Abstract
The conjugative, chromosomally integrating element R391 is the archetype of the IncJ class of mobile genetic elements. Originally found in a South African Providencia rettgeri strain, R391 carries antibiotic and mercury resistance traits, as well as genes involved in mutagenic DNA repair. While initially described as a plasmid, R391 has subsequently been shown to be integrated into the bacterial chromosome, employing a phage-like integration mechanism closely related to that of the SXT element from Vibrio cholerae O139. Analysis of the complete 89-kb nucleotide sequence of R391 has revealed a mosaic structure consisting of elements originating in bacteriophages and plasmids and of transposable elements. A total of 96 open reading frames were identified; of these, 30 could not be assigned a function. Sequence similarity suggests a relationship of large sections of R391 to sequences from Salmonella, in particular those corresponding to the putative conjugative transfer proteins, which are related to the IncHI1 plasmid R27. A composite transposon carrying the kanamycin resistance gene and a novel insertion element were identified. Challenging the previous assumption that IncJ elements are plasmids, no plasmid replicon was identified on R391, suggesting that they cannot replicate autonomously.
Horizontal gene transfer, the intraspecies and interspecies exchange of genetic information, plays an important role in the evolution of bacteria (10, 24, 26, 39, 65). Three major mechanisms, transformation, transduction, and conjugation (9), provide bacterial populations with access to a “horizontal gene pool,” enabling them to rapidly respond to environmental challenges (16, 34).
The most important contributor to horizontal gene transfer is the heterogeneous group of mobile genetic elements that includes plasmids, insertion (IS) elements, transposons, integrons, phages, and genomic islands (10, 17). They are “selfish” elements that promote their own maintenance and distribution and, in addition, can function as vectors for accessory DNA elements. These accessory elements commonly consist of genes that are nonessential for survival but confer a phenotype, which is advantageous under particular environmental conditions. Prominent examples of such traits are antibiotic and heavy metal resistances, degradation of xenobiotic compounds, symbiosis and virulence determinants, resistance to radiation, and increased mutation frequency (15).
A group of related conjugative DNA elements has been identified as carriers of antibiotic resistance genes in members of the γ-Proteobacteria from dispersed global locations. R391, the archetype of this group, was originally detected in a South African Providencia rettgeri isolate (8); for many years, it was believed to be a plasmid and was assigned to a new incompatibility group, IncJ. Subsequently, other IncJ elements, conferring the same phenotype as that conferred by R391 (19, 20) and possibly of clonal origin, were identified in Providencia spp. (R748 and R749) and Proteus vulgaris (R705 and R706) from South Africa, Vibrio cholerae (pJY1; phenotype, Cmr Smr Sur) from the Philippines (72), Proteus vulgaris (R997; phenotype, Apr Smr Sur) from India (32), and Shewanella putrefaciens (pMERPH; mercury resistance) from the United Kingdom (48). Previous studies have examined R391's ability to mobilize chromosomal markers (38), its mutagenic DNA repair function (27), and its unusual UV-sensitizing phenotype (46, 47).
More recently, the plasmid nature of these elements was challenged by the discovery of their transfer and stable integration into the host chromosome in recombination-deficient (recA) backgrounds, suggesting that they are conjugative transposons (37). Furthermore, Hochhut et al. (21) have since reported a close relationship between R391 and the self-transmissible SXT element from Vibrio cholerae O139, which carries multiple antibiotic resistance cassettes (Sur Tmr Smr). Both code for a nearly identical phage-like integrase, which mediates site-specific integration into the 5′ end of the prfC gene of the Escherichia coli chromosome (23).
In this study, the complete nucleotide sequence of R391 has been determined and analyzed. The main objective of the analysis was to elucidate the nature of R391 and, by inference, of related elements, to identify the genetic systems responsible for both the chromosomal integration of R391 and its conjugative transfer, and to determine whether R391 carries a recognizable autonomous replicon. Additionally, given that the chromosomal association of these elements makes detection difficult, it is possible that such elements are representatives of a much larger and important group of mobile genetic elements, potentially acting as vectors of antibiotic resistances and other phenotypes in the γ-Proteobacteria. Thus, this study aimed to provide an enhanced understanding of the archetype of this group, to broaden our molecular understanding of its genetic structure, regulatory systems, and evolutionary origins, to identify important accessory functions, and to facilitate the future molecular detection of further related elements.
MATERIALS AND METHODS
Isolation and purification of DNA.
An extrachromosomal circular intermediate of R391 can be generated and visualized when it is transferred to an E. coli recA recipient strain harboring the related element R997 integrated in the chromosome (45). For the purpose of subcloning R391, the circular form was extracted from E. coli AB2463 (R997 and R391) with a Qiagen plasmid maxikit purification protocol for plasmids with a very low copy number. To minimize contamination with chromosomal DNA, the preparation was purified further by cesium chloride density gradient centrifugation in accordance with standard protocols (58).
Shotgun cloning and DNA sequencing.
Shotgun cloning of R391 was performed with a TOPO shotgun subcloning kit (Invitrogen) in accordance with the manufacturer's protocol. Briefly, DNA was sheared with compressed air by using a nebulizer, generating fragments of about 1 to 2 kb in size. After blunt-end repair and dephosphorylation, the fragments were ligated to the vector pCR4Blunt-TOPO and transformed into E. coli TOP10, with subsequent blue/white screening used to detect transformants carrying an insert. Plasmid DNA from clones was isolated with a Qiaprep8 miniprep kit (Qiagen). Library subclones were sequenced by the chain termination method (59) with sequencing kits provided by Applied Biosystems, Amersham, or Cambio. Electrophoresis and analysis were carried out on an ABI Prism 310 genetic analyzer (Applied Biosystems) or a Licor IR4200. The complete nucleotide sequence of R391 has been determined by sequencing 750 shotgun clones, yielding a fourfold coverage in combination with primer walking for the closure of gaps and regions of insufficient coverage. Primers for gap filling were designed with PrimerSelect software (Lasergene) and used either directly for sequencing on shotgun clones or to sequence PCR amplicons. PCR amplification with the extrachromosomal form of R391 as the template DNA was performed according to standard protocols.
Annotation and phylogenetic analysis.
Sequences were assembled by using SeqMan software (Lasergene). ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was employed to search the complete R391 sequence for open reading frames (ORFs), initially with a cutoff value of 50 amino acids, by using the bacterial genetic code. Subsequently, the ORFs were compared to sequences in the public sequence databases of GenBank and EMBL by using the BLAST (1) and FASTA (44) search tools.
Of the original 885 ORFs identified, only those that had a minimum length corresponding to 60 amino acids and were not overlapping other ORFs with significant identity to ORFs in the databases were included in the results. Significant identity was defined as being greater than 20% identity over at least 60% of the ORF or, alternatively, over at least 50 corresponding amino acids. ORF products were further analyzed for the presence of conserved protein domains by using the Web-based tools Cognitor (66), PFAM Search (3), and ProDom (1).
