Abstract
The bacterial sigma factor RpoS is strongly induced under a variety of stress conditions and during growth into stationary phase. Here, we used rpoS-lac fusions in Escherichia coli to investigate control acting at the level of RpoS synthesis, which is especially evident when cells approach stationary phase in rich medium. Previous work has shown that the small molecule ppGpp is required for normal levels of RpoS in stationary phase. Despite the attraction of a model in which the ppGpp level controls stationary-phase induction of RpoS, careful measurement of rpoS-lac expression in a mutant lacking ppGpp showed similar effects during both exponential growth and stationary phase; the main effect of ppGpp was on basal expression. In addition, a modest regulatory defect was associated with the mutant lacking ppGpp, delaying the time at which full expression was achieved by 2 to 3 h. Deletion analysis showed that the defect in basal expression was distributed over several sequence elements, while the regulatory defect mapped to the region upstream of the rpoS ribosome-binding site (RBS) that contains a cis-acting antisense element. A number of other genes that have been suggested as regulators of rpoS were tested, including dksA, dsrA, barA, ppkx, and hfq. With the exception of the dksA mutant, which had a modest defect in Luria-Bertani medium, none of these mutants was defective for rpoS stationary-phase induction. Even a short rpoS segment starting at 24 nucleotides upstream of the AUG initiation codon was sufficient to confer substantial stationary-phase regulation, which was mainly posttranscriptional. The effect of RBS-proximal sequence was independent of all known trans-acting factors, including ppGpp.
The rpoS gene encodes a sigma factor, σS or RpoS, which is required for expression of a large number of genes in response to various stresses, including nutrient limitation and osmotic challenge and during growth into stationary phase (see references 15 and 21 for reviews). The rpoS gene has been found in a variety of gram-negative bacteria, and its function and regulation have been studied extensively in the enteric species Escherichia coli and Salmonella enterica serovar Typhimurium (here referred to as S. enterica). RpoS is also a virulence factor for S. enterica (13), and its expression is induced when these bacteria enter mammalian host cells (9). It is not clear how information about stress, nutrient limitation, and host environment is used to control RpoS. Increased RpoS abundance has been reported to be regulated at many levels, including transcription initiation and elongation (17, 18, 30, 34), translation (19, 22, 24), and protein stability (19, 31, 35). RpoS protein activity is also regulated (32). No in vitro system that mimics any aspect of in vivo control of RpoS synthesis has been described.
Genetic analysis has led to the idea that some, perhaps most, regulation of RpoS synthesis occurs at the posttranscriptional level via an inhibitory mRNA secondary structure (7, 20, 23). An upstream antisense element has been localized through computer analysis of RNA folding and identification of compensatory mutations (7; our unpublished data); the antisense element can pair with the ribosome-binding site (RBS) region and inhibit rpoS translation. This proposed RNA structure is not yet supported by physical evidence. It is, however, strongly supported by genetic analysis of the dsrA RNA, a small untranslated RNA which acts as an anti-antisense RNA, increasing rpoS expression (23). dsrA RNA is important for expression of rpoS in E. coli at growth temperatures at or below 30°C (33, 39) but is not required in S. enterica (unpublished data). It is not yet clear whether the antisense element functions in other regulatory inputs to RpoS.
Mutations in more than 20 genes have been identified as affecting RpoS synthesis alone. Many of these “regulators” exhibit highly pleiotropic phenotypes, and it seems unlikely that most act directly on rpoS expression. Often, such mutations cause changes in the shape of the growth curve even in rich medium. Thus, their effects on RpoS may be a secondary consequence of altered growth rates and early or prolonged entry into stationary phase. There are clearly strong selective forces both for RpoS activity (in early stationary phase) and against it (in both the late stationary and exponential phases). Given these forces, it is more than a formal possibility that uncharacterized strain differences may influence the observed regulation. Known examples include the wild-type S. enterica strain LT2, which is defective in the RpoS protein turnover mechanism (3, 11), and the widely used E. coli strain MC4100, which is a relA mutant and is often used despite the reported role of ppGpp in RpoS regulation (14). Thus, even more than for most regulatory systems, the results observed may depend on which strain was used and how the cells were grown.
Here, we investigate the induction of RpoS that occurs in the wild-type E. coli strain MG1655 as cells are grown to stationary phase in Luria-Bertani (LB) medium, usually at 37°C. This medium was chosen because the induction ratio (stationary-phase expression to exponential-phase expression) is particularly high under these conditions, ca. 35-fold as measured with an rpoS-lac protein fusion. Previous work showed that RpoS abundance is greatly reduced in an MG1655 ΔrelA ΔspoT mutant, which lacks ppGpp (14). (This genetic background is referred to below as ppGppo for convenience.) Artificially increasing ppGpp levels by synthesis of a truncated RelA protein also leads to a very large and rapid increase in RpoS abundance, while substantially increased RpoS abundance can also be observed in certain spoT mutants that have modestly elevated ppGpp levels (14).
Another study concluded that the main effect of ppGpp is on transcription elongation across the rpoS leader (17). This conclusion was based on apparently normal stationary-phase and ppGpp regulation of plasmid-borne rpoS-lac fusions (to codon 23 of rpoS) which had been deleted for the known rpoS promoters. However, the source of this low-level residual transcription was not identified. There is also an apparent conflict between this conclusion and experiments with ppGpp overproduction (8), which found that rpoS mRNA abundance is not elevated by ppGpp overproduction during exponential phase, pointing to translation-level control of rpoS by ppGpp.
To further investigate these questions, we employed a set of lacUV5 promoter substitution and deletion derivatives of rpoS-lac, which allow sensitive, quantitative measurement of rpoS expression in LB medium in different mutant backgrounds.
MATERIALS AND METHODS
Bacterial strains and construction.
Strains used in this study (Table 1) for physiological experiments are derived from the wild-type E. coli K-12 strain MG1655. The parental strain was CF7968, which is MG1655 that has been corrected to rph+ (16) and deleted for lacIZ, obtained from M. Cashel. This lac deletion extends between the MluI sites in lacI and lacZ and was constructed by D. Vinella. Many of the lac fusions used in this work have been described previously (6, 7, 10). These fusions are placed in the E. coli trp operon, as described previously (12). Phage P1 vir was used for transduction; P1 growth and transduction were carried out by standard methods (37).
TABLE 1.
