5 h) (Fig. 5A). Mutant 25-1 has previously been found to exhibit a similar behavior during growth on lactate (1). Growth of mutant HB1 on pyruvate under air plus 10% (vol/vol) CO2 ceased after a shift to air but returned to the initial rate upon resupply of CO2 (Fig. 5B). This effect was most evident when the cultures were shifted at low cell densities (OD436 of up to about 0.4). Shifts to air at higher densities simply led to decreased growth or had no detectable effect (data not shown). In these cases the metabolically generated CO2 presumably was sufficient to compensate for the high CO2 requirement of the mutant. High concentrations of metabolic CO2, occurring at high cell densities, seem to mask the need for CA activity in the HCR mutant. Changes in the CO2 content of the atmosphere did not affect the growth of wild-type H16 (Fig. 5B), although R. eutropha has been shown to require elevated initial CO2 concentrations to shorten the lag phase of low-density cultures (47). It is conceivable that low CO2 concentrations present in such cultures due to limiting metabolic CO2 do not allow the cells to convert sufficient CO2 into HCO3− before CO2 diffuses out (35). Bicarbonate is essential for some carboxylation reactions. Carbonic anhydrase would support the provision of bicarbonate to these reactions. However, the CO2 demand of the wild type during the lag phase is different from the general high CO2 requirement of the can mutants, since the latter could not be suppressed by added metabolites. The lag-phase CO2 demand might contribute to the substrate-dependent variations in the CO2 requirement of the can mutants.
FIG. 5.
Growth of R. eutropha wild-type strain H16 and can mutant HB1 in pyruvate-mineral medium under different aeration. (A) Constant aeration with air or air plus 10% (vol/vol) CO2. (B) Shift from air plus 10% (vol/vol) CO2 to air and back to air plus 10% (vol/vol) CO2.
Phenotypic complementation of HCR mutant HB1 by heterologous CA genes.
Mutant HB1 was subjected to complementation analyses to check whether expression of heterologous CA genes encoding different types of CA could alleviate the HCR phenotype. For this purpose, the β-CA gene cynT from E. coli (pMP-cynT) or the human α-CA gene CAII (pMP-HCAII) was transferred into HB1. Both resulting transconjugants regained the ability to grow under air, indicating that sufficient CA activity is apparently required for R. eutropha to grow at ambient CO2 concentrations. Can is not specifically needed, as its function can be replaced by other CAs which may belong to different families.
The cynT gene is part of the cyn operon of E. coli, which enables the organism to utilize cyanate as the sole nitrogen source. Inactivation of cynT caused an HCR phenotype of the mutant when it was growing in the presence of cyanate (24). Similar to the case for the R. eutropha can mutants, the E. coli cynT mutant was unable to grow under air unless the cyanate inhibition was overcome by elevated CO2 concentrations or complementation by the human CAII (37). The phenotype of the cynT mutant has been attributed to inhibition by cyanate of a metabolic function involving DIC, rather than to a specific involvement of CynT in cyanate degradation (36, 37) as had been proposed previously (24). Since R. eutropha is also able to utilize cyanate as the sole nitrogen source (data not shown), Can might allow growth of the organism in presence of cyanate in much the same way that CynT does in E. coli. However, in contrast to the case for the E. coli cynT mutant, the HCR phenotype of mutant HB1 is not correlated to the presence of cyanate. This notion gained support by the phenotypic complementation of the E. coli cynT mutant expressing the can gene of R. eutropha (data not shown). It is likely that YadF, a second β-CA in E. coli (13) sharing high similarity with Can, might be the target of the CO2-suppressible cyanate inhibition in vivo. The following observations support this conclusion: (i) the cynT mutant did not exhibit the HCR phenotype in the absence of cyanate (35), (ii) the CA activity of YadF but not that of CynT is strongly inhibited by cyanate in vitro (36; F. von Götz, B. Kusian, and B. Bowien, unpublished data), (iii) a yadF-deficient mutant showed an HCR phenotype except in the presence of cyanate (von Götz et al., unpublished data), and (iv) can and yadF complemented the yadF mutant and mutant HB1 of R. eutropha, respectively (von Götz et al., unpublished data). Therefore, the physiological role of YadF in E. coli seems to correspond to that of Can in R. eutropha. CynT would replace the function of YadF in the presence of cyanate.