Multiple sequence alignments and bootstrap neighbor-joining phylogenies (12, 56) were generated with ClustalX software (67). Gaps in the alignment were excluded from the calculation of the phylogenetic tree. The G+C plot was created with the program Freak from the EMBOSS package (Human Genome Mapping Project Resource Centre, Cambridge, United Kingdom) with a window size setting of 300 nucleotides (nt) and a step size setting of 10.
Nucleotide sequence accession number.
The nucleotide sequence of R391 has been deposited in the GenBank database under accession number AY090559.
RESULTS AND DISCUSSION
General properties of the nucleotide sequence of R391.
Assembly of the subclone sequences resulted in a circular DNA sequence of 88,532 nt. Ninety-six ORFs, corresponding to a total coding region of 90%, were identified (Table 1 Fig. 1). Six translated ORFs (6%) were found to show no significant similarity to protein sequences in the databases, while another 24 putative proteins (25%) are similar only to hypothetical proteins of unknown function. Putative genes which code for the backbone functions of R391 (integration/excision and conjugative transfer) and the phenotypic traits (kanamycin and mercury resistance) have been identified, as have a number of transposable elements.
TABLE 1.
Homology between ORFs in R391 and corresponding representative prokaryotic protein sequences
| ORFa | ORF product size (amino acids) | ORF location (start, stop)b | ORF product exhibits homology to: | Source | Identityc
|
Accession no. | ||
|---|---|---|---|---|---|---|---|---|
| No. | Name | % | Amino acids | |||||
| 1 | 107 | 442, 119 | ABC transporter permease protein | Lactococcus lactis | 27 | 25/92 | NP 267026 | |
| 2 | 106 | 790, 1110 | Y4dL; hypothetical 21.8-kDa protein | Rhizobium sp. strain NGR234 | 50 | 44/87 | NP 443821 | |
| 3 | 404 | 1103, 2317 | Y4dM; putative protein | Rhizobium sp. strain NGR234 | 55 | 224/405 | NP 443822 | |
| 4 | 66 | 2528, 2728 | Possible DNA-binding protein | Mycobacterium leprae | 34 | 17/50 | np 302574 | |
| 5 | int | 413 | 3986, 2745 | Integrase | SXT element; Vibrio cholerae | 99 | 410/413 | AAC72397 |
| 6 | 89 | 4257, 3988 | Soluble lytic murein transglycosylase | Brucella melitensis | 28 | 20/71 | np 540005 | |
| 7 | 324 | 5234, 4260 | StbA; putative plasmid partitioning protein | Plasmid R27; Salmonella enterica serovar Typhi | 24 | 62/252 | Q9RGU9 | |
| 10 | rumB | 435 | 7460, 6153 | SamB; UV protection protein | S. enterica serovar Typhi CT18 | 61 | 256/419 | np 458681 |
| 11 | rumA | 149 | 7878, 7429 | RumA | SXT element; Vibrio cholerae | 98 | 147/149 | AAK64594 |
| 13 | 302 | 8518, 9426 | DNA polymerase III (epsilon subunit) | Agrobacterium tumefaciens strain C58 | 39 | 117/300 | AAL43906 | |
| 14 | 104 | 9500, 9814 | Polynucleotide phosphorylase | Deinococcus radiodurans | 33 | 18/53 | np 295786 | |
| 15 | 308 | 9883, 10809 | NMA1157; hypothetical protein | Neisseria meningitidis | 35 | 93/261 | np 283925 | |
| 16 | tnpIS15 | 238 | 11635, 10919 | P22 protein | IS15; Escherichia coli | 100 | 234/234 | Q56369 |
| 17 | 279 | 11096, 11935 | P11 protein | IS15; Escherichia coli | 100 | 197/197 | Q56370 | |
| 18 | 202 | 12560, 11952 | P11 protein | IS15; Escherichia coli | 100 | 197/197 | Q56370 | |
| 19 | tnpIS15 | 238 | 12021, 12737 | P22 protein | IS15; Escherichia coli | 100 | 234/234 | Q56369 |
| 20 | aph | 271 | 12877, 13692 | Aminoglycoside phosphotransferase | Klebsiella pneumoniae | 100 | 271/271 | S16630 |
| 21 | tnpIS15 | 238 | 14588, 13872 | P22 protein | IS15; Escherichia coli | 100 | 234/234 | Q56369 |
| 22 | 199 | 14049, 14648 | P11 protein | IS15; Escherichia coli | 100 | 197/197 | Q56370 | |
| 23 | 210 | 14567, 15199 | Putative inner membrane protein | S. enterica serovar Typhimurium LT2 | 41 | 76/182 | NP 463357 | |
| 24 | 194 | 15196, 15780 | Putative cytoplasmic protein | S. enterica serovar Typhimurium LT2 | 61 | 116/188 | NP 463356 | |
| 25 | 947 | 15799, 18642 | Putative ATPase involved in DNA repair | S. enterica serovar Typhimurium LT2 | 62 | 585/943 | NP 463355 | |
| 26 | 317 | 18524, 19477 | Putative ATPase involved in DNA repair | S. enterica serovar Typhimurium LT2 | 43 | 123/280 | NP 463355 | |
| 27 | 171 | 19634, 20149 | Putative type II restriction enzyme, methylase subunit | S. enterica serovar Typhimurium LT2 | 56 | 96/171 | NP 463354 | |
| 28 | tnp391A | 225 | 20708, 21385 | OrfB of IS911 | IS911; Escherichia coli | 88 | 198/224 | AAC70114 |
| 29 | 1,083 | 21303, 24554 | Putative type II restriction enzyme, methylase subunit | S. enterica serovar Typhimurium LT2 | 59 | 632/1060 | NP 463354 | |
| 30 | 894 | 24554, 27238 | Putative cytoplasmic protein | S. enterica serovar Typhimurium LT2 | 50 | 455/896 | NP 463351 | |
| 31 | 746 | 27251, 29491 | Putative ATP-dependent Lon protease | S. enterica serovar Typhimurium LT2 | 85 | 576/672 | NP 463350 | |
| 32 | 883 | 29535, 32186 | Orf74 | Plasmid pB171; Escherichia coli | 25 | 235/920 | NP 053136 | |
| 33 | traI | 716 | 32341, 34491 | Hypothetical protein | Xylella fastidiosa 9a5c | 26 | 94/353 | NP 299042 |
| 34 | 606 | 34540, 36360 | TraG | Plasmid R27; S. enterica serovar Typhi | 30 | 186/615 | NP 458648 | |
| 35 | 186 | 36370, 36930 | Large repetitive protein | S. enterica serovar Typhi CT18 | 25 | 20/78 | NP 458558 | |
| 36 | 214 | 36908, 37552 | Hypothetical protein | Plasmid R27; S. enterica serovar Typhi | 29 | 29/98 | NP 058330 | |
| 37 | 195 | 38166, 37579 | Hypothetical protein | Listeria monocytogenes | 24 | 30/123 | NP 464171 | |
| 39 | traL | 93 | 38455, 38736 | Sex pilus assembly and synthesis protein | Plasmid pNL1; Novosphingobium aromaticivorans | 25 | 18/72 | NP 049168 |
| 40 | traE | 130 | 38967, 39359 | HtdE | Plasmid R478; Serratia marcescens | 25 | 29/115 | AAL27019 |
| 41 | traK | 300 | 39340, 40242 | HtdP | Plasmid R478; Serratia marcescens | 26 | 66/251 | AAL27020 |