Bacterial strains
| Strain | Genotype | Previous namea |
|---|---|---|
| CF1693 | ΔrelA251::kan ΔspoT207::cat | |
| CF3032 | argA::Tn10 ΔrelA252::kan | |
| CF7968 | MG1655 Δ(lacIZ) rph+ | |
| DDS724 | MC4100 cpsB-lac ΔdsrA5 zed::Tn10d-Tet | |
| DY330 | W3110 lacU169 [λ cI857ts Δ(cro-bioA)] | |
| TE8184 | CF7968 trpDC700::putPA1303::Kanr-rpoS-lac [pr] | A |
| TE8197 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-rpoS-lac [pr] | A |
| TE8199 | TE8197 ΔrelA252::kan ΔspoT207::cat | |
| TE8222 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-rpoS-lac [op] | B |
| TE8224 | TE8222 ΔrelA252::kan ΔspoT207::cat | |
| TE8226 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] | K |
| TE8228 | TE8226 ΔrelA252::kan ΔspoT207::cat | |
| TE8230 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [op] | N |
| TE8232 | TE8230 ΔrelA252::kan ΔspoT207::cat | |
| TE8260 | TE8184 ΔbarA::tet | |
| TE8263 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-lac [op] | O |
| TE8265 | TE8230 ΔrelA252::kan ΔspoT207::cat | |
| TE8378 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (+1) | |
| TE8383 | TE8378 ΔrelA252::kan ΔspoT207::cat | |
| TE8380 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-rpoS-lac [pr] (codon 8) | E |
| TE8382 | TE8380 ΔrelA252::kan ΔspoT207::cat | |
| TE8267 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ1) | L |
| TE8269 | TE8267 ΔrelA252::kan ΔspoT207::cat | |
| TE8271 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ2) | M |
| TE8273 | TE8271 ΔrelA252::kan ΔspoT207::cat | |
| TE8344 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ3) | |
| TE8340 | TE8344 ΔrelA252::kan ΔspoT207::cat | |
| TE8345 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ4) | |
| TE8341 | TE8345 ΔrelA252::kan ΔspoT207::cat | |
| TE8275 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-rpoS-lac [pr] (C469G) | |
| TE8277 | TE8275 ΔrelA252::kan ΔspoT207::cat | |
| TE8279 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-rpoS-lac [pr] (G549C) | |
| TE8281 | TE8279 ΔrelA252::kan ΔspoT207::cat | |
| TE8283 | CF7968 argA::Tn10 trpDC700::putPA1303::Kanr-rpoS-lac [pr] (C469G/G549C) | |
| TE8285 | TE8283 ΔrelA252::kan ΔspoT207::cat | |
| TE8266 | CF7968 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ1) | |
| TE8316 | CF7968 trpDC700::putPA1303::Kanr-rpoS-lac [pr] dsrA+zed::Tn10d-Tet | |
| TE8317 | CF7968 trpDC700::putPA1303::Kanr-rpoS-lac [pr] ΔdsrA5 zed::Tn10d-Tet | |
| TE8318 | TE8316 ΔrelA252::kan ΔspoT207::cat | |
| TE8319 | TE8317 ΔrelA252::kan ΔspoT207::cat | |
| TE8372 | TE8266 ΔdksA::tet | |
| TE8270 | CF7968 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ2) | |
| TE8373 | TE8270 ΔdksA::tet | |
| TE8314 | CF7968 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ3) | |
| TE8374 | TE8314 ΔdksA::tet | |
| TE8315 | CF7968 trpDC700::putPA1303::Kanr-lacUV5p-rpoS-lac [pr] (Δ4) | |
| TE8375 | TE8315 ΔdksA::tet | |
| TE8363 | CF7968 trpDC700::putPA1303::bla-rpoS-lac [pr] | |
| TE8377 | TE8363 hfq-1::Ω-Km | |
| TE8387 | CF7968 trpDC700::putPA1303::bla-lacUV5p-rpoS-lac [pr] (Δ1) | |
| TE8388 | TE8387 hfq-1::Ω-Km | |
| TE8391 | CF7968 trpDC700::putPA1303::bla-lacUV5p-rpoS-lac [pr] (Δ2) | |
| TE8392 | TE8391 hfq-1::Ω-Km | |
| TE8405 | TE8184 Δppkx::tet | |
| TE8419 | CF7968 trpDC700::putPA1303::tet-lacUV5p-rpoS-lac [pr] (+1, codon 8) | |
| TE8420 | CF7968 trpDC700::putPA1303::tet-lacUV5p-rpoS-lac [pr] (541, codon 8) | |
| TE8421 | CF7968 trpDC700::putPA1303::tet-lacUV5p-rpoS-lac [op] (+1, codon 8) | |
| TE8422 | CF7968 trpDC700::putPA1303::tet-lacUV5p-rpoS-lac [op] (541, codon 8) | |
| TE8448 | CF7968 trpDC700::putPA1303::tet-lacUV5p-lac RBS-rpoS-lac [pr] (565, codon 8) |
Some constructs were reported previously and given alphabetical designations (10). These include an rpoS-lac [pr] (protein) fusion expressed from the native rpoS promoters (fusion A); an rpoS-lac [op] (operon) fusion expressed from the native rpoS promoters (fusion B); an rpoS-lac [pr] fusion expressed from the lacUV5 promoter at the KpnI site in nlpD (fusion K); an rpoS-lac [op] fusion expressed from the lacUV5 promoter at the KpnI site in nlpD (construct N); an rpoS-lac [pr] fusion expressed from the native rpoS promoters, with rpoS codon 8 fused to lac (fusion E); and a lac [op] fusion expressed from the lacUV5 promoter (fusion O). The rpoS-lac [pr] fusion expressed from the lacUV5 promoter placed near +1 and carried by strain TE8378 is described in Materials and Methods.
Media and growth conditions.
Bacteria were grown at 37°C (with one exception as noted below) in LB medium (37) and on nutrient agar plates containing 5 g of NaCl per liter, except where indicated. Minimal agar was prepared with NCE medium containing 0.2% glucose (4). Antibiotics were added to final concentrations in selective plates as follows: 20 μg of tetracycline hydrochloride/ml (10 μg/ml for minimal medium), 20 μg of chloramphenicol/ml, 50 μg of kanamycin sulfate/ml (except for hfq crosses, as noted below), and 30 μg of sodium ampicillin/ml (100 μg/ml when selecting for plasmids).
New mutations affecting putative trans-acting factors.
New insertion mutations were made by the method of Yu et al. (43) employing host strain DY330 (E. coli W3110 ΔlacU169 gal490 [λcI857ts Δ(cro-bioA)]). Primers containing 20 nucleotides (nt) of tet homology at the 3′ end were used to amplify the tetAR genes from plasmid pWM7 (25).
PCR was performed with Taq polymerase (Qiagen), as suggested by the manufacturer. Amplified DNA was purified by a QIAquik PCR purification kit (Qiagen); residual template DNA was then removed by digestion with DpnI, which cuts specifically at methylated GATC sites, followed by repurification of the PCR product and elution in a volume equal to that of the original PCR. Heat induction and transformation of DY330 were done as described previously (43) with 5 μl of DNA; transformants were selected at 30°C.
Primers for tet amplification were as follows (tet homology regions are italic): dksA, ATGCAAGAAGGGCAAAACCGTAAAACATCGTCCCTGAGTATTCTCGCCATCTCTTGGGTTATCAAGAGGG and TTAGCCAGCCATCTGTTTTTCGCGAATTTCAGCCAGCGTTTTGCAGTCGAACTCGACATCTTGGTTACCG; barA, CTTTCTCAATTTAACAGTGTGACCTTAATTGTCCCATAACGCTCTTGGGTTATCAAGAGGG and CCAGCGTCATAAAAAGCCGATTGCTACTCGACAAGACATCCATTAACTCGACATCTTGGTTACCG; and ppkx, GGTCAGGAAAAGCTATACATCGAAAAAGAGCTCAGTTGGTCTCTTGGGTTATCAAGAGGG and TCGTCGGCCCGCAAAGTATTAAGCGGCGATTTCTGGTGTAACTCGACATCTTGGTTACCG.
The resulting deletion-insertion mutations result in loss of target gene sequence as follows: dksA, codons 18 through 136; barA, 7 bp upstream of codon 1 through the termination codon; and ppkx, codon 15 of ppk through codon 507 of ppx. Strains were checked for the insertion or deletion by PCR with flanking primers. The sequences of these primers are available on request. Only a small number of candidate insertions were checked for each gene knockout experiment; in every case, a PCR product of the predicted size was observed.
The same general method was used to substitute the bla (Ampr) gene for Kanr of certain lac fusions. The primers used have bla homology at their 3′ ends (italic), and the template was pBR322: TCATGAACAATAAAACTGTCTGCTTACATAAACAGTAATATGAAGACGAAAGGGCCTCGTGATAC and GCGTAATGCTCTGCCAGTGTTACAACCAATTAACCAATTCTTACCAATGCTTAATCAGTGAGGCAC.