Concluding remarks.
The wide distribution and multiple occurrence of CAs in bacteria suggest a fundamental physiological significance of these enzymes in DIC metabolism by cells. Our results indicate that growth of R. eutrophus at air levels of CO2 is principally dependent on sufficient CA activity, which is provided by the β-CA Can. In E. coli the β-CA YadF seems to serve this function. Further evidence suggests that growth of other organisms in air also requires CA activity. Although the involvement of CA in autotrophic growth of cyanobacteria at low CO2 concentrations has been known for a long time, this is the first report relating obligate high-CO2 requirements of heterotrophs directly to the lack of CA. In view of these findings, CAs must be assigned an essential role in DIC metabolism, at least at low CO2 levels in the environment. However, the metabolic functions depending on CA activity under these conditions still remain to be identified. In this context, the availability of CO2-HCO3− for carboxylation reactions, pH homeostasis, and possibly DIC-directed gene regulation are potential areas of interest.
Acknowledgments
This work was supported by a grant from the Ministerium für Wissenschaft und Kultur des Landes Niedersachsen.
Some strains and plasmids used in this study were kindly provided by Cecilia Forsman (Umea University, Umea, Sweden) and James A. Fuchs (University of Minnesota, St. Paul). Preliminary genome sequence data (A. ferrooxidans) were obtained from The Institute for Genomic Research. We thank Dimitar Dushkov, Plamena Entcheva, Martina Meister, Kerstin Röske, Gertrud Stahlhut, Mladen Tzvetkov, Silke Walburg, and Tanja Wendt for their efforts at various stages of this project.
REFERENCES
- 1.Ahrens, W., and H. G. Schlegel. 1972. Carbon dioxide requiring mutants of Hydrogenomonas eutropha strain H 16. 1. Growth and CO2-fixation. Arch. Mikrobiol. 85:142-152. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T.-L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
- 4.Badger, M. R., and G. D. Price. 1989. Carbonic anhydrase activity associated with the cyanobacterium Synechococcus PCC7942. Plant Physiol. 89:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowien, B., and H. G. Schlegel. 1981. Physiology and biochemistry of aerobic hydrogen-oxidizing bacteria. Annu. Rev. Microbiol. 35:405-452. [DOI] [PubMed] [Google Scholar]
- 6.Broadbent, J. A., and H. P. Charles. 1965. Some carbon dioxide requiring mutants of Neurospora crassa. J. Gen. Microbiol. 39:63-74. [DOI] [PubMed] [Google Scholar]
- 7.Brown, O. R., and H. F. Howitt. 1969. Growth inhibition and death of Escherichia coli from CO2 deprivation. Microbios 3:241-246. [Google Scholar]
- 8.Burnell, J. N. 2000. Carbonic anhydrases of higher plants: an overview, p. 501-518. In W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (ed.), The carbonic anhydrases: new horizons. Birkhäuser, Basel, Switzerland. [DOI] [PubMed]
- 9.Burns, B. P., S. L. Hazell, and G. L. Mendz. 1995. Acetyl-CoA carboxylase activity in Helicobacter pylori and the requirement of increased CO2 for growth. Microbiology 141:3113-3118. [DOI] [PubMed] [Google Scholar]
- 10.Charles, H. P., and G. A. Roberts. 1968. Carbon dioxide as a growth factor for mutants of Escherichia coli. J. Gen. Microbiol. 51:211-224. [DOI] [PubMed] [Google Scholar]
- 11.Chegwidden, W. R., S. J. Dodgson, and I. M. Spencer. 2000. The roles of carbonic anhydrase in metabolism, cell growth and cancer in animals, p. 343-364. In W. R.Chegwidden, N. D. Carter, and Y. H. Edwards (ed.), The carbonic anhydrases: new horizons. Birkhäuser, Basel, Switzerland. [DOI] [PubMed]