| 42 | traB | 429 | 40242, 41531 | TrhB; putative transfer protein | Plasmid R27; S. enterica serovar Typhi | 40 | 84/209 | NP 058242 |
| 43 | htdD | 216 | 41528, 42178 | HtdD | Plasmid R478; Serratia marcescens | 28 | 45/158 | AAD01915 |
| 44 | traA | 128 | 42175, 42561 | TraA | Plasmid R27; S. enterica serovar Typhi | 24 | 21/85 | NP 058247 |
| 45 | 106 | 42922, 42602 | Putative acetyltransferase | S. enterica serovar Typhimurium LT2 | 45 | 41/90 | NP 463183 | |
| 46 | 88 | 43365, 43099 | Hypothetical protein | Escherichia coli O157:H7 | 44 | 40/89 | NP 289961 | |
| 47 | 125 | 43920, 43543 | SMA0431, hypothetical protein | Rhizobium meliloti | 27 | 31/114 | Q930G9 | |
| 48 | tnp391B | 305 | 45323, 44406 | Transposase | Listonella anguillarum | 66 | 201/301 | AAA81774 |
| 49 | 107 | 46010, 45687 | Glycosyl transferase | Streptomyces fradiae | 22 | 21/95 | AAF00214 | |
| 50 | 82 | 46700, 46452 | Hypothetical protein G | Tn1404 Pseudomonas sp. strain R9 | 60 | 15/25 | AAD47995 | |
| 51 | 508 | 45742, 47268 | Sulfate permease | Tn1404 Pseudomonas sp. strain R9 | 80 | 401/497 | AAD47994 | |
| 52 | 283 | 47280, 48131 | Hypothetical protein B | Tn1404 Pseudomonas sp. strain R9 | 38 | 109/283 | AAD47991 | |
| 53 | 149 | 48604, 48155 | Conserved hypothetical protein | Pseudomonas aeruginosa | 36 | 53/146 | NP 250418 | |
| 54 | 169 | 49144, 48635 | Conserved hypothetical protein | Pseudomonas aeruginosa | 35 | 39/109 | NP 249266 | |
| 56 | htdT | 230 | 49588, 50280 | HtdT; putative transfer protein | Plasmid R27; S. enterica serovar Typhi | 26 | 53/197 | NP 058240 |
| 57 | traC | 799 | 50280, 52679 | TraC | Plasmid R27; S. enterica serovar Typhi | 27 | 239/869 | NP 058238 |
| 59 | trhF | 163 | 53024, 53515 | TrhF | Plasmid R27; S. enterica serovar Typhi | 28 | 36/127 | NP 058222 |
| 60 | traW | 375 | 53523, 54650 | Putative transfer protein, pilus formation | Plasmid R27; S. enterica serovar Typhi | 27 | 84/303 | NP 058221 |
| 61 | traU | 342 | 54634, 55662 | TraU | Plasmid pNL1; Novosphingobium aromaticivorans | 36 | 125/339 | NP 049157 |
| 62 | traN | 1,230 | 55665, 59357 | TraN | Genomic island STI1; S. enterica serovar Typhimurium | 33 | 418/1253 | AAK02035 |
| 64 | 220 | 59951, 60613 | DNA ligase | Escherichia coli O157:H7 | 25 | 31/124 | NP 288973 | |
| 65 | 200 | 61306, 60704 | Lipoprotein | Haemophilus ducreyi | 21 | 30/139 | AAB48950 | |
| 66 | 112 | 61662, 62000 | AnsG oxidoreductase | Streptomyces collinus | 27 | 19/68 | AAD31836 | |
| 67 | 139 | 62016, 62435 | Single-strand binding protein | Plasmid pLP231a; Escherichia coli | 38 | 52/135 | C38487 | |
| 68 | 272 | 62515, 63333 | Recombination protein Bet | Bacteriophage 933W; Escherichia coli O157:H7 | 55 | 108/195 | NP 049474 | |
| 69 | 338 | 63621, 64637 | Hypothetical protein | Xylella fastidiosa 9a5c | 32 | 51/155 | NP 298936 | |
| 70 | 358 | 64730, 65806 | Probable cobalamin biosynthesis protein | Sinorhizobium meliloti | 30 | 87/281 | NP 386778 | |
| 71 | 255 | 65806, 66573 | Molybdate metabolism regulator-related protein | Deinococcus radiodurans | 31 | 33/105 | NP 386778 | |
| 72 | 317 | 66672, 67625 | Hypothetical protein | Plasmid R27; S. enterica serovar Typhi | 20 | 41/205 | NP 058418 | |
| 74 | 551 | 68197, 69852 | Hypothetical protein | Plasmid R27; S. enterica serovar Typhi | 21 | 138/636 | NP 058417 | |
| 75 | 165 | 69938, 70435 | RadC; putative DNA repair protein | Vibrio cholerae | 56 | 86/151 | NP 231421 | |
| 78 | 357 | 70867, 71940 | Hypothetical protein | Plasmid R27; S. enterica serovar Typhi | 30 | 113/375 | NP 058415 | |
| 79 | 235 | 72030, 72737 | Hypothetical protein | SXT element; Vibrio cholerae | 80 | 61/76 | AAK95994 | |
| 80 | 180 | 73088, 73630 | Probable oxidoreductase | Agrobacterium tumefaciens | 27 | 35/126 | NP 396434 | |
| 81 | traF | 314 | 74368, 75312 | TraF | SXT element; Vibrio cholerae | 97 | 306/314 | AAK 95986 |
| 82 | traH | 462 | 75315, 76703 | TraH | Genomic island STI1; Salmonella enterica serovar Typhimurium | 42 | 189/449 | AAK02038 |
| 83 | traG | 1,189 | 76707, 80276 | Putative pilus assembly protein | Genomic island STI1; Salmonella enterica serovar Typhimurium | 30 | 375/1214 | AK02037 |
| 84 | merR | 129 | 80374, 80763 | MerR | IncJ element pMERPH; Shewanella putrefaciens | 100 | 557/557 | |
| 85 | merT | 107 | 80848, 81171 | MerT | IncJ element pMERPH; Shewanella putrefaciens | 97 | 99/102 | Q54462 |
| 86 | merP | 91 | 81256, 81531 | MerP | IncJ element pMERPH; Shewanella putrefaciens | 98 | 90/91 | Q54463 |
| 87 | merC | 137 | 81541, 81954 | MerC | IncJ element pMERPH; Shewanella putrefaciens | 98 | 135/137 | CAA89056 |
| 88 | merA | 557 | 81992, 83665 | MerA; mercuric reductase | IncJ element pMERPH; Shewanella putrefaciens | 100 | 557/557 | Q54465 |
| 89 | 143 | 84202, 83771 | Hypothetical protein | Listeria innocua | 26 | 23/86 | CAC42036 | |
| 90 | 177 | 84790, 84257 | FlhC; flagellar transcriptional activator | S. enterica serovar Typhimurium LT2 | 27 | 41/150 | AAB96640 | |
| 91 | 99 | 85086, 84787 | FlhD protein | Serratia liquefaciens | 28 | 28/97 | S61276 | |
| 92 | 182 | 85631, 85083 | Hypothetical protein | S. enterica serovar Typhi CT18 | 32 | 39/121 | NP 458645 | |
| 93 | 202 | 86226, 85618 | Hypothetical transmembrane protein | Ralstonia solanacearum | 39 | 26/66 | CAD17270 | |
| 94 | 289 | 87136, 86267 | Unknown protein | Cryptic prophage CP-933M; Escherichia coli O157:H7 | 35 | 27/76 | NP 286859 | |
| 96 | 215 | 87561, 88208 | Putative prophage repressor protein; similar to cI repressor | Yersinia pestis | 38 | 87/227 | NP 404832 | |
ORFs not showing significant homology to corresponding proteins in the public databases are not shown. Nucleotide position from start to stop codon in the R391 sequence. Identity in percentage and amino acids (number identical relative to total number examined) as determined with BLAST and FASTA.