In contrast to the transformations used to construct tet insertions, for the bla substitutions it was found that only a minority of Ampr transformants had lost the Kanr marker, as predicted for events of the desired type (the frequency of the correct event ranged from 1 to 25%). However, the Ampr marker of the desired class (Kans) showed 100% linkage to Lac+ upon backcross.
Other mutations affecting trans-acting factors.
The relA deletion used was from strain CF3032 (ΔrelA252::kan argA::Tn10) (26). Since most of the lac fusions used here are marked with Kanr, the ΔrelA marker was introduced by cotransduction with argA::Tn10, selecting Tetr. Transductants were screened for the Rel− phenotype by testing sensitivity to SMGL (serine, methionine, glycine, and leucine) on minimal glucose plates with tetracycline. When comparing wild-type cells with ppGppo (ΔrelA ΔspoT) strains, the relA+ spoT+ control strains also carried argA::Tn10 (with one exception noted below). The spoT deletion used was from strain CF1693 (ΔrelA251::kan ΔspoT207::cat) (42). The dsrA deletion used was from strain DDS724 (ΔdsrA5 with linked Tn10), obtained from D. Sledjeski. This deletion is described in reference 39. The dsrA deletion was introduced by cotransduction with the linked Tn10. For comparison with ΔdsrA strains, wild-type dsrA+ strains also carried this linked Tn10. To construct strains for the epistasis test of dsrA and relA spoT, the ΔrelA252::kan marker from CF3032 was introduced by linkage to argA::Tn10, and then the Tn10 was removed by subsequent transduction to Arg+.
The hfq insertion used was from strain TX2822 (hfq-1::Ω-Km), obtained from M. Winkler (40). This insertion is at codon 41 of the 102-codon hfq gene. We encountered difficulty with Kanr to select for transfer of hfq-1::Ω-Km. In fact, all the parental strains carrying hfq-1::Ω-Km that we obtained grew very poorly when streaked out on either LB or NB agar containing 50 μg of kanamycin/ml, showing a typical pattern of variably sized single colonies in the primary streak at a position where dense growth would normally be expected (suggesting suppression). This behavior is not understood. Growth was normal in the absence of kanamycin. To construct the needed strains, selection for transductants carrying hfq-1::Ω-Km was carried out on LB agar with kanamycin at 25 μg/ml at room temperature, and transductants were then purified on LB agar without kanamycin at 37°C. Successful introduction of the hfq insertion (and all other deletions) was confirmed by PCR.
rpoS-lac fusions.
We have previously described the detailed methods used to make the rpoS-lac constructs employed for most of the experiments in the present work (6, 10). They use the general system originally designed by Simons et al. (38), as modified (12). The relevant gene segments include (in order) an upstream Kanr element, tandem transcriptional terminators, and the promoter or regulatory sequence under investigation, followed by the lac operon. This assembly is placed in single copy in the bacterial chromosome (at trp); therefore, all strains also carry a wild-type copy of rpoS. Most lac fusions used in this study have lacZ placed to form either an operon or a protein fusion at the EagI site at codon 73 of rpoS. A different set of fusions, to codon 8 of rpoS, was used for the last set of experiments as described below.
Some constructs carried the native rpoS promoters (6). In the others, including the KpnI construct as well as the numbered deletions, rpoS-lac is expressed from lacUV5p with a constant lac-derived leader of 36 nt plus several restriction sites, followed by different amounts of rpoS sequence; these constructs vary only in the extent of the deletion that removes rpoS material from the upstream side. The lacUV5 promoter is derived from pRS476 (38); it includes only one of the cyclic AMP receptor protein (CRP) half-sites on the upstream side and the lac operator on the downstream side.
The reference KpnI site construct (construct K) and the Δ1 and Δ2 constructs have been described previously (10). The Δ3 and Δ4 deletions were made by PCR in exactly the same way as Δ1 and Δ2; the PCR-amplified segments were resequenced to ensure that no unwanted changes had been introduced. The deletion endpoints for Δ2 through Δ4 are illustrated with respect to the sequence in Fig. 1. All deletion endpoints are numbered starting from the first transcribed nucleotide for transcripts initiated from the rpoS promoter. We have taken this transcript sequence to begin with GGGUGAACAG (the first G is nt 1) (17). The coordinates of the first base pair that is still present in each construct are as follows: construct K, nt 73; Δ1, nt 344; Δ2, nt 454; Δ3, nt 477; and Δ4, nt 541. The rpoS ATG initiation codon is at nt 565.
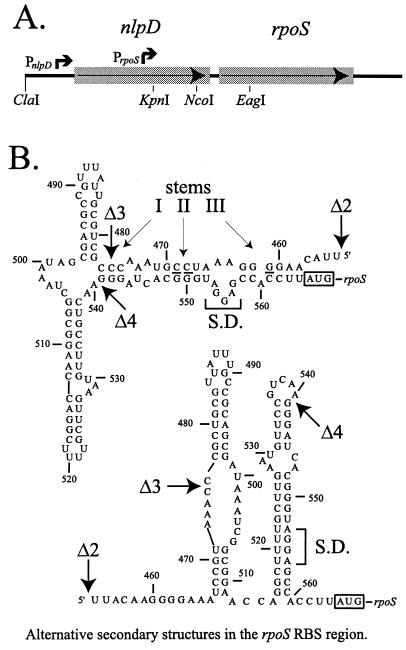
FIG. 1.
Map and sequence of the rpoS region. (A) Partial restriction map of the DNA encompassing the nlpD and rpoS open reading frames of E. coli, which consist of 321 and 330 codons, respectively. The function of nlpD (encoding a lipoprotein) is not related to that of rpoS. Arrows indicate the orientation of the two open reading frames and the known promoters. The upstream promoter cluster (PnlpD), which is thought not to be regulated, serves both nlpD and rpoS. The downstream promoter (PrpoS), is regulated and serves only rpoS; its transcript includes an untranslated leader of 564 nt. The first nucleotide of the transcript from PrpoS (as specified in the text) is taken as the basis for numbering used here (nt 1). The numbers used previously by us (7, 10) can be converted to this system by adding an offset of 341 nt. Most lac fusions used in this study have lacZ placed to form either an operon or protein fusion at the EagI site within rpoS (codon 73). Other fusions, to codon 8 of rpoS, are so indicated in the text. (B) Two possible secondary structures for RNA including the end of the nlpD coding sequence and the short intergenic region up to the AUG start codon of rpoS. The UAA stop codon terminating nlpD lies at nt 500 to 502. The top structure is that proposed previously by us (7) and includes an upstream antisense element with three stems that can pair with a complementary sequence within the RBS, directly upstream of the rpoS AUG start codon. The Shine-Dalgarno (S.D.) sequence complementary to 16S rRNA is also indicated. The lacUV5 promoter was used to drive expression of various constructs of two general types, as detailed in the text. The end points of sequence derived from nlpD-rpoS included in these fusions can be described with reference to this figure as follows. Fusions for which the promoter is shown as rpoS include DNA starting from the ClaI site (A). Other fusions contain substitutions of the lacUV5 promoter followed by DNA starting from nt 1 of the PrpoS transcript, the KpnI site, Δ1 (nt 344), Δ2 (nt 454), Δ3 (nt 477), or Δ4 (nt 541). For Δ2 through Δ4, included leader DNA sequences are shown in panel B.