- 12.Chirica, L. C., B. Elleby, and S. Lindskog. 2001. Cloning, expression and some properties of α-carbonic anhydrase from Helicobacter pylori. Biochim. Biophys Acta 1544:55-63. [DOI] [PubMed] [Google Scholar]
- 13.Cronk, J. D., J. A. Endrizzi, M. R. Cronk, J. W. O'Neill, and K. Y. J. Zhang. 2001. Crystal structure of E. coli β-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci. 10:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehority, B. A. 1971. Carbon dioxide requirement of various species of rumen bacteria. J. Bacteriol. 105:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon, N. M., and D. B. Kell. 1989. The inhibition by CO2 of the growth and metabolism of microorganisms. J. Appl. Bacteriol. 67:109-136. [DOI] [PubMed] [Google Scholar]
- 16.Forsman, C., G. Behravan, A. Osterman, and B. H. Jonsson. 1988. Production of active human carbonic anhydrase II in E. coli. Acta Chem. Scand. B 42:314-318. [DOI] [PubMed] [Google Scholar]
- 17.Forster, R. E., and S. J. Dodgson. 2000. Membrane transport and provision of substrates for carbonic anhydrase, p. 263-280. In W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (ed.), The carbonic anhydrases: new horizons. Birkhäuser, Basel, Switzerland. [DOI] [PubMed]
- 18.Freter, A., and B. Bowien. 1994. Identification of a novel gene, aut, involved in autotrophic growth of Alcaligenes eutrophus. J. Bacteriol. 176:5401-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 20.Fukuzawa, H., E. Suzuki, and S. Miyachi. 2000. Algal carbonic anhydrase, p. 535-546. In W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (ed.), The carbonic anhydrases: new horizons. Birkhäuser, Basel, Switzerland.
- 21.Fukuzawa, H., E. Suzuki, Y. Komukai, and S. Miyachi. 1992. A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc. Natl. Acad. Sci. USA 89:4437-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gladstone, G. P., P. Fildes, and G. M. Richardson. 1935. Carbon dioxide as an essential factor in the growth of bacteria. Br. J. Exp. Pathol. 16:335-348. [Google Scholar]
- 23.Götz, R., A. Gnann, and F. K. Zimmermann. 1999. Deletion of the carbonic anhydrase-like gene NCE103 of the yeast Saccharomyces cerevisiae causes an oxygen-sensitive growth defect. Yeast 15:855-864. [DOI] [PubMed] [Google Scholar]
- 24.Guilloton, M. B., A. F. Lamblin, E. I. Kozliak, M. Gerami-Nejad, C. Tu, D. Silverman, P. M. Anderson, and J. A. Fuchs. 1993. A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J. Bacteriol. 175:1443-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewett-Emmett, D. 2000. Evolution and distribution of the carbonic anhydrase gene families, p. 29-76. In W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (ed.), The carbonic anhydrases: new horizons. Birkhäuser, Basel, Switzerland. [DOI] [PubMed]
- 26.Hewett-Emmett, D., and R. E. Tashian. 1996. Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol. Phylogenet. Evol. 5:50-77. [DOI] [PubMed] [Google Scholar]
- 27.Huang, S., Y. Xue, E. Sauer-Eriksson, L. Chirica, S. Lindskog, and B.-H. Jonsson. 1998. Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide. J. Mol. Biol. 283:301-310. [DOI] [PubMed] [Google Scholar]
- 28.Hutner, S. H. 1972. Inorganic nutrition. Annu. Rev. Microbiol. 26:313-346. [DOI] [PubMed] [Google Scholar]
- 29.Jeffke, T., N.-H. Gropp, C. Kaiser, C. Grzeszik, B. Kusian, and B. Bowien. 1999. Mutational analysis of the cbb operon (CO2 assimilation) promoter of Ralstonia eutropha. J. Bacteriol. 181:4374-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan, A., and L. Reinhold. 1999. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:539-570. [DOI] [PubMed] [Google Scholar]