Nucleotide position from start to stop codon in the R391 sequence.
Identity in percentage and amino acids (number identical relative to total number examined) as determined with BLAST and FASTA.
FIG. 1.
Linear map of R391 showing all ORFs and their orientations in the chromosomally integrated state, from the left junction attL to the right junction attR. The boxes above and below the axis represent ORFs in the forward and reverse frames, respectively. ORF numbers, names, and sequence features are indicated; the scale bar is given in kilobases. See also Table 1.
The total G+C content of R391 is 46%. The G+C content varies significantly within R391, displaying a number of peaks and troughs (Fig. 2). These peaks and troughs correspond to different functional units, such as transposable elements and conjugative transfer elements, suggesting that R391 has a mosaic structure composed of elements of different origin. In particular, four putative transposase genes and the mercury resistance (mer) operon, which are visible as peaks in Fig. 2, have a notably higher G+C content than their surroundings. Four transfer regions, TRA I to IV, are homogeneous in G+C content but clearly differ from the adjacent regions. A region corresponding to a putative restriction enzyme (ORF 29) and a transposon-like region between TRA II and III show a lower G+C contents, with the exception of a putative transposase gene located within this region.
FIG. 2.
G+C plot of the DNA sequence of R391. Regions indicated are the transfer regions TRA I to IV (I to IV), the kanamycin resistance transposon (K), the IS element (IS), the putative restriction enzyme methylase subunit ORF 27 (R), and the mercury resistance operon (M). Asterisks indicate putative transposase genes.
One-third of the ORFs found on R391 have homologs in Salmonella enterica serovar Typhi and Salmonella enterica serovar Typhimurium, and 21 of those are associated with the multiple antibiotic resistance plasmid R27 (63) (see “Conjugative transfer” below), the remaining 12 being chromosomally associated ORFs. These homologies suggest evolutionary relatedness and a possible origin of large sections of R391 from these or closely related ancestral strains.
Remarkably, 67 of the ORFs (70%) are transcribed in the three forward frames (Table 1; Fig. 1). The exceptions are, first, two clusters of eight and six ORFs lying, respectively, upstream and downstream of the attachment site attP, which is located at the end of the linear map (Fig. 1). Second, a few ORFs belonging to the IS elements, as well as a number of dispersed ORFs, also lie on the reverse strand. This unidirectionality is even more surprising considering the different origins of this element.
Given that it has the ability to transfer by conjugation and that numerous antibiotic resistance markers are carried by plasmids, R391 had initially been thought to be a plasmid. However BLASTP analysis of the 96 ORFs failed to show similarity to any known plasmid replication gene; likewise, no similarity was found between R391 and any known origin of replication, suggesting the absence of a plasmid replicon on R391. It is likely, however, that major parts of R391 have been derived from a plasmid, as suggested by the high number of homologs to ORFs from Salmonella plasmids. If the ancestral R391 did possess a plasmid replicon, it might have been lost upon acquiring the integration mechanism.
Integration and excision.
Integration of R391 into the host chromosome and its excision from it share similarities to the integration and excision of bacteriophages and genomic islands. Integration into the prfC gene is mediated by a phage-like integrase and results in short direct repeats at both ends of the element and in the reconstitution of prfC by the replacement of its 5′ end (20, 22). However, this mechanism differs notably from those of the majority of genomic islands described to date in its use of an integration site that is not located in, or near, a tRNA gene and in the replacement of the 5′ end of the interrupted gene. Other mobile elements using phage-like integrases normally carry a 3′ terminus of the interrupted gene, presumably because upstream regulatory sequences such as the promoter region are more complex and difficult to replace (74).
The integrase gene int (ORF 5) is located from 2.7 to 4 kb from the left end of R391 (Fig. 1) and is translated towards the left junction attL of the element, unlike genomic islands, where the integrase gene is typically located directly adjacent to the junction with the chromosome and is translated divergently from the junction.
The IncJ elements R997 and pMERPH probably possess an integration mechanism closely related to those of R391 and the SXT element. In both R997 and pMERPH PCR, amplification generates a PCR product of the expected size for the integrase gene and the integration sites are nearly identical (B. McGrath and J.T. Pembroke, unpublished data). In addition, restriction digestion analysis revealed many restriction fragments common to both R391 and R997 (45). R391 and the SXT element can integrate into the same chromosome in tandem fashion, with an incoming element probably using one of the junction sites of the resident element with the chromosome attL or attP as the integration site (21). Conversely, in recA mutant strains containing both R391 and R997, the incoming element could be isolated in the extrachromosomal circular form (45). The existence of a circular extrachromosomal form raises the possibility of mutual exclusion from integration between these two elements and the probable requirement for extrachromosomal replication. However, this study found no indication of a plasmid replicon, suggesting that the circular form may be a nonreplicative transposition or transfer intermediate.
Evolutionary relationships between phage-related integrases.
