Abstract
Bacteriophage N4 middle genes are transcribed by a phage-coded, heterodimeric, rifampin-resistant RNA polymerase, N4 RNA polymerase II (N4 RNAPII). Sequencing and transcriptional analysis revealed that the genes encoding the two subunits comprising N4 RNAPII are translated from a common transcript initiating at the N4 early promoter Pe3. These genes code for proteins of 269 and 404 amino acid residues with sequence similarity to the single-subunit, phage-like RNA polymerases. The genes encoding the N4 RNAPII subunits, as well as a synthetic construct encoding a fusion polypeptide, have been cloned and expressed. Both the individually expressed subunits and the fusion polypeptide reconstitute functional enzymes in vivo and in vitro.
Sequence and structural analysis of DNA-dependent RNA polymerases (RNAPs) conclusively supports the existence of two enzyme families. One family consists of the large, evolutionarily related, multisubunit polymerases of eubacteria, archaea, and the nuclear and chloroplast polymerases of eukaryotes (reference 56 and references therein). Single-subunit enzymes with sequence homology to the T3/T7 phage polymerases belong to the second family of DNA-dependent RNAPs (38).
Transcription of coliphage N4 middle mRNAs requires the activities of three early proteins: p17, p7, and p4 (53, 70, 72). Two of these gene products, p7 (30 kDa) and p4 (40 kDa), are soluble, have been purified to homogeneity, and constitute a heterodimeric, rifampin-resistant RNAP, N4 RNAPII (71). However, this heterodimer does not transcribe promoter-containing, double-stranded DNAs (dsDNA) and utilizes single-stranded DNAs (ssDNA) with low efficiency and no specificity. At least one additional phage-coded protein, p17, is essential in vivo for N4 middle RNA synthesis but is not sufficient in vitro for utilization of N4 middle promoters (1, 70; R. H. Carter, unpublished data). Recent in vitro characterization of p17 indicates that p17 is a ssDNA binding protein that recruits N4 RNAPII to ssDNA templates (R. H. Carter and A. Demidenko, unpublished data).
In the present study, the genes encoding the p7 and p4 subunits of N4 RNAPII were sequenced and cloned. Both genes are transcribed from the same phage early promoter. The two subunits display sequence similarity to separate, nonoverlapping regions of single-subunit, T7-like RNAPs, suggesting that gene fusion or gene splitting events have occurred during the evolution of this class of polymerases. An N4 fusion polypeptide was generated that is active in vitro and complements phages carrying mutations in either of the two subunits when expressed in vivo.
MATERIALS AND METHODS
Bacterial strains and phages.
Escherichia coli K-12 strains W3350 [F− galK2 galT22 λ− IN(rrnD-rrnE)1] and W3350supF were used in N4 wild-type and N4am mutant phage infections, respectively (53). Strain TG1 [Δ(lac-proAB) supE thi Δ(hsdM-mcrB)5/F′ traD36 proA+B+ lacIq ΔlacZM15] was used for isolation of single-stranded and RF M13 DNA. HMS174 (F− hsdR rK12− mK12+ recA1) was used in the construction of expression vectors. Protein expression was carried out in either BL21(DE3) (62) or W3350(DE3)pLysS (11). Phage were grown as described previously (53).
Preparation and manipulation of DNAs.
N4 DNA was prepared according to the work of VanderLaan et al. (64). DNA amplification reaction mixtures contained 1 μg of template DNAs, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.01% gelatin, 0.2 mM deoxynucleoside triphosphates, 1 μM primers, and 2.5 U of AmpliTaq polymerase (Perkin-Elmer Cetus) in a final volume of 100 μl. DNA sequences of PCR-generated products were confirmed by sequencing.
DNA sequencing and sequence analysis.
DNA fragments generated by HpaI digestion of N4 genomic DNA and cloned into pBR322 (35) were transferred to M13mp18 or M13mp19. Viral ssDNA was isolated and sequenced by the chain termination technique using Sequenase T7 DNA polymerase (version 2.0) and reagents purchased from United States Biochemical Corp. (USB). DNA sequences were determined for both strands. Oligonucleotides used for sequencing and cloning were synthesized at the University of Chicago Howard Hughes Medical Institute Oligonucleotide Facility or purchased from Operon Technologies, Inc. (Alameda, Calif.).