- 32.Kimber, M. S., and E. F. Pai. 2000. The active site architecture of Pisum sativum β-carbonic anhydrase is a mirror image of that of α-carbonic anhydrases. EMBO J. 19:1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klumpp, D. J., D. W. Plank, L. J. Bowdin, C. S. Stueland, T. Chung, and D. C. LaPorte. 1988. Nucleotide sequence of aceK, the gene encoding isocitrate dehydrogenase kinase/phosphatase. J. Bacteriol. 170:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolmar, H., K. Friedrich, J. Pschorr, and H. J. Fritz. 1990. Hybrids of circular DNA single strands as intermediates in DNA cloning, sequence analysis and directed mutagenesis. Technique 2:237-245. [Google Scholar]
- 35.Kozliak, E. I., J. A. Fuchs, M. B. Guilloton, and P. M. Anderson. 1995. Role of bicarbonate/CO2 in the inhibition of Escherichia coli growth by cyanate. J. Bacteriol. 177:3213-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozliak, E. I., M. B. Guilloton, J. A. Fuchs, and P. M. Anderson. 2000. Bacterial carbonic anhydrases, p. 547-565. In W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (ed.), The carbonic anhydrases: new horizons. Birkhäuser, Basel, Switzerland. [DOI] [PubMed]
- 37.Kozliak, E. I., M. B. Guilloton, M. Gerami-Nejad, J. A. Fuchs, and P. M. Anderson. 1994. Expression of proteins encoded by the Escherichia coli cyn operon: carbon dioxide-enhanced degradation of carbonic anhydrase. J. Bacteriol. 176:5711-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kusian, B., J.-G. Yoo, R. Bednarski, and B. Bowien. 1992. The Calvin cycle enzyme pentose-5-phosphate 3-epimerase is encoded within the cfx operons of the chemoautotroph Alcaligenes eutrophus. J. Bacteriol. 174:7337-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusian, B., R. Bednarski, M. Husemann, and B. Bowien. 1995. Characterization of the duplicate ribulose-1,5-bisphosphate carboxylase genes and cbb promoters of Alcaligenes eutrophus. J. Bacteriol. 177:4442-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maren, T. H., and G. Sanyal. 1983. The activity of sulfonamides and anions against the carbonic anhydrases of animals, plants, and bacteria. Annu. Rev. Pharmacol. Toxicol. 23:439-459. [DOI] [PubMed] [Google Scholar]
- 41.Mitsuhashi, S., and S. Miyachi. 1996. Amino acid sequence homology between N- and C-terminal halves of a carbonic anhydrase in Porphyridium purpureum, as deduced from the cloned cDNA. J. Biol. Chem. 271:28703-28709. [DOI] [PubMed] [Google Scholar]
- 42.Mitsuhashi, S., T. Mizushima, E. Yamashita, M. Yamamoto, T. Kumasaka, H. Moriyama, T. Ueki, S. Miyachi, and T. Tsukihara. 2000. X-ray structure of β-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO2 hydration. J. Biol. Chem. 275:5521-5526. [DOI] [PubMed] [Google Scholar]
- 43.Palmqvist, K., J.-W. Yu, and M. R. Badger. 1994. Carbonic anhydrase activity and inorganic carbon fluxes in low- and high Ci-cells of Chlamydomonas reinhardtii and Scenedesmus obliquus.Physiol. Plant. 90:537-547. [Google Scholar]
- 44.Price, G. D., and M. R. Badger. 1989. Isolation and characterization of high CO2-requiring mutants of the cyanobacterium Synechococcus PCC7942. Plant. Physiol. 91:514-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ratledge, C. 1982. Nutrition, growth and metabolism, p. 181-271. In C. Ratledge and J. Stanford (ed.), The biology of mycobacteria. Academic Press, London, United Kingdom.