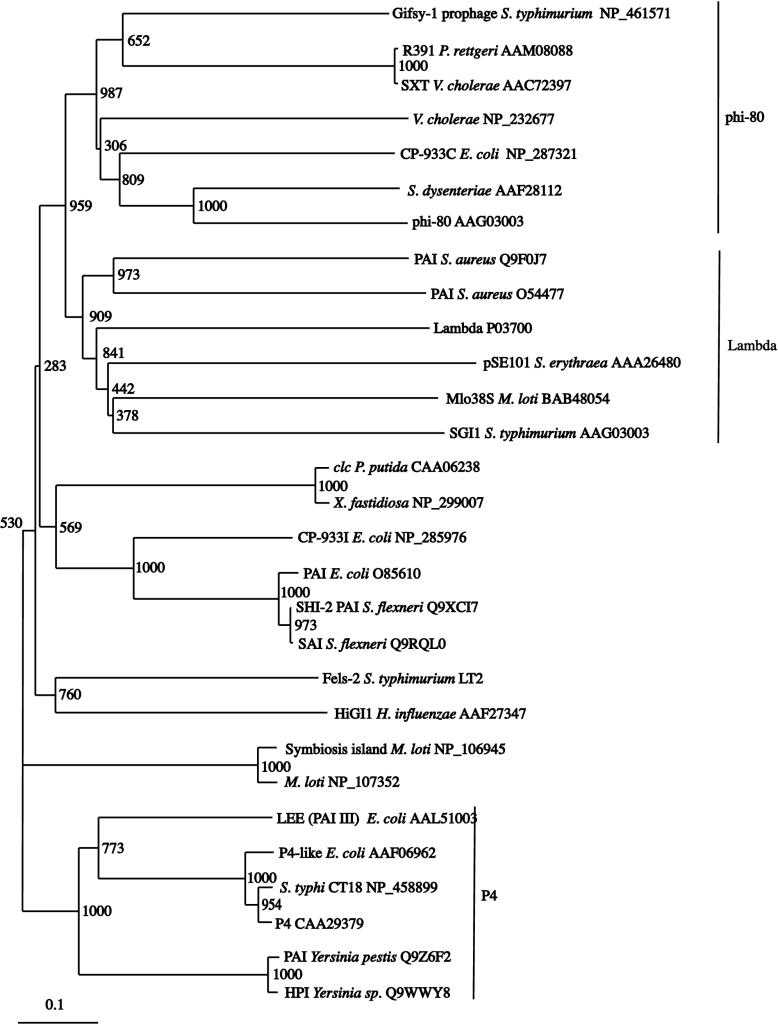
The R391 integrase belongs to a diverse family of integrase proteins within a wider group of tyrosine recombinases with a range of functions in DNA metabolism (71; http://mywebpages.comcast.net/domespo/trhome.html). It has previously been reported that the closely related integrase from the SXT element was itself closely related to that from bacteriophage φ80 (23). BLAST analysis of the R391 integrase showed it to be most closely related (excluding SXT) to that from the Gifsy-1 prophage from Salmonella enterica serovar Typhimurium LT2 (36% identity over 413 amino acids).
To further investigate the evolutionary relationships within the integrase family, representative integrases from R391, the SXT element, phages, and pathogenicity and other genomic islands that were identified by a BLAST similarity search were aligned and a phylogenetic tree was constructed (Fig. 3). From the several distinct clusters identified, the integrases from R391 and the SXT element belong to a group containing the prophage φ80 (29) and other cryptic prophages. This φ80 group branches next to a cluster of integrases which is related to phage lambda (28) and which contains two Staphylococcus pathogenicity islands (13, 30) and the Salmonella genomic island SGI1 (6). The relatively large evolutionary distances within both groups suggest early and considerable divergence of most of their members. A third distinct subgroup contains the satellite phage P4 (49) and a number of pathogenicity islands from Yersinia spp. (18, 61) and E. coli. A number of pathogenicity islands from E. coli, Shigella flexneri (35, 69), and Haemophilus influenzae (7), as well as the clc element from Pseudomonas putida (54) and a symbiosis island from Mesorhizobium loti (64), cannot be assigned unambiguously to any of these three groups, but the relatively smaller evolutionary distances suggest a relationship to the P4 subgroup.
FIG. 3.
Bootstrap neighbor-joining tree of integrases from R391 and SXT, including genomic islands (SGI1, clc, and HiGI), pathogenicity islands (PAI, SHI, SAI, LEE, and HPI), phages (Gifsy1, CP-933C, φ80, lambda, Fels-2, and P4), and uncharacterized putative integrases. Host organisms, accession numbers of protein sequences, and bootstrap values (1,000 replicates) are indicated.
These results indicate multiple origins of mobile integrating elements, such as pathogenicity islands and other genomic islands, by independent acquisition of the integration module of a prophage.
Conjugative transfer.
A total of 17 putative transfer genes, located in four clusters (TRA I to IV) spanning 24 kb of R391, were identified (Fig. 1). Given the lack of consistency in the nomenclature of transfer genes among different incompatibility (Inc) groups, in this study the putative R391 transfer genes were named by their homologs in the IncF plasmids (14) and, for ORFs not having close relatives in F plasmids, according to their closest relative.
The first cluster, TRA I, is 4 kb in size and is located 32 kb from the left end of the R391 sequence, following a region containing mainly ORFs whose putative function is related to DNA repair or modification. TRA I codes for two putative transfer proteins. The first (encoded by ORF 33) is a homolog of the traI product from plasmid R27 (63), which is thought to be a relaxase, responsible for nicking the DNA during conjugative transfer. ORF 34 is related to a putative coupling protein from R27. Such proteins usually contain a transmembrane domain and an ATP-binding domain. The R391 homolog similarly carries motifs of this protein family.
Separated from TRA I by four ORFs of unknown function, the second cluster, TRA II, is also about 4 kb in size and contains homologs of traL, traE, traK, traB, htdD, and traA (ORFs 39 to 44, respectively), which are probably all involved in pilus synthesis and assembly.
TRA II and TRA III are separated by a 7-kb region containing a number of ORFs which are related to those found on transposons and which possibly represent the remnants of such an element. TRA III spans nearly 10 kb and contains homologs of htdT (ORF 56), traC (ORF 57), trhF (ORF 59), traW (ORF 60), traU (ORF 61), and traN (ORF 62). From the function of their closest relatives, it can be assumed that the first five of these ORFs are involved in pilus synthesis and assembly while traN is probably involved in aggregate stability. Additionally, this cluster contains an ORF (ORF 58) of unknown function.
The fourth cluster, TRA IV, is about 6 kb in size and is separated from TRA III by a 15-kb region. It contains homologs of traF (ORF 81) and traH (ORF 82), again putatively involved in pilus synthesis and assembly, and of traG (ORF 83), putatively involved in pilus assembly and mating pair stabilization.
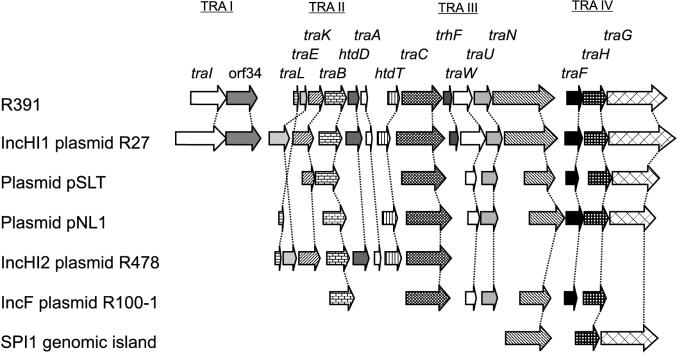
Generally, in terms of individual homology and overall structure, the R391 transfer genes are most closely related to homologs from large plasmids from Salmonella strains (Fig. 4). Of the 17 transfer genes identified, 16 have homologs (average identity, 27%) in the Salmonella enterica serovar Typhi plasmid R27 (63) and its derivative pHCM1 (43). Both of these plasmids are very large (180 and 218 kb, respectively) and carry multiple antibiotic resistance genes. The order and structural organization of transfer genes, however, are significantly different in R391 and R27. The transfer genes might have been derived from a common ancestral plasmid, having subsequently diverged and undergone rearrangements. Other conjugative plasmids sharing transfer gene homologs with R391 are the 90-kb Salmonella enterica serovar Typhimurium plasmid pSLT (33), with nine homologs, and the 184-kb plasmid pNL1 from Novosphingobium aromaticivorans (55), having 10 homologs, all of them located within clusters TRA II, III, and IV of R391. The F plasmid derivative R100 (2) shares seven homologs within these clusters. The IncHI2 plasmid R478 from Serratia marcescens (42) has a total of eight homologs, including all of the putative transfer genes from TRA II and the first two of TRA III. Furthermore, two genomic islands from Salmonella enterica serovar Typhimurium (6) and Neisseria gonorrhoeae (11) have three and two homologs, respectively.