DNA sequences were translated and compared to available sequence databases by using the BlastP program (2). Further regions of sequence similarity were identified by comparison of the p7 and p4 polypeptide sequences to an alignment of 26 T7-like single-subunit RNAPs generated in this laboratory by using the ClustalX program (22).
Construction of N4 RNAPII expression plasmids.
All cloning and transformations followed standard techniques (54). A DNA fragment corresponding to nucleotides (nt) 4606 to 6674 of the N4 genome, which comprise ORF15 (p7 subunit) and ORF16 (p4 subunit), was generated by PCR amplification using the 5′ oligonucleotide primer 5′-GGAATTCCATGGGGTCTACTATCGAACATCAG-3′ and the 3′ primer 5′-CGGGATCCCGTTAAGATAGAGCGTATTCGG-3′. The 5′ primer introduces EcoRI and NcoI sites upstream of the p7 coding region and a single glycine codon (GGG) between the first two naturally occurring codons of the p7 gene. The 3′ primer introduces a BamHI site downstream of the p4 coding region. The PCR product was digested with EcoRI and BamHI (New England Biolabs) and ligated into M13mp18. After confirmation of the DNA sequence, M13 RF DNA was digested with NcoI and BamHI to release the fragment containing ORF15 and ORF16, which was cloned into the pET11d expression vector (Novagen) under the control of a T7/lacO promoter.
Three different expression plasmids were constructed by using the above oligonucleotides and either wild-type or mutant N4 phage genomic DNAs as PCR templates. Plasmid pSH+ contains a fragment generated by using wild-type N4 phage DNA as a template, pSHam15 contains a fragment generated by using N4am15 DNA as a template, and pSHam23 contains a fragment generated by using N4am23 DNA as a template.
To create a p7/p4 fusion polypeptide, mutagenesis was performed on M13mp18 containing the wild-type N4 PCR fragment by using the T7-Gen in vitro mutagenesis kit (USB). The wild-type N4 PCR fragment includes the p7 and p4 coding sequences separated by a 47-bp intergenic region. This intergenic region and the termination codon for ORF15 were precisely deleted. The oligonucleotide used to create the deletion was complementary to nt 5398 to 5409 and 5460 to 5478 of the N4 genome and had the sequence 5′-GGGCAGTAAAAGTTTGCATATCAATTACTTC-3′. Removal of the intergenic region from M13mp18 was verified by restriction digestion. The desired fragment was isolated and cloned into pET11d to create plasmid pSHF.
Labeling of proteins after induction.
E. coli W3350(DE3)/pLysS bearing the N4 RNAPII expression vectors described above was grown at 37°C to an optical density at 620 nm of 0.5 in minimal salts medium (40) supplemented with 4% Lenox L Broth (LB) (Gibco-BRL), 1% Casamino Acids (Difco), and 0.01 mg of thiamine/ml. Cells were collected by centrifugation and resuspended in fresh medium. After addition of 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce expression of T7 RNAP and subsequent transcription of the N4 genes, followed by incubation for 30 min at 37°C, 200 μg of rifampin/ml was added to inhibit host transcription. After incubation for 90 min at 37°C, 50-μl samples were labeled with 5 μl of Tran35S-label (>1,000 Ci/mmol; ICN Radiochemicals, Irvine, Calif.) for 10 min at 37°C. Cells were collected by centrifugation and processed as described previously (14). Samples were analyzed by electrophoresis on sodium dodecyl sulfate (SDS)-12% polyacrylamide gels followed by autoradiography.
Enzyme purification.