Another construct in which lacUV5p drives rpoS expression starting from “+1” of rpoS (nt 1) was constructed by PCR on an rpoS-lac template with a lac-specific oligonucleotide together with the following oligonucleotide (the lacUV5 mutation in the promoter's −10 region is shown in bold; rpoS homology is shown by italics): CGCGAATTCAGGCTTTACACTTTATGCTTCCGGCTCGTATAATGTGTGGAATTGGGTGAACAGAGTGCTAACAAAATG.
Transcripts originating from lacUV5p in this construct are predicted to contain the 5′ sequence AAUUGGGUGAACAGAGTGCTAACAAAATG, where the underlined nucleotides are derived from rpoS sequence. This construct does not include the lac operator. The PCR product was substituted as an EcoRI-KpnI fragment in several steps, and the substituted segment was sequenced to make sure that no unwanted mutations had been introduced by the PCR step. The strain with this fusion carried in the bacterial chromosome was named TE8378.
Second method for making promoter fusions.
We subsequently developed a convenient method for making constructs in which rpoS (or any gene) can be expressed from lacUV5p by employing the lambda lysogenic strain background and technique of Yu et al. (43). The general transformation method is the same as described in the section above on tet insertions.
For this purpose, we first placed tetAR upstream of lacUV5p, replacing Kanr (38) in the standard Δ1 fusion by PCR with the following two oligonucleotides: ATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGAATTCACTCGACATCTTGGTTACCG (tetR homology in italic, lacUVp homology elsewhere) and TCATGAACAATAAAACTGTCTGCTTACATAAACAGTAATACTCTTGGGTTATCAAGAGGG (tetA homology in italic, kan homology elsewhere).
In a second step, the marked lacUV5 promoter was joined to each of the desired target sites by PCR amplifying the tetAR-lacUV5p segment with an oligonucleotide which included appropriate rpoS homology attached 5′ to the lacUV5p-specific sequence: AATTCCACACATTATACGAG. As in the first step, on the upstream side the oligonucleotide for PCR was chosen so that the tetAR-lacUV5p substitution will replace the kan gene of the standard fusion. The resulting constructs are marked with Tetr, join lacUV5p directly to the desired target sequence, and do not include the lac operator. With this method, we made a new set of deletions extending to various positions directly upstream of the rpoS ATG initiation codon as described above in the text. Depending on the strain used for lambda red-mediated transformation, the resulting constructs are lac [op] or [pr] fusions at codon 8 of rpoS. The full lacUV5p sequence is GAATTCTCACTCATTAGGCACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATAATGTGTGGAATT.
Constructs made by this method were confirmed genetically as Kans and physically by PCR with primers specific to tetR and lac, followed by DNA sequencing across the lacUV5 promoter and the first 200 to 300 nt at the joint to rpoS. After verification, the fusion constructs were transduced into the CF7968 background by selecting for Tetr.
The lac fusions marked with Kanr, as designed by Simons et al. (38), carried tandem insertions of a terminator between the drug resistance cassette and the site where test segments are joined to lac; terminators were not explicitly included in the fusions marked with Tetr made by the new method. This was considered unlikely to be necessary because of the weak activity of the tetR promoter and its more than 20-fold dependence on tetracycline for induction. Indeed, assays of constructs grown in the presence and absence of tetracycline showed that induced transcription from tetR accounted for ≈10% of lac transcription from exponential-phase cells; the contribution from uninduced transcription (i.e., our standard growth conditions) is therefore negligible.
Assay of β-galactosidase.
Cells were centrifuged and resuspended in Z-buffer (100 mM NaPO4 [pH 7.0], 10 mM KCl, 1 mM MgSO4) and then permeabilized by treatment with sodium dodecyl sulfate (SDS) and chloroform (27). The samples from exponential-phase time points were concentrated before assay to be approximately equal in density to samples from later times. Assays were performed in Z-buffer containing 50 mM β-mercaptoethanol by a kinetic method with a plate reader (Molecular Dynamics). Activities (change in optical density at 420 nm [OD420] per minute) were normalized to actual cell density (OD650) and were always compared to appropriate controls assayed at the same time. All β-galactosidase assays were performed within 3 h of the time of sampling. The values shown are averages of at least three experiments with standard deviations of less than 20% except for the very low values from exponential phase for the ppGppo mutant, for which the standard deviations were less than 30%. We should point out that comparisons of the relative defect in stationary-phase induction involve ratios of experimental values, with a corresponding increase in the uncertainty.
RESULTS
Expression of rpoS-lac along the growth curve.
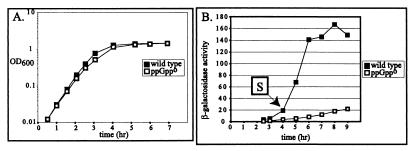
Cells for the β-galactosidase assay were grown at 37°C (with one exception noted below) in LB medium. Cultures were started by a 1:500 dilution, starting from overnight cultures grown under the same conditions. In LB medium at 37°C, the generation time of the wild-type (relA+ spoT+) strain was 24 min, while the generation time of the otherwise isogenic ppGppo mutant (ΔrelA ΔspoT) was 33 min (Fig. 2A). For each culture, the time at which stationary phase begins (designated S below and in tables) was arbitrarily defined as 1 h after the time at which the OD600 reached 0.5. This definition compensates for the slower growth rate of the ppGppo mutant; it also allows the times for stationary-phase sampling to be fixed while the culture is still in exponential phase. The point on the growth curve at S is very close to the inflection point between the lines for the exponential and stationary phases.
FIG. 2.
Expression of rpoS-lac [pr] as a function of growth phase. (A) Growth curve of the wild-type strain carrying rpoS-lac [pr] (TE8197, solid squares) and its ppGppo mutant derivative (TE8199, open squares) in LB medium at 37°C. As described in the text, stationary phase (S) was defined as 1 h past the time at which the OD600 reached 0.5. In this experiment, for the wild-type strain, S = 222 min, and for the ppGppo strain, S = 234 min. (B) Cultures were grown as in panel A and sampled at various times for assay of β-galactosidase. The first point for each curve corresponds to the time at which the OD600 was 0.25. The x axes of the plots have been shifted slightly to align the points corresponding to an OD600 of 0.5 at the 3-h mark. Each subsequent sample was taken at 1-h intervals.
In a preliminary experiment, we found that wild-type cells carrying the rpoS-lac [pr] fusion, taken at densities between an OD600 of 0.01 and 0.25, showed nearly the same β-galactosidase activity (data not shown). To maximize recovery, an OD600 of 0.25 was chosen as the reference density for exponential phase, before the increase characterizing the transition into stationary phase. Plots of β-galactosidase activity determined as cells achieved stationary phase are shown in Fig. 2B. The times shown in this panel are slightly different from those shown in panel A: the x axis of the plot of the ppGppo mutant has been shifted to align it with the wild-type data at an OD600 of 0.5 (as well as S and subsequent points).
Our data are consistent with results reported in previous studies (14, 17). The wild-type strain with rpoS-lac [pr] showed low but significant activity during exponential phase (Table 2), which then rapidly increased during the approach to and in early stationary phase. Maximum activity was achieved by S + 2 h and was not increased by overnight growth (data not shown). The overall induction ratio was 35- to 40-fold. In contrast to wild-type cells, the ppGppo mutant showed much lower expression of rpoS-lac at all times. Expression in the mutant was about one-sixth that of the wild type even during exponential phase (Table 2), while in late stationary phase (S + 5), the values were 150 U for the wild type and 22 U for the mutant. Thus, the ratio of activity in the mutant compared to that in the wild type is approximately the same during exponential phase and late stationary phase. It is only during early stationary phase that the ppGppo mutant seems to be delayed in comparison to the increase seen in rpoS-lac expression for the wild type (Table 2). For the mutant, there was a gradual threefold increase in β-galactosidase activity at late times, between S + 2 h and S + 5 h (Fig. 2B).