- 46.Repaske, R., A. C. Repaske, and R. D. Mayer. 1974. Carbon dioxide control of lag period and growth of Streptococcus sanguis. J. Bacteriol. 117:552-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Repaske, R., C. A. Ambrose, A. C. Repaske, and M. L. De Lacy. 1971. Bicarbonate requirement for elimination of the lag period of Hydrogenomonas eutropha. J. Bacteriol. 107:712-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, R., U. Priefer, and A. Pühler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 49.Simpson, A. J. G., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 50.Slater, S., K. L. Houmiel, M. Tran, T. A. Mitsky, N. B. Taylor, S. R. Padgette, and K. J. Gruys. 1998. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 180:1979-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, K. S., and J. G. Ferry. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335-366. [DOI] [PubMed] [Google Scholar]
- 52.So, A. K. C., and G. S. Espie. 1998. Cloning, characterization and expression of carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Plant Mol. Biol. 37:205-215. [DOI] [PubMed] [Google Scholar]
- 53.Soltes-Rak, E., M. E. Mulligan, and J. R. Coleman. 1997. Identification and characterization of a gene encoding a vertebrate-type carbonic anhydrase in cyanobacteria. J. Bacteriol. 179:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 55.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrook-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. M. Lim, K. A. Smith, D. H. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 56.Strop, P., K. S. Smith, T. M. Iverson, J. G. Ferry, and D. C. Rees. 2001. Crystal structure of the “cab”-type β class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J. Biol. Chem. 276:10299-10305. [DOI] [PubMed] [Google Scholar]
- 57.Sültemeyer, D. 1998. Carbonic anhydrase in eukaryotic algae: characterization, regulation, and possible function during photosynthesis. Can. J. Bot. 76:962-972. [Google Scholar]
- 58.Sültemeyer, D., G. D. Price, J. W. Yu, and M. R. Badger. 1995. Characterisation of carbon dioxide and bicarbonate transport during steady-state photosynthesis in the marine cyanobacterium Synechococcus strain PCC7002. Planta 197:597-607. [Google Scholar]
- 59.Swenson, E. R. 2000. Respiratory and renal roles of carbonic anhydrase in gas exchange and acid-base regulation, p. 281-341. In W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (ed.), The carbonic anhydrases: new horizons. Birkhäuser, Basel, Switzerland. [DOI] [PubMed]
- 60.Talley, R. S., and C. L. Baugh. 1975. Effects of bicarbonate on growth of Neisseria gonorrhoeae: replacement of gaseous CO2 atmosphere. Appl. Microbiol. 29:469-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 63.Tripp, B. C., K. S. Smith, and J. G. Ferry. 2001. Carbonic anhydrases: new insights for an ancient enzyme. J. Biol. Chem. 276:48615-48618. [DOI] [PubMed] [Google Scholar]
- 64.Vivian, A., and H. P. Charles. 1970. The occurrence and genetics of some CO2 mutants in Streptomyces coelicolor. J. Gen. Microbiol. 61:263-271. [DOI] [PubMed] [Google Scholar]
- 65.Windhövel, U., and B. Bowien. 1990. On the operon structure of the cfx gene clusters in Alcaligenes eutrophus. Arch. Microbiol. 154:85-91. [DOI] [PubMed] [Google Scholar]
- 66.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 67.Yu, J. W., G. D. Price, L. Song, and M. R. Badger. 1992. Isolation of a putative carboxysomal carbonic anhydrase gene from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 100:794-800. [DOI] [PMC free article] [PubMed] [Google Scholar]