Recombinant N4 RNAPII subunits were expressed from plasmid pSH+ in E. coli BL21(DE3) cells. Cells were grown in a Lab-Line Hi-Density fermentor at 37°C with agitation and aeration to an optical density at 600 nm of 0.6, at which time IPTG was added to 0.4 mM. Induction proceeded for 3 h. Cells were collected by centrifugation and resuspended in 50 mM NaCl-50 mM Tris-HCl (pH 7.9)-2 mM EDTA-1 mM dithiothreitol (DTT)-1 mM phenylmethylsulfonyl fluoride (Sigma). All the following procedures were carried out at 4°C. Cells were lysed by sonication after one cycle of freezing and thawing. After removal of cell debris by centrifugation, chromosomal DNA was precipitated with 0.6% polyethylenimine-0.6 M NaCl. Proteins precipitating between 30 and 65% ammonium sulfate were resuspended in buffer A-10 (10 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 1 mM EDTA, 1 mM DTT, and 10% [vol/vol] glycerol) and applied to a heparin Sepharose CL-6B column (25 ml of resin/liter of cell culture; Amersham-Pharmacia) equilibrated in the same buffer. After a wash with 2 column volumes of 150 mM NaCl in A-10 buffer, bound protein was eluted with a 7-column-volume linear gradient of 150 to 450 mM NaCl in buffer A-10. Fractions containing RNAP activity were pooled and applied to a Biogel HTP hydroxyapatite column (8 ml of resin/liter of cell culture; Bio-Rad). Bound protein was eluted with a 10-column-volume linear gradient of 10 to 80 mM sodium phosphate buffer (pH 7.9)-10 mM MgCl2-1 mM EDTA-1 mM DTT-10% (vol/vol) glycerol. Transcriptionally active fractions were pooled and applied to a Q-Sepharose Fast Flow column (15 ml/liter of cell culture; Amersham-Pharmacia). Bound protein was eluted with a 9-column-volume linear gradient of 50 to 500 mM NaCl in buffer A-10. Fractions were pooled, dialyzed overnight against buffer A-10 containing 50% glycerol, and stored at −80°C.
The fusion polymerase, expressed from plasmid pSHF, was purified in the same manner as heterodimeric N4 RNAPII, with the exception that the pooled hydroxyapatite fractions were purified by using a similar gradient profile on a Mono-Q HR5/5 column (Amersham-Pharmacia) instead of the Q-Sepharose Fast Flow column.
In vitro N4 RNAPII assays.
The in vitro transcription assay used to detect N4 RNAPII activity during purification was performed as described previously (1, 70). In the presence of rifampin, this assay allows for the specific identification of RNAPII during purification from lysates that also contain E. coli RNAP. The DNA/membrane complex isolated from N4am15/23-infected cells provides template-bound p17 required for efficient transcription (1).
In vitro runoff transcription reaction mixtures contained 10 mM MgCl2, 0.1 mM EDTA, 1 mM (each) ATP, CTP, and GTP, 0.1 mM UTP, [α-32P]UTP (1,000 Ci/mmol; Amersham-Pharmacia), 100 μg of bovine serum albumin/ml, 20 mM Tris-HCl (pH 8.0), 75 nM poly(dC)-tailed template, and either purified heterodimeric or fused N4 RNAPII. Reaction mixtures were preincubated at 37°C for 5 min before addition of the four ribonucleoside triphosphates. Reactions were allowed to proceed for 5 min at 37°C before quenching by addition of 1.5 volumes of Sequenase Stop Buffer (USB) and analysis on 8% polyacrylamide-8 M urea gels.
To construct poly(dC)-tailed templates, DNA including the HpaI-HaeIII Mc fragment of the N4 genome (35) was generated by PCR amplification. One of the amplification primers used contains a KpnI site that, after digestion of the PCR product with KpnI, provides a preferred substrate for tailing by terminal transferase. Addition of poly(dC) tails was performed in a 50-μl reaction mixture containing 2 mM CoCl2, 0.2 mM DTT, 0.1 M potassium cacodylate (pH 7.2), 15 pmol of KpnI-PCR product, 20 μM dCTP, and 30 U of terminal transferase (Gibco-BRL). Reaction mixtures were incubated at 37°C for 1 h and terminated by phenol extraction and ethanol precipitation. The precipitated template was resuspended in RNase-free 10 mM Tris-HCl (pH 8.0)-1 mM EDTA buffer for use in transcription reactions. The resulting dC-tailed template contains a 294-bp dsDNA segment.
Plating efficiency.
Bacterial strains were grown in brain heart infusion broth (Becton Dickinson) to a density of 3 × 108 cells/ml. One hundred microliters of N4 phage stocks diluted in 15 mM Tris-HCl (pH 8.0)-10 mM MgCl2 was mixed with 0.1 ml of bacterial culture. The mixtures were incubated for 10 min at 37°C to allow for phage adsorption, plated on LB agar, and incubated at 37°C. In work with bacterial strains containing expression vectors, IPTG (0.4 mM) and ampicillin (0.2 mg/ml) were added to the top agar at the time of plating. Phage mutations were considered rescued when the wild-type PFU/total PFU ratio (efficiency of rescue) obtained after plating on a nonsuppressing host was 100- to 1,000-fold higher than the ratio obtained by using mutant phage stocks grown in a host containing a control plasmid (35).