TABLE 2.
Effect of ppGpp on rpoS-lac and lac expression in vivoa
|
lac fusion
|
Fusion type | β-Galactosidase activity (U)
|
Activity ratio, mutant/wild type
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
Mutant (ppGpp°)
|
||||||||||
| Promoter | Reporter | E | S + 3 | S + 3/E | E | S + 3 | S + 3/E | E | S + 3 | S + 3/E | |
| rpoS | rpoS-lac | Protein | 3.4 | 120 | 35 | 0.54 | 5.1 | 9.4 | 0.16 | 0.04 | 0.27 |
| rpoS | rpoS-lac | Operon | 35 | 510 | 15 | 11 | 50 | 4.5 | 0.31 | 0.10 | 0.31 |
| lacUV5 | rpoS-lac | Protein | 5.3 | 71 | 13 | 2.1 | 7.6 | 3.6 | 0.40 | 0.11 | 0.27 |
| lacUV5 | rpoS-lac | Operon | 100 | 330 | 3.3 | 64 | 130 | 2.0 | 0.64 | 0.39 | 0.61 |
| lacUV5 | rpoS-lac (+1) | Protein | 10 | 130 | 13 | 3.2 | 25 | 7.8 | 0.32 | 0.19 | 0.60 |
| rpoS | rpoS-lac (codon 8)b | Protein | 4 | 130 | 32 | 0.62 | 13 | 21 | 0.15 | 0.10 | 0.64 |
| lacUV5 | lac | Operon | 120 | 160 | 1.3 | 81 | 80 | 0.99 | 0.67 | 0.50 | 0.74 |
Exponential-phase (E) and stationary-phase (S + 3) samples are defined in the text. Values are averages with a variation of <20%, with one exception noted in the text. The S + 3/E induction ratio was calculated as the activity of the S + 3 sample divided by that of the E sample.
All other fusions were to codon 73 of rpoS.
The defect in stationary-phase rpoS-lac expression seen for the ppGppo mutant was reported previously by others (17). However, we also found a quantitatively similar exponential-phase defect in rpoS-lac expression in the same mutant. We would distinguish between the defect in basal (or constitutive, nonregulated) expression and a true regulatory defect. The ppGppo mutant shows a basal defect in rpoS-lac expression of about sixfold, while the regulatory defect for stationary-phase induction seen at S + 3 h was approximately threefold. These results indicate that stationary-phase induction of rpoS is not due simply to increased ppGpp during stationary phase, because it still occurs normally even in the complete absence of ppGpp. Accumulation of RpoS in the ppGppo mutant after overnight incubation in stationary phase was seen in previous studies (14, 17). Of course, these experiments do not exclude the possibility that ppGpp might play a quantitatively larger regulatory role under other conditions.
Promoter substitution.
Expression of rpoS-lac was measured from constructs in which the lacUV5 promoter was substituted for the native rpoS promoters (10). The lacUV5 promoter was chosen because its activity was previously reported to be completely independent of ppGpp (1, 2); also, its activity is only slightly increased during stationary phase (see the last entry in Table 2). When expression of rpoS-lac [pr] was driven by lacUV5p, a strong stationary-phase induction still occurred (induction ratio of 13-fold; Table 2). The stationary-phase induction was smaller by a factor of 3 than that seen with the native promoters, suggesting that the native promoters contribute to stationary-phase induction to this extent.
We also compared the effect of stationary phase on expression of rpoS-lac protein and operon fusions driven by either native or lacUV5 promoters. The combined results support the idea that stationary phase controls multiple stages in rpoS expression. (i) A role for translational regulation is suggested by the larger induction ratio when [pr] fusions are compared with [op] fusions, whether driven by the native promoters or by lacUV5p; (ii) a role for the native rpoS promoters is suggested by the smaller induction ratio when lacUV5p is substituted in either [op] or [pr] fusion contexts; and (iii) a role for postinitiation transcriptional control is suggested by the residual stationary-phase induction seen in the lacUV5p-rpoS-lac [op] construct.
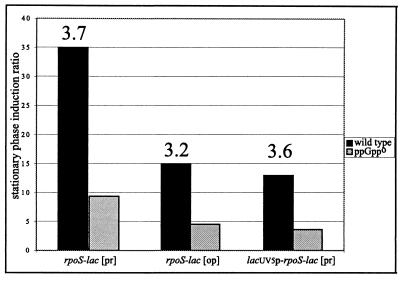
Expression of rpoS-lac [pr] from the lacUV5 promoter was also decreased in the ppGppo mutant background (Table 2). The time course for both mutant and wild-type (data not shown) cells was similar to that seen when the construct was driven by the native promoters (Fig. 2B). Expression was lower for the mutant at all times but with a gradual increase in expression in the mutant until S + 5 h. To detect the regulatory defect of the mutant, we chose S + 3 h as the time of sampling for stationary phase (Table 2). Selected stationary-phase induction ratios shown in Table 2 are plotted in Fig. 3. It can be seen that the regulatory defect of the ppGppo mutant is similar for rpoS-lac protein and operon fusions as well as for protein fusions whether driven by the native promoters or lacUV5p. This suggests that neither the promoter nor translation-level control is a significant target for the regulatory effect of ppGpp.
FIG. 3.
Nature of the ppGppo regulatory defect. Various rpoS-lac fusion strains were grown and assayed for β-galactosidase as reported in Table 2. The stationary-phase induction ratio (expression at S + 3 divided by that at E1) is plotted in bar format for both mutant and ppGppo versions of each fusion. The ratio (value in the wild type divided by that in the ppGppo mutant) of the stationary-phase induction ratios is shown above the bar for each wild-type strain. The strains used are listed in Table 1.
In contrast to its regulatory effects, the defect in basal expression of the ppGppo mutant seems to be distributed over several elements. For example, whether comparing exponential- or stationary-phase values, the biggest defect was shown by rpoS-lac [pr] driven by the native promoters. The relative defect was less for either a lacUV5 promoter substitution or an operon fusion. The former result is consistent with direct measurement of rpoS mRNA 5′ ends (17). We also found that lacUV5 promoter activity was somewhat lower in a ppGppo strain, by a factor of 2 in stationary phase (Table 2). This effect of ppGpp in the control, while small, makes the significance of other small effects uncertain (in particular, the effect of ppGpp in the lacUV5p-rpoS-lac [op] construct).
The reference lacUV5p constructs were made with a convenient KpnI site (10); this strategy removed 72 nt of rpoS leader sequence. Therefore, an additional lacUV5p construct which included all nucleotides of the native transcript (denoted +1) was made; it is regulated identically to the reference construct in stationary phase, although it seems to be somewhat less sensitive to ppGpp during stationary phase. The significance of this difference is not understood at present. Finally, one fusion in which lac is joined to rpoS at codon 8 (rather than to codon 73, as in all other rpoS fusions used to this point) was tested. The codon 8 fusion (driven by the native rpoS promoters) was regulated normally by stationary phase and was highly responsive to ppGpp.
Deletion mutants.