FIG. 4.
Relationship between the putative transfer genes of R391 and homologs present in related plasmids and genomic islands. The names of the R391 transfer genes and transfer regions are indicated. Homologous proteins are depicted by arrows with the same shading and connected by dotted lines. Clustered genes are shown as linked. Note that the structural organization and gene order in the different plasmids and genomic islands differ from those of R391. Both traI and orf34 have no homologs with significant similarity in plasmids except R27. orf34 is a homolog of traG on R27 (based on IncP nomenclature); however, in R391, ORF 83 has been designated traG, in accordance with the IncF nomenclature.
The location of the origin of transfer (oriT), at which a single-strand nick is introduced and transfer to the recipient is initiated, could not be located precisely. Five families of homologous oriT core nick sites have been identified in different plasmid groups (73). None of the signature sequences of these oriT families could be found in R391, apart from the 8-bp consensus sequence of the IncP plasmid family (YATCCTGY), which is present in eight copies. Six of these copies are placed within coding regions and are thus unlikely to be the oriT. Of the remaining two copies, one is located in an untranslated region upstream of TRA IV, although this is not sufficient evidence for it being the oriT. No regulatory element of the putative transfer genes has been identified, and for that reason it remains unclear if the expression of the transfer genes is constitutive or linked to excision of R391 from the chromosome.
Phenotypic traits. (i) Kanamycin resistance.
R391 confers a kanamycin resistance phenotype. This trait is conferred by an aminoglycoside phosphotransferase gene (ORF 20) which is 100% identical to a number of such kanamycin resistance genes from different strains (Table 1). The putative gene is carried by a transposon flanked by one copy of IS15-Δ1 downstream and two copies upstream, as similarly reported by Hochhut et al. (22). The composite transposon, which is about 3.5 kb in size, is inserted in a region containing several ORFs that are related to genes involved in DNA modification and DNA repair.
(ii) The mercury resistance (mer) operon.
Narrow-spectrum mercury resistance is mediated by a 3.5-kb mer operon consisting of four structural genes, merT, merP, merC, and merA (ORFs 85 to 88) and the regulatory gene merR (ORF 84). The structural genes of the mer operon of R391 were found to be highly similar (97 to 100% nucleotide identity) to those of the IncJ element pMERPH from Shewanella putrefaciens (40). The merR gene of pMERPH had not been sequenced previously, but PCR amplification and sequence analysis of the pMERPH merR gene in this study proved that the predicted amino acid sequence is 100% identical to the R391 MerR sequence (D. Böltner and A. M. Osborn, unpublished data). Unusually for a mer operon from a gram-negative organism, there is no indication of the operon being carried by a transposon, and furthermore, the merR gene is transcribed unidirectionally with the structural genes, a characteristic of gram-positive mer operons (41).
(iii) Mutagenic DNA repair and UV sensitization.
Previously, R391 had been shown to carry genes involved in mutagenic DNA repair. These genes, rumA and rumB (ORFs 11 and 10, respectively), had been cloned, sequenced, and shown to be homologs of the E. coli umuDC genes and to function in damage-inducible mutagenic DNA repair (27). In contrast to these results, R391 more recently has been reported to confer a UV sensitizing phenotype, apparently compromising the host's DNA repair capability and leading to decreased postirradiation survival (46, 47, 70). The UV-sensitizing phenotype is probably conferred by ORF 13, located adjacent to rumA, which encodes a putative exonuclease domain similar to the DNA polymerase III epsilon subunit. This protein is likely to interact with RumA and/or RumB, altering their activity and dramatically reducing the DNA repair capacity (C. MacMahon and J. T. Pembroke, unpublished data).
Sequence analysis of R391 revealed a number of additional ORFs whose homologs are involved in DNA modification or repair or which contain motifs corresponding to such proteins: ORF 23 (DNA mismatch repair), ORFs 25 and 26 (putative ATPases involved in DNA repair), ORFs 27 and 29 (putative type II restriction enzyme), ORF 64 (DNA ligase), ORF 67 (single-strand binding protein), and ORF 75 (DNA repair protein RadC). While the restriction enzyme is probably not functional due to insertion of an IS element (see “Transposable elements” below), the role of the other ORFs remains unclear. Homology of ORFs 23 to 27 and 29 to 31 to a 10-kb region from Salmonella enterica serovar Typhimurium LT2 suggests that they may be derived from the same origin.
Transposable elements.
A number of transposable elements have been identified on R391. The kanamycin resistance gene (ORF 20) is flanked by two copies of IS15-Δ1 (68) in inverse orientation upstream and one copy downstream in inverse orientation to the second element. Each of the three identical copies of IS15-Δ1 is 820 nt in size. The 5′ end of the adjacent ORF 23 spans the right end of the transposon, and thus it is likely to be a pseudogene. IS15-Δ1 and the nearly identical IS26 have been found in a number of enteric bacteria, including Salmonella and Proteus spp. (http://www-is.biotoul.fr/is.html).
A novel IS element related to IS911 (82% identity) from Shigella dysenteriae (50) has been identified. Located 20 kb from the start of the sequence, the IS element is 1,223 nt in length and is flanked by 26-nt imperfect inverted repeats. The two ORFs upstream and downstream of the IS element (ORFs 27 and 29) are both related to the same putative restriction modification enzyme from Salmonella enterica serovar Typhimurium (GenBank accession no. AAL23313). Insertion of the IS element has probably interrupted the original ORF in R391. Although the right end of the IS element contains a new start codon for ORF 29, both ORF 27 and 29 are probably pseudogenes. Between the transfer regions TRA II and TRA III is a region containing the gene for a putative transposase, tnp391B (ORF 48, with 66% identity to the IS5 transposase gene) and, additionally, three ORFs (ORFs 50 to 52), one of which encodes a sulfate permease (ORF 51) and which is related to the Pseudomonas sp. strain R9 transposon Tn1404 (60). This region might be a transposon or the remnant of such an element, although no inverted repeats, marking the ends of transposons, were found.
Putative regulatory elements.