Nucleotide sequence accession numbers.
The nucleotide sequences presented in this paper can be found in GenBank under accession numbers AY074660 and AY074661.
RESULTS
Identification of N4 ORFs encoding the subunits of N4 RNAPII.
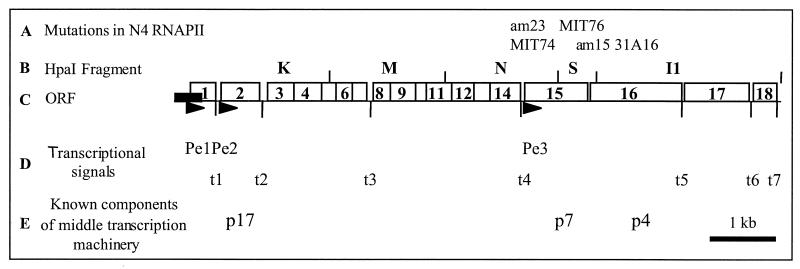
Restriction fragments comprising the majority of the 70.6-kbp N4 genome have been cloned into vector pBR322 or pOP203 (35). We sequenced the region spanning nt 960 to 7997, which is downstream from the previously sequenced XbaI F fragment (42). Together these sequences code for 18 potential open reading frames (ORFs), which are diagrammed in Fig. 1. The locations of the three early promoters, Pe1, Pe2, and Pe3 (19), and of transcriptional terminators t1 through t7 are indicated. Mutations known to lie within RNAPII subunits p7 and p4 were mapped within this region by a marker rescue assay, which measures the ability of mutant phage to recombine with an episomal copy of a N4 DNA fragment to produce wild-type progeny (Table 1). Three amber mutations in p7 were analyzed. N4am23 and N4MIT74 phages were rescued after growth in E. coli W3350 bearing plasmid pBR(N), while N4MIT76 phage was rescued by pBR(S), indicating that the gene encoding p7 spans N4 fragments HpaI N and HpaI S. ORF15 also spans these two fragments, strongly suggesting that p7 is encoded by ORF15 (Fig. 1). Two amber mutations in p4, am15 and 31A16, were rescued by plasmids pBR(S) and pBR(I1), respectively, indicating that ORF16 must encode p4 (Fig. 1).
FIG. 1.
Organization of the leftmost 8 kb of the bacteriophage N4 genome. (A) Mutations within RNAPII subunits. (B) HpaI restriction map of the region. (C) Positions of the potential 18 ORFs predicted by sequence analysis. The solid rectangle indicates the position and length of the genomic terminal repeat. (D) Positions of the early promoters Pe1, Pe2, and Pe3 are indicated by solid arrowheads. Transcription terminators are designated t1 to t7. (E) Polypeptides required for middle gene transcription.
TABLE 1.
Rescue of amber mutant phage by cloned fragments of N4 DNA
| Plasmid | Fragment | Mutation | EORa | RFb (10−5) |
|---|---|---|---|---|
| pBR (I1) | HpaI I1 | 31A16 | 5.0 × 10−3 | 1.0 |
| pBR (N) | HpaI N | am23 | 4.3 × 10−2 | 3.1 |
| pBR (N) | HpaI N | MIT74 | 2.0 × 10−2 | 5.4 |
| pBR (S) | HpaI S | am15 | 8.6 × 10−3 | 7.0 |
| pBR (S) | HpaI S | MIT76 | 1.0 × 10−2 | 3.0 |
EOR, efficiency of rescue, calculated as described in Materials and Methods.
RF, reversion frequency, defined as the ratio of the phage titer on W3350 to the phage titer on W3350supF.
A polycistronic transcript initiating at promoter Pe3 (19) that extends through ORFs 15 and 16 has been observed (L. L. Haynes, unpublished data). ORF15 encodes a 269-amino-acid (aa) polypeptide with a calculated molecular mass of 31,501 Da, as expected based on this protein's migration on denaturing gels. A ribosome-binding site with the sequence 5′-GGAG-3′ is located 8 bp upstream of the translation start site (AUG). The am23 mutation was identified by sequencing as a CAG (Gln)-to-TAG (amber) transition at nt 73 (aa 25) in ORF15, verifying that this ORF encodes p7. ORF16 encodes a 404-aa polypeptide with a calculated molecular mass of 46,225 Da, which is larger than the apparent 40-kDa molecular mass determined by SDS-polyacrylamide gel electrophoresis (PAGE). An intergenic region of 47 nt lies between the end of ORF15 and the start of ORF16. A ribosome binding site (5′-GAGGA-3′) is located 7 bp upstream of the translation start site (AUG) of ORF16. The am15 mutation was identified as a TGG (Trp)-to-TAG transition at nt 176 (aa 59) of the ORF, verifying that ORF16 encodes p4.