Deletion analysis was used to define the region(s) of the rpoS leader that is required for stationary-phase induction and the response to wild-type ppGpp levels (Table 3). Cell samples from wild-type and ppGppo backgrounds were collected in both exponential and stationary phase, and stationary-phase induction ratios were calculated. For each construct, the promoter was lacUV5p. All constructs have a common transcribed leader of 60 nt (partially derived from lac); the only difference between constructs is the amount of rpoS sequence that is retained. The first entry in Table 3 shows the reference construct (substitution at the KpnI site, construct K from reference 10). The deletions (Δ1 through Δ4) extend progressively closer to the rpoS ATG initiation codon, as shown in Fig. 1. The Δ4 construct retains only 24 nt of rpoS sequence upstream of the rpoS ATG initiation codon.
TABLE 3.
Role of the antisense element in stationary-phase induction and ppGpp effects on rpoS-laca
|
lac fusion
|
β-Galactosidase activity (U)
|
Activity ratio, mutant/wild type
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
Mutant (ppGpp°)
|
|||||||||
| Promoter | Reporter | E | S + 3 | S + 3/E | E | S + 3 | S + 3/E | E | S + 3 | S + 3/E |
| lacUV5 | rpoS-lac | 5.7 | 82 | 14 | 2.1 | 6.7 | 3.2 | 0.37 | 0.08 | 0.22 |
| rpoS-lac, Δ1 | 4.7 | 65 | 14 | 1.8 | 7.5 | 4.2 | 0.38 | 0.12 | 0.30 | |
| rpoS-lac, Δ2 | 3.1 | 34 | 11 | 1.6 | 5.7 | 3.6 | 0.52 | 0.17 | 0.32 | |
| rpoS-lac, Δ3 | 13 | 73 | 5.6 | 8.4 | 62 | 7.4 | 0.65 | 0.85 | 1.3 | |
| rpoS-lac, Δ4 | 8.3 | 40 | 4.8 | 5.2 | 30 | 5.8 | 0.63 | 0.75 | 1.2 | |
| rpoS | rpoS-lac | 3.1 | 120 | 39 | 0.66 | 8.8 | 13 | 0.21 | 0.07 | 0.34 |
| rpoS-lac C469G | 7.7 | 210 | 27 | 1.4 | 47 | 34 | 0.18 | 0.22 | 1.23 | |
| rpoS-lac G549C | 14 | 360 | 26 | 3.3 | 29 | 8.8 | 0.24 | 0.08 | 0.34 | |
| rpoS-lac C469G G549C | 6.5 | 170 | 26 | 1.4 | 7.9 | 5.6 | 0.22 | 0.05 | 0.22 | |
All fusions listed in this table are protein fusions. See Table 2, footnote a, for definitions.
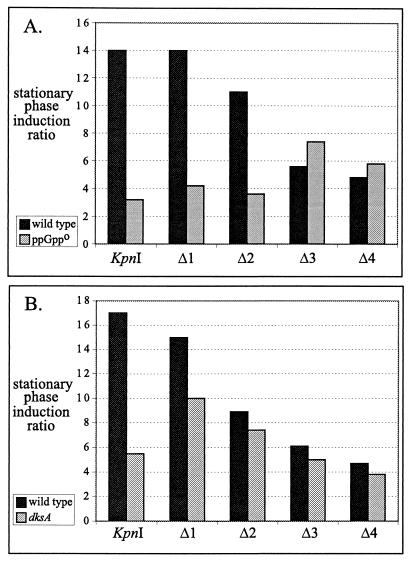
There was no significant difference in either induction ratio or ppGpp response between the K construct and Δ1, and there was only a slight decrease in induction ratio as the deletion was extended in Δ2. However, a clear breakpoint can be seen between Δ2 and Δ3. It is striking that the two deletions that extend farther downstream (Δ3 and Δ4) lack the ppGpp dependence shown by the reference construct and upstream deletions. Selected data from Table 3 highlighting the effect of ppGpp on the induction ratio are shown in a bar plot in Fig. 4A. The plot clearly shows that the Δ3 and Δ4 deletions retained substantial stationary-phase induction but lost the ppGpp response. The region between the Δ2 and Δ3 deletion endpoints required for this facet of the ppGpp response includes the upstream antisense element. There was also an approximately 50% decrease in the stationary-phase induction ratio between Δ2 and Δ3. However, a substantial (fivefold) stationary-phase induction was retained in both the Δ3 and Δ4 deletions despite the fact that they did not respond to ppGpp.
FIG. 4.
Deletion mapping of sequences required for ppGppo and dksA mutation effects on rpoS expression. Various rpoS-lac fusion strains, all driven by the lacUV5 promoter but with various amounts of the upstream rpoS leader, were assayed for β-galactosidase during exponential phase and stationary phase (S + 3 h). Stationary-phase induction ratios were calculated and are plotted here. The underlying data for the ppGppo strain are from Table 3. The strains used are listed in Table 1.
In a previous study, we described a number of mutations that affect the upstream antisense element or its complementary sequence within the RBS of rpoS mRNA, immediately upstream of the rpoS ATG initiation codon (7). Two of these mutations were examined here, C469G and G549C, which alter the antisense (top) and sense (bottom) components of stem II, respectively (see the legend to Fig. 1 for details). We chose these two mutations because, as tested in S. enterica, the resulting rpoS-lac expression phenotype in the double mutant is indistinguishable from that in the wild type and very different from that in each single mutant, providing strong genetic support for the proposed inhibitory mRNA secondary structure. Furthermore, mutations at adjacent positions also confer these properties (unpublished data).
Here, it is striking that the C469G mutation allowed high stationary-phase expression and restored a normal induction ratio of rpoS in the ppGppo background (Table 3). This lesion clearly relieved most of the ppGpp requirement of rpoS. Furthermore, the C469G/G549C double mutation restored ppGpp dependence, which is consistent with the model that ppGpp dependence can be counteracted by blocking interaction of the antisense element with the RBS. However, since the G549C single mutation should loosen the secondary structure as effectively as C469G, the model predicts that G549C should relieve the ppGpp requirement of rpoS, and yet this prediction was not fulfilled.
Additionally, when comparing E. coli with S. enterica, the phenotype of the compensatory double mutant was not always as clear-cut in E. coli. Particularly in a wild-type (relA+ spoT+) strain background, the C469G/G549C double mutation did not confer a fully wild-type phenotype but rather one intermediate between those of the single C469G mutant and the wild type (Table 3), giving 120 U of β-galactosidase activity in the wild type, 170 U in the double mutant, and 210 U in the C469G mutant. On the other hand, at stationary phase in the ppGppo background, the phenotype of the double mutant was more convincingly like that of the wild type: 8.8 U of β-galactosidase activity for the wild type, 7.9 U for the double mutant, and 47 U for the C469G and 29 U for the G549C single mutants. The partial suppression observed for the double mutant in a wild-type background leaves some question of whether the observed phenotypes result simply from a failure of antisense element interaction with the RBS. They might reflect an additional sequence-specific interaction. Nevertheless, it is clear that C469G eliminates a requirement for ppGpp for normal stationary-phase induction and the requirement for ppGpp is restored in the C469G/G549C double mutant.
Effect of dksA.
A dksA insertion in S. enterica was reported to decrease stationary-phase induction of an rpoS-lac protein fusion in supplemented minimal medium (41). In other work, it was found that a dksA deletion in E. coli almost completely blocks induction of rpoS by elevated ppGpp levels during exponential growth in LB medium (8). Therefore, we tested whether the effect of dksA on rpoS-lac [pr] expression exhibits the same sequence requirements as observed for the regulatory defect of ppGppo.