ORF 96, located upstream of the right end of R391, is related to both the cI repressor from phage lambda and the lexA repressor. Both these repressors carry a DNA-binding helix-turn-helix motif and an autoprotease domain, which cleaves the protein in the presence of RecA and ATP. In the case of the cI repressor, cleavage inactivates the protein, resulting in prophage induction (25). LexA controls the expression of a number of DNA repair proteins and is likewise inactivated upon cleavage. The rumA gene carried by R391 (ORF 11) is one such DNA repair gene, and it is preceded by a LexA binding site (27). Interestingly, RumA itself also contains an autoprotease domain, but in this case the protein is activated when cleaved (25). The putative repressor encoded by ORF 96 could possibly control the expression of the integrase and inhibit excision, given that both the repressor and the integrase are related to phage lambda proteins. However, the presence of a LexA binding site may also suggest a role in regulation of the rumAB operon. Alternatively, the repressor could belong to the remains of a coresident phage, for which a number of phage-like ORFs on R391 might be an indication (see below). At present, the exact role of ORF 96 remains unknown but clearly deserves further attention.
While being most closely related to ORFs of unknown function from Rhizobium sp. strain NGR234, ORFs 2 and 3 also show similarity to hipB and hipA, respectively, which in E. coli act as a pair of transcriptional regulators that affect lethality due to inhibition of peptidoglycan or DNA synthesis (4, 5). ORFs 90 and 91 are homologous, respectively, to flhC and flhD,the transcriptional regulators of flagellar synthesis. These form a master operon at the top of the regulatory hierarchy of flagellar synthesis while themselves being regulated by a number of molecules, including the cyclic AMP-catabolite activator protein complex and OmpR. flhD is also a repressor of cell division (31, 51-53). ORF 15 may be another transcriptional regulator, since it contains a motif encoding such proteins.
Conclusions.
Sequence analysis of R391 revealed a composite structure of elements of different origins (phage, plasmid, and transposable elements). While the integrase is clearly phage related, there are a number of further phage-related ORFs, including the cI repressor homolog (ORF 96), an upstream ORF (ORF 94) whose function is unclear, ORF 67 (related to single-strand binding proteins, which are common to phages and plasmids), ORF 68 (related to phage recombination proteins), and ORF 75 (related to DNA primases). Since these ORFs are dispersed within a 30-kb region, it is unclear if all or some of them are the remains of one prophage or if they are of multiple origins. The conjugative transfer functions are related to those from a number of plasmids and probably share a common origin with the plasmids R27, pHCM1, and pSLT from Salmonella and pNL1 from Novosphingobium. One-third of the 96 ORFs of R391 are related to homologs from Salmonella, in particular the transfer regions and a further 15-kb region containing 8 ORFs which is homologous to a chromosomal region of Salmonella enterica serovar Typhimurium LT2. Further elements contributing to the mosaic nature of R391 are the transposable elements and the mercury resistance operon, whose origin remains unknown because of the versatility and widespread distribution of these elements.
R391 and the related IncJ elements were originally described as transmissible resistance factors (8) and were initially assumed to be plasmids. They were reclassified as conjugative transposons (CTns) on the basis of their RecA-independent transfer and chromosomal integration (36, 37). More recently, the demonstration of the chromosomal integration of R391 by its use of a phage-like integration system closely related to that of the SXT element from Vibrio cholerae led to a proposal that they should be included in the new group of constins (conjugative, self-transmitting, integrating elements) (21).
R391 and the SXT element share functional and structural similarities to genomic islands, conjugative transposons, and bacteriophages. R391 and its relatives carry specific phenotypes that are also found in genomic islands (including symbiosis and pathogenicity islands). Moreover, many genomic islands similarly carry phage-related integrase determinants and terminal direct repeats (15, 16). R391 likewise shares a number of common features with CTns (for example, Tn916 and the Bacteroides CTnDot). CTns represent a diverse group of self-transmissible elements that are normally integrated into the chromosome but are capable of excision, integration, and transfer by conjugation. While exhibiting a transposon-like phenotype in terms of their ability to excise from, and integrate into, the chromosome, these elements have molecular mechanisms that clearly differ from those of classical transposons such as Tn5 and Tn10 and are instead related to those of lambdoid phages (57, 62). With conjugative transfer functions related to those of plasmids, CTns are thus hybrid elements comprising phage integration and plasmid conjugation functions. The primary difference between CTns and R391 is that the latter shows a strong specificity with regard to integration site, integrating into the prfC gene of E. coli while CTns integrate relatively randomly into the chromosome (57, 62). While genomic islands exhibit nonrandom target integration, as does R391, their preferred integration targets are tRNA genes (15, 16). Finally, in contrast to prophages, R391 lacks the genes that are essential for the formation of a bacteriophage. Our future analysis will further investigate the relationships between, and evolutionary origins of, these different classes of mobile genetic elements.
Acknowledgments
Our collaboration on this study benefited from the support of the EU concerted action BIO4-CT-0099 on “Mobile Genetic Elements' Contribution to Bacterial Adaptability and Diversity (MECBAD).”
We thank Barry McGrath for inspiring discussions and Angela Rosin for her contribution to the DNA sequencing of R391.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, K. G., P. Kathir, D. Moore, K. Ippen-Ihler, and L. S. Frost. 1996. Analysis of the traLEKBP sequence and the TraP protein from three F-like plasmids: F, R100-1, and ColB2. J. Bacteriol. 178:3194-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, D. S., B. Irwin, and H. S. Moyed. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, E. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, C.-C., J. R. Gilsdorf, V. J. DiRita, and C. F. Marrs. 2000. Identification and genetic characterization of Haemophilus influenzae genetic island 1. Infect. Immun. 68:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coetzee, J. N., N. Datta, and R. W. Hedges. 1972. R factors from Proteus rettgeri. J. Gen. Microbiol. 72:543-552. [DOI] [PubMed] [Google Scholar]
- 9.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 10.de la Cruz, F., and J. Davies. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128-133. [DOI] [PubMed] [Google Scholar]
- 11.Dillard, J. P., and H. S. Seifert. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263-277. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald, J. R., S. R. Monday, T. J. Foster, G. A. Bohach, P. J. Hartigan, W. J. Meaney, and C. J. Smyth. 2001. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 183:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F-sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity—a Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 17.Hall, R. M., and H. W. Stokes. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115-132. [DOI] [PubMed] [Google Scholar]
- 18.Hare, J. M., A. K. Wagner, and K. A. McDonough. 1999. Independent acquisition and insertion into different chromosomal locations of the same pathogenicity island in Yersinia pestis and Yersinia pseudotuberculosis. Mol. Microbiol. 31:291-303. [DOI] [PubMed] [Google Scholar]
- 19.Hedges, R. W. 1975. R factors from Proteus mirabilis and Proteus vulgaris. J. Gen. Microbiol. 87:301-331. [DOI] [PubMed] [Google Scholar]
- 20.Hedges, R. W. 1974. R factors from Providence. J. Gen. Microbiol. 81:171-181. [DOI] [PubMed] [Google Scholar]
- 21.Hochhut, B., J. W. Beaber, R. Woodgate, and M. K. Waldor. 2001. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 183:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 24.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemper, B. 1999. Bacteriophages as models for differentiation, p. 602-626. In J. W. Lengeler, G. Drews, and H. G. Schlegel (ed.), Biology of prokaryotes. Blackwell Science, Oxford, United Kingdom.