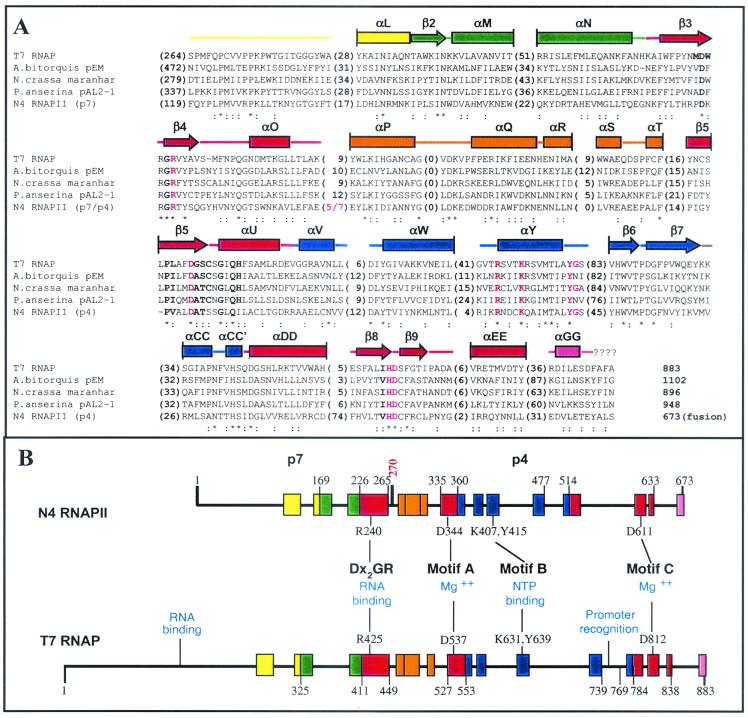
The deduced amino acid sequences encoded by ORFs 15 and 16 were independently compared to the National Center for Biotechnology Information database by using the BlastP program (2). Both p7 and p4 sequences display similarity to separate, nonoverlapping regions of single-subunit, DNA-directed RNAPs. These RNAPs include phage-encoded enzymes, nuclear-encoded mitochondrial enzymes, and linear-plasmid-encoded enzymes. The p7 and p4 sequences were next joined to create a hypothetical “fusion polypeptide” of 673 aa. The fusion joins these sequences in the order in which ORFs 15 and 16 are found in the N4 genome. When the hypothetical fusion polypeptide sequence was subjected to BlastP analysis, significance scores obtained for matches to single-subunit RNAPs were much better than those obtained by using the individual subunits.
The N4 RNAPII sequence was compared to an alignment of 26 single-subunit RNAPs in order to identify additional regions of sequence similarity. An alignment of the p7/p4 fusion polypeptide sequence to that of T7 RNAP and the three highest-scoring (by BlastP) linear-plasmid-encoded enzymes (Fig. 2A) reveals 13 blocks of conserved sequence (Fig. 2B). These correspond approximately to blocks II to X defined for this family of enzymes by Li et al. (33). No match was detected to the most amino-terminal block I. The alignment includes four conserved motifs (DxxGR, A, B, and C) essential for polymerase activity (59). The N4 RNAPII sequence contains all invariant residues located within these motifs, which in T7 RNAP are R425, D537, R627, K631, Y639, H811, and D812 (Fig. 2A) (59).
FIG. 2.