Expression of rpoS-lac [pr] in the ΔdksA::tet mutant was not detectably different from that in the wild type during exponential phase but was one-third to two-thirds of the wild-type level in stationary phase, measured either with the wild-type fusion or with lacUV5p constructs (data not shown). The regulatory defect of dksA, as reflected in the stationary-phase induction ratio, was larger for fusions carrying more upstream sequence (the KpnI fusion in Fig. 4B). Only a small regulatory effect of dksA was seen for Δ1, and this was nearly completely lost for Δ2 and with reporters deleted of the antisense element (Δ3 and Δ4). This pattern is similar to that for hfq, where its effect on rpoS expression requires upstream sequences (10; also see below) and differs from the requirements for ppGpp effects as described above.
We have found similar results for dksA in S. enterica in LB medium (unpublished data). A much more substantial effect for dksA can be observed in S. enterica in minimal medium supplemented with amino acids (41; unpublished data); this difference may be related to the multiple auxotrophy of dksA mutants of both E. coli and S. enterica.
Other trans-acting factors.
In addition to the above studies in the ppGppo background (ΔrelA ΔspoT) and a dksA mutation, we also examined the effects on stationary-phase induction of rpoS-lac for mutations in four other genes. The mutations tested were newly constructed insertions or deletions of barA (29) or ppkx (36) and existing mutations affecting dsrA (deletion [39]) and hfq (Ω-Km insertion [6, 28, 40]).
Previously, a barA insertion (of λplacMu53) in MC4100 was shown to reduce RpoS by Western blot and rpoS RNA by Northern blot (29). A ppk ppx mutant in the JM101 background was reported to have decreased catalase (hydroperoxidase II [HPII], under RpoS control), and plasmid-encoded yeast PPX1 polyphosphatase interfered with katE and rpoS induction, as measured by Western blot and lac operon fusions, consistent with positive regulation of rpoS by polyphosphate (36).
For the first two mutations (barA and ppkx), we found that the effects on rpoS-lac expression during growth in LB medium were small (data not shown). No significant effect of ΔbarA::tet was observed for katE-lac [op] expression in either the MC4100 or MG1655 background (<20% variation); rpoS-lac [pr] expression was either unchanged (MC4100) or increased twofold (MG1655). The Δppkx::tet strain expressed rpoS-lac [pr] at about 75% of the wild-type level in stationary phase, whereas the ppGppo derivative level was about 10% of the wild-type level in the same experiment. The Δppkx::tet mutant showed an rpoS-lac [pr] induction ratio of 26-fold, versus 34-fold for the wild type. These small effects are difficult to interpret.
The dsrA gene encodes a small, untranslated RNA which is proposed to base pair with the upstream antisense element of rpoS, thereby freeing the RBS to be engaged by ribosomes for translation. Strong genetic evidence supports this model, including the behavior of several sets of compensatory mutations (23). Sledjeski et al. showed that in wild-type cells, RpoS abundance and rpoS-lac expression increase dramatically at lower growth temperatures. One major effect of the dsrA mutation is that this low temperature response is lost (39). At 37°C, a dsrA mutation has no effect on rpoS-lac expression in either the exponential or stationary phase, whether in the MC4100 (33) or the MG1655 (our unpublished results) background. dsrA RNA is not required for stationary-phase induction under these conditions.
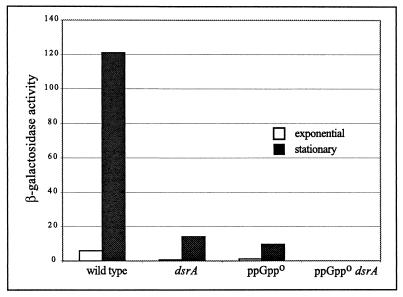
In order to see the effect of loss of dsrA, cells were grown at 30°C. At this temperature, wild-type cells with the standard fusion (construct A) showed normal stationary-phase induction (Fig. 5). Both the dsrA mutant and the ppGppo mutant showed a substantial reduction in rpoS-lac expression, but only modest effects were observed on the stationary-phase induction ratio. A mutant lacking both dsrA and ppGpp showed a more severe phenotype, in which rpoS-lac expression was completely absent. Because both the ppGppo and dsrA backgrounds have severe effects on rpoS-lac, we suggest that during growth at 30°C, most rpoS expression requires both ppGpp and dsrA RNA (whether directly or indirectly). We interpret the additive effect of the combined mutations by suggesting that the two effectors act independently, at least in part—ppGpp does not act simply by changing DsrA level or activity. However, we emphasize the finding that during growth at 37°C, dsrA function did not affect rpoS expression, and even at 30°C, dsrA mutants showed a stationary-phase induction ratio of about 23-fold.
FIG. 5.
Additive effect of ppGppo and a dsrA mutation. β-Galactosidase activities of exponential-phase (OD600 = 0.25) and stationary-phase (S + 3 h) cultures are given. The activity of the ppGppo dsrA strain was below the limit of detection (<0.2 U) under both conditions. Stationary-phase induction ratios were as follows: wild type, 35-fold; dsrA, 23-fold; and ppGppo, 20-fold. The strains used are listed in Table 1.
We also compared the effect of ΔdsrA in Δ1 through Δ4 (data not shown). The model of DsrA acting as an anti-antisense RNA predicts that mutants with deletions of the antisense element should not respond to lack of DsrA. As predicted, the ΔdsrA deletion did not affect Δ3 and Δ4.
The last trans-acting regulator investigated here was hfq. We found that, as in S. enterica (10), the Δ2 construct did not respond to loss of hfq and the Δ1 construct showed only a modest effect (data not shown). Furthermore, the stationary-phase induction ratio of an hfq mutant was very similar to that of the hfq+ control whether the fusion was rpoS-lac [pr] driven by the native promoters or the Δ1 or Δ2 constructs driven by lacUV5p (data not shown). Therefore, Hfq cannot be a mediator of stationary-phase induction. Finally, many mutations identifying “regulators” of rpoS expression have been reported to have no effect in an hfq mutant background. This set includes hns (28); stpA; galU, pgi, and pgm (5); and oxyS (44). Similarly, a leuO mutation makes no difference in the absence of dsrA (16). Since stationary-phase induction is normal in an hfq mutant and these mutations presumably affect RpoS secondarily through their effects on hfq, these factors cannot regulate stationary-phase induction.
Stationary-phase regulation localized to the rpoS RBS region.
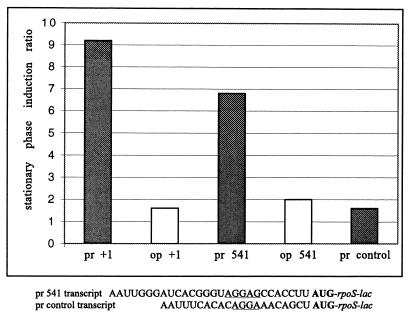
As shown in Table 3 and Fig. 4, substantial stationary-phase induction can be observed even with the Δ4 construct, driven by the lacUV5 promoter, which contains only 24 nt upstream of the rpoS ATG initiation codon. All lac fusion constructs used so far have contained rpoS coding sequences extending to codon 73. We wondered whether this stationary-phase response would be retained with lac fusions to upstream sites and whether operon fusions would show a difference from protein fusions. To make the new constructs, we used the λred recombination method (43) to position the lacUV5 promoter precisely at various sites. We chose to retain the first 4 nt of the lac transcript (AAUU) in these fusions. Otherwise identical operon and protein fusion constructs were made by simply transforming the fusion fragment into a different host strain (see Materials and Methods for details). The results (Fig. 6) show that this aspect of the stationary-phase response was seen with protein but not with operon fusions, indicating a translation-level defect, and did not require rpoS sequences downstream of codon 8. In the fusion labeled pr control in Fig. 6, the lac RBS (not including operator sequences) was substituted for the rpoS RBS.
FIG. 6.