- 26.Krishnapillai, V. 1996. Horizontal gene transfer. J. Genet. 75:219-232. [Google Scholar]
- 27.Kulaeva, O. I., J. C. Wootton, A. S. Levine, and R. Woodgate. 1995. Characterization of the umu-complementing operon from R391. J. Bacteriol. 177:2737-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon, H. J., R. Tirumalai, A. Landy, and T. Ellenberger. 1997. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science 276:126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leong, J. M., S. E. Nunesduby, A. B. Oser, C. F. Lesser, P. Youderian, M. M. Susskind, and A. Landy. 1986. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J. Mol. Biol. 189:603-616. [DOI] [PubMed] [Google Scholar]
- 30.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 31.Liu, X. Y., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthew, M., R. W. Hedges, and J. T. Smith. 1979. Types of β-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 138:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Y. Du, S. F. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 34.Merlin, C., J. Mahillon, J. Nešvera, and A. Toussaint. 2000. Gene recruiters and transporters: the modular structure of bacterial mobile elements, p. 363-409. In C. M. Thomas (ed.), The horizontal gene pool—bacterial plasmids and gene spread. Harwood Academic, Amsterdam, The Netherlands.
- 35.Moss, J. E., T. J. Cardozo, A. Zychlinsky, and E. A. Groisman. 1999. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 33:74-83. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, D. B., and J. T. Pembroke. 1999. Monitoring of chromosomal insertions of the IncJ elements R391 and R997 in Escherichia coli K-12. FEMS Microbiol. Lett. 174:355-361. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, D. B., and J. T. Pembroke. 1995. Transfer of the IncJ plasmid R391 to recombination deficient Escherichia coli K12: evidence that R391 behaves as a conjugal transposon. FEMS Microbiol. Lett. 134:153-158. [DOI] [PubMed] [Google Scholar]
- 38.Nugent, M. E. 1981. A conjugative ‘plasmid' lacking autonomous replication. J. Gen. Microbiol. 126:305-310. [DOI] [PubMed] [Google Scholar]
- 39.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 40.Osborn, A. M., K. D. Bruce, D. A. Ritchie, and P. Strike. 1996. The mercury resistance operon of the IncJ plasmid pMERPH exhibits structural and regulatory divergence from other Gram-negative mer operons. Microbiology 142:337-345. [DOI] [PubMed] [Google Scholar]
- 41.Osborn, A. M., K. D. Bruce, P. Strike, and D. A. Ritchie. 1997. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operons. FEMS Microbiol. Rev. 19:239-262. [DOI] [PubMed] [Google Scholar]
- 42.Page, D. T., K. F. Whelan, and E. Colleran. 1999. Mapping studies and genetic analysis of transfer genes of the multiresistant IncHI2 plasmid, R478. FEMS Microbiol. Lett. 179:21-29. [DOI] [PubMed] [Google Scholar]
- 43.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. G. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 44.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 45.Pembroke, J. T., and D. B. Murphy. 2000. Isolation and analysis of a circular form of the IncJ conjugative transposon-like elements, R391 and R997: implications for IncJ incompatibility. FEMS Microbiol. Lett. 187:133-138. [DOI] [PubMed] [Google Scholar]
- 46.Pembroke, J. T., and E. Stevens. 1984. The effect of plasmid R391 and other IncJ plasmids on the survival of Escherichia coli after UV irradiation. J. Gen. Microbiol. 130:1839-1844. [DOI] [PubMed] [Google Scholar]
- 47.Pembroke, J. T., E. Stevens, J. A. Brandsma, and P. Van de Putte. 1986. Location and cloning of the ultraviolet-sensitizing function from the chromosomally associated IncJ group plasmid, R391. Plasmid 16:30-36. [DOI] [PubMed] [Google Scholar]
- 48.Peters, S. E., J. L. Hobman, P. Strike, and D. A. Ritchie. 1991. Novel mercury resistance determinants carried by IncJ plasmids pMERPH and R391. Mol. Gen. Genet. 228:294-299. [DOI] [PubMed] [Google Scholar]
- 49.Pierson, L. S., and M. L. Kahn. 1987. Integration of satellite bacteriophage P4 in Escherichia coli—DNA sequences of the phage and host regions involved in site-specific recombination. J. Mol. Biol. 196:487-496. [DOI] [PubMed] [Google Scholar]
- 50.Prère, M.-F., M. Chandler, and O. Fayet. 1990. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J. Bacteriol. 172:4090-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prüβ, B. M., D. Markovic, and P. Matsumura. 1997. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J. Bacteriol. 179:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prüβ, B. M., and P. Matsumura. 1997. Cell cycle regulation of flagellar genes. J. Bacteriol. 179:5602-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prüβ, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravatn, R., S. Studer, A. J. B. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romine, M. F., L. C. Stillwell, K.-K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 57.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L.-Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 59.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schubert, S., A. Rakin, D. Fischer, J. Sorsa, and J. Heesemann. 1999. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol. Lett. 179:409-414. [DOI] [PubMed] [Google Scholar]
- 62.Scott, J. R., and G. G. Churchward. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49:367-397. [DOI] [PubMed] [Google Scholar]
- 63.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500 kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Syvanen, M. 1994. Horizontal gene-transfer—evidence and possible consequences. Annu. Rev. Genet. 28:237-261. [DOI] [PubMed] [Google Scholar]
- 66.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trieu-Cuot, P., and P. Courvalin. 1984. Nucleotide sequence of the transposable element IS15. Gene 30:113-120. [DOI] [PubMed] [Google Scholar]
- 69.Vokes, S. A., S. A. Reeves, A. G. Torres, and S. M. Payne. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33:63-73. [DOI] [PubMed] [Google Scholar]
- 70.Wang, T. C., B. de Saint Phalle, K. L. Millman, and R. G. Fowler. 1996. The ultraviolet-sensitizing function of plasmid R391 interferes with a late step of postreplication repair in Escherichia coli. Mutat. Res. 362:219-226. [DOI] [PubMed] [Google Scholar]
- 71.Yang, W., and K. Mizuuchi. 1997. Site-specific recombination in plane view. Structure 5:1401-1406. [DOI] [PubMed] [Google Scholar]
- 72.Yokota, T., and S. Kuwahara. 1977. Temperature-sensitive R plasmid obtained from naturally isolated drug-resistant Vibrio cholerae (biotype El Tor). Antimicrob. Agents Chemother. 11:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool—bacterial plasmids and gene spread. Harwood Academic, Amsterdam, The Netherlands.
- 74.Zhao, S. H., and K. P. Williams. 2002. Integrative genetic element that reverses the usual target gene orientation. J. Bacteriol. 184:859-860. [DOI] [PMC free article] [PubMed] [Google Scholar]