N4 RNAPII is a member of the single-subunit family of RNAPs. (A) Alignment of N4 RNAPII sequence to conserved sequence domains of single-subunit RNAPs. Alignment to three linear-plasmid-encoded fungal RNAPs that are most closely related to N4 RNAPII (as determined by BlastP analysis) and to T7 RNAP is presented. Sequences are ordered relative to their degree of similarity, with the Podospora anserina enzyme being most similar to N4 RNAPII. Conserved sequence blocks correspond approximately to those previously defined by McAllister and Raskin (38) and Li et al. (33). Numbers in parentheses are numbers of residues occurring between sequence blocks. The p7/p4 junction is marked in red. Asterisks indicate positions at which the identical amino acid is found in all five sequences. Colons indicate positions at which amino acids possessing similar physicochemical properties are found in at least four of the sequences. The sequences of motifs essential for polymerase activity are boldfaced. Residues experimentally shown to be important for polymerase activity are in red (6, 25, 44). The corresponding secondary-structure elements, as determined from the T7 RNAP crystal structure (27), are shown above the alignment. Rectangles, alpha helices; arrows, beta strands; lines, coil regions. Structure elements are color coded as in the work of Jeruzalmi and Steitz (27). Vertical lines mark elements that continue beyond the sequence blocks shown. The RNAP sequences shown have the following accession numbers: T7, CAA24390; Agaricus bitorquis, P33539; Neurospora crassa, P33540; P. anserina, S26945; N4 RNAPII p7, AY074660; N4 RNAPII p4, AY074661. (B) Comparison of N4 RNAPII and T7 RNAP. Relative locations of conserved sequence blocks (see panel A) are schematized. Numbers indicate the known boundaries of structural domains in T7 RNAP (27) and inferred boundaries in N4 RNAPII. Numbering in N4 RNAPII reflects the positions of these blocks in the p7/p4 fusion polymerase sequence. Residue 270 corresponds to aa 1 of p4. Blocks are colored according to the designations of Jeruzalmi and Steitz (27): yellow, N-terminal domain; green, thumb; red, palm; orange, palm insertion domain; blue, fingers; pink, foot module. The locations and functional roles of polymerase motifs and catalytically important residues within them are indicated.
Characterization of recombinant N4 RNAPII and of a fusion polypeptide.
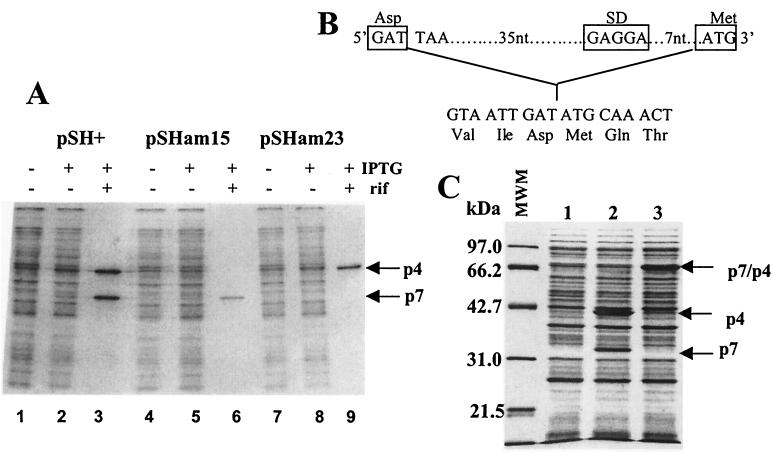
Four plasmids were constructed to express various combinations of wild-type and mutant p7 and p4 subunits. All plasmids contain the p7 and p4 coding region cloned downstream of a T7 promoter and the Shine-Dalgarno (SD) sequence of phage T7 gene 10. Translation initiation of p7 is directed by the vector-coded SD sequence, whereas translation initiation of p4 is directed by its naturally occurring SD sequence. Two polypeptides of approximately 31 and 40 kDa are visible in cells carrying pSH+, which encodes wild-type copies of both subunits, after induction of T7 RNAP expression and subsequent transcription of the N4 genes (Fig. 3A, lane 3). pSHam15 encodes a p4 subunit containing the am15 mutation; only the lower-molecular-weight polypeptide, p7, is expressed from this plasmid (Fig. 3A, lane 6). Conversely, pSHam23 encodes a p7 subunit containing the am23 mutation, and consequently, only the higher-molecular-weight polypeptide, p4, is expressed upon induction (Fig. 3A, lane 9). Plasmid pSHF encodes a synthetic fusion of the p7 and p4 genes (ORF15 and ORF16) in the order in which they occur in the genome. A 50-bp intergenic region containing the ORF15 termination codon and the SD sequence upstream of ORF16 was deleted to create a fusion junction that lies between the terminal residue of ORF15 (aa 269) and the initiating methionine of ORF16 (Fig. 3B). Expression of a 70-kDa polypeptide is clearly observed in IPTG-induced cells (Fig. 3C, lane 3).
FIG. 3.
Expression of plasmid-borne N4 ORF15 and ORF16. (A) Recombinant RNAPII subunits were synthesized in vivo in the presence of [35S]methionine and analyzed by denaturing gel electrophoresis and autoradiography. Lanes 1 to 3, cell lysates from W3350(DE3)pLysS carrying pSH+; lanes 4 to 6, lysates from W3350(DE3)pLysS carrying pSHam15; lanes 7 to 9, lysates from W3350(DE3)pLysS cells carrying pSHam23. Arrows indicate the positions of p4 and p7. (B) Sequence of the ORF15 and ORF16 fusion junction. A diagram of the intergenic region removed by site-specific mutagenesis is shown at the top. (C) Coomassie-stained gel of recombinant protein expression. Lane 1, W3350(DE3)pLysS cells containing pET11d; lane 2, expression of p7 and p4 from pSH+; lane 3, expression of the p7/p4 fusion polymerase from pSHF. Sizes of protein standards are indicated at the left of the gel. Arrows indicate the positions of proteins of the expected sizes.
N4 phages containing mutations within N4 RNAPII were tested for growth on E. coli cells expressing the recombinant p7 and p4 subunits (Table 2). In some cases, the presence of a plasmid in the host cells reduces the efficiency of plating of N4. This depression of phage production in plasmid-carrying cells may result from competition by the abundantly produced, chromosomally encoded T7 RNAP for a limited nucleotide pool. Complementation of the phage mutations is apparent despite this underlying effect. N4am15 phage cannot grow in E. coli carrying either the empty expression vector (pET11d) or a plasmid expressing only p7 (pSHam15); the plating efficiencies are <10−7 and <10−9, respectively. N4am15 phage do plate on cells expressing p7 and p4 (pSH+), p4 alone (pSHam23), or the fusion polymerase (pSHF). Similarly, N4am23 phage cannot grow in E. coli carrying pET11d or a plasmid expressing only p4, as evidenced by plating efficiencies of <10−6 and <10−7, respectively. N4am23 phage plate on cells expressing p7 and p4 (pSH+), p7 alone (pSHam15), or the fusion polymerase (pSHF). When N4am15 or N4am23 phage are grown on bacteria bearing pSH+, wild-type progeny phage are recovered at frequencies of approximately 10−3 (data not shown). Therefore, the much larger increases in plating efficiency observed in this assay are due to complementation rather than recombination between the mutant phage and expression plasmids. Furthermore, complementation occurs even if one of the polymerase subunits is produced by the phage and its partner is produced from a plasmid. Therefore, the independently produced subunits are still able to associate and reconstitute active enzyme.
TABLE 2.
Plating efficiencies of N4 phages on various E. coli strains
| Strain/plasmid | Efficiency of platinga
|
|||
|---|---|---|---|---|
| N4 (WT) | N4am15 | N4am23 | N4am15/23 | |
| W3350supF | 1.00 | 1.00 | 1.00 | 1.00 |
| W3350 | 0.42 | 1.5 × 10−7 | 6.3 × 10−7 | 1.4 × 10−10 |
| W(DE3)plysS/pET11d | 0.24 | 3.9 × 10−8 | 7.1 × 10−7 | 1.3 × 10−10 |
| W(DE3)plysS/pSH+ | 0.25 | 0.27 | 0.19 | 0.13 |
| W(DE3)plysS/pSHam15 | 0.22 | <8.6 × 10−10 | 0.25 | <1.3 × 10−9 |
| W(DE3)plysS/pSHam23 | 0.31 | 0.25 | <1.1 × 10−8 | <1.3 × 10−9 |
| W(DE3)plysS/pSHF | 0.05 | 0.16 | 0.16 | 0.03 |
Calculated by dividing the number of PFU obtained on a given strain by the number of PFU obtained on W3350supF. WT, wild type.
The double mutant N4am15/23 phage cannot grow in E. coli carrying either pET11d or plasmids expressing the polymerase subunits singly (pSHam15 or pSHam23); the plating efficiencies observed are <10−9. These phage plate only on cells producing p7 and p4 (pSH+) or the fusion polymerase (pSHF). These results confirm that both RNAPII subunits are essential for N4 phage growth, and they demonstrate that the two subunits retain activity when combined in a single polypeptide in vivo. Therefore, the fusion polymerase is able to provide all functions necessary for production of viable phage.
The N4 RNAPII fusion polymerase is active in vitro.