Stationary-phase regulation requires the wild-type rpoS RBS. Various rpoS-lac fusion strains driven by the lacUV5 promoter were grown and assayed for β-galactosidase. The stationary-phase induction ratio (expression at S + 3 divided by that at E1) is plotted in bar format for protein (pr) and operon (op) versions of each fusion. Fusions marked +1 contained the entire 564-nt rpoS leader; those marked 541 contained only 24 nt upstream of the AUG start codon. The pr control transcript sequence is compared to the pr 541 transcript below the bar graph. In each fusion, codon 8 of the rpoS sequence is joined to lac. The sequence between the AUG initiation codon and the joint to lac was AUG AGU CAG AAU ACG CUG AAA GUU CAG GAT CCG (non-rpoS nucleotides underlined). The strains used are listed in Table 1.
DISCUSSION
Ever since the discovery by Gentry et al. that mutants lacking ppGpp are deficient in RpoS after growth to stationary phase in rich medium (14), a role for ppGpp in RpoS regulation has been an attractive unifying hypothesis. Thus, ppGpp control of RpoS might explain not only induction of RpoS during growth into stationary phase, but also growth rate regulation, induction by limitation for single nutrients and by other stresses such as challenge with high salt, and even induction during growth within eukaryotic host cells. Here, the sensitivity obtained by using lac as a reporter for rpoS expression allows a clear demonstration that ppGppo mutants are just as defective during exponential as in late stationary phase, and therefore, we suggest that ppGpp should be considered mainly as a basal and not a regulatory factor.
This conclusion should be qualified. First, ppGppo mutants do show a delay in the rate of increase in RpoS during stationary phase, so that relatively early (at the time we define as S + 3 h), there is a modest regulatory defect which is made up for by a slow, late accumulation. Second, it is conceivable that ppGpp is regulatory in stationary phase in wild-type cells but a redundant mechanism operates in the ppGppo mutant. Also, the delay of the mutant in achieving stationary phase and halting protein synthesis could allow more time for RpoS to accumulate or might trigger some type of compensatory increase. Finally, a regulatory role for ppGpp under other conditions is not ruled out. Preliminary experiments indicate that osmotic shock can still induce rpoS normally even in a ppGppo host (unpublished data), but ppGpp control might explain the large difference between rpoS expression in rich and minimal medium. The complex nutritional defect of ppGppo mutants makes that idea difficult to test.
Analysis of rpoS-lac operon and protein fusions, as well as promoter substitutions and deletions, should allow us to suggest one or more mechanisms of ppGpp action in this system. The total effect is robust: expresssion of rpoS-lac [pr] was ca. 25-fold higher in the wild type than in the ppGppo mutant at S + 3 h (Table 2). However, this large effect seemed to be distributed in small installments over several targets. The basal increase in expression involves (i) an effect lost when transcription is from the lacUV5 promoter (consistent with the primer extension results in reference 17), (ii) an effect seen with a protein fusion but not an operon fusion, and (iii) a target, probably transcription elongation, which is distinct from the first two. In contrast, the regulatory effect of ppGpp apparently involves the antisense element (Table 3 and Fig. 4) yet paradoxically is still visible with an operon fusion and thus presumably involves transcription elongation (Fig. 3). The distributed nature of ppGpp's targets complicates analysis by combining several small, presumably multiplicative effects.
Another area of uncertainty involves the question of whether high levels of ppGpp, achieved by inducing a truncated ribosome-independent RelA protein during exponential growth, alter rpoS expression in the same way as the effects seen here by use of a ppGppo mutant (14). Recent work shows that overproduction of ppGpp dramatically increases RpoS protein synthesis with little change in the amount of rpoS RNA (8). Our results, showing only a small role for translational control, seem inconsistent with this. One resolution is to suppose that ppGpp overproduction affects rpoS by a different mechanism. In support of this interpretation, the sequences required for dksA to affect rpoS expression in our experiments are different from those required for ppGpp regulation (Fig. 4).
The most surprising conclusion from these experiments is that much of stationary-phase regulation is retained in lacUV5p-driven fusions that retain only 24 nt upstream of the rpoS AUG initiation codon and an additional seven codons downstream (Fig. 6). This effect of sequences close to the RBS is independent of the antisense element, ppGpp, and all known trans-acting regulators of rpoS, including dsrA and dksA.
Acknowledgments
We are grateful to Mike Cashel for many helpful discussions and to individuals cited in the text for bacterial strains.
REFERENCES
- 1.Barker, M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 2.Barker, M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin, W. H., Jr., X. Wu, and W. E. Swords. 1996. The predicted amino acid sequence of the Salmonella typhimurium virulence gene mviA+ strongly indicates that MviA is a regulator protein of a previously unknown S. typhimurium response regulator family. Infect. Immun. 64:2365-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutants. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohringer, J., D. Fischer, G. Mosler, and R. Hengge-Aronis. 1995. UDP-glucose is a potential intracellular signal molecule in the control of expression of σS and σS-dependent genes in Escherichia coli. J. Bacteriol. 177:413-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, L., and T. Elliott. 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 178:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, L., and T. Elliott. 1997. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 179:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, L., D. Gentry, T. Elliott, and M. Cashel. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 9.Chen, C. Y., L. Eckmann, S. J. Libby, F. C. Fang, S. Okamoto, M. F. Kagnoff, J. Fierer, and D. G. Guiney. 1996. Expression of Salmonella typhimurium rpoS and rpoS-dependent genes in the intracellular environment of eukaryotic cells. Infect. Immun. 64:4739-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunning, C., L. Brown, and T. Elliott. 1998. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J. Bacteriol. 180:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunning, C., and T. Elliott. 1999. RpoS synthesis is growth rate regulated in Salmonella typhimurium, but its turnover is not dependent on acetyl phosphate synthesis or PTS function. J. Bacteriol. 181:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative σ factor KatF (RpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σS is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 16.Jensen, K. F. 1993. The Escherichia coli K-12 “wild-types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Klauck, E., J. Bohringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 17.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 19.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at levels of transcription, translation and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 20.Lease, R. A., and M. Belfort. 2000. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38:667-672. [DOI] [PubMed] [Google Scholar]
- 21.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 22.Loewen, P. C., I. Von Ossowski, J. Switala, and M. R. Mulvey. 1993. KatF (σS) synthesis in Escherichia coli is subject to posttranscriptional regulation. J. Bacteriol. 175:2150-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCann, M. P., C. D. Fraley, and A. Matin. 1993. The putative sigma factor KatF is regulated posttranscriptionally during carbon starvation. J. Bacteriol. 175:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 26.Metzger, S., G. Schreiber, E. Aizenman, M. Cashel, and G. Glaser. 1989. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J. Biol. Chem. 264:21146-21152. [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Muffler, A., D. Fischer, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10:1143-1151. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay, S., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 30.Mulvey, M. R., J. Switala, A. Borys, and P. C. Loewen. 1990. Regulation of transcription of katE and katF in Escherichia coli. J. Bacteriol. 172:6713-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratt, L. A., and T. J. Silhavy. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225-1236. [DOI] [PubMed] [Google Scholar]
- 33.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schellhorn, H. E., and V. L. Stones. 1992. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J. Bacteriol. 174:4769-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweder, T., K-H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σS) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiba, T., K. Tsutsumi, H. Yano, Y. Ihara, A. Kameda, K. Tanaka, H. Takahashi, M. Munekata, N. N. Rao, and A. Kornberg. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. USA 94:11210-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 39.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 40.Tsui, H. C., H. C. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli. K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 41.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112-123. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 43.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17:6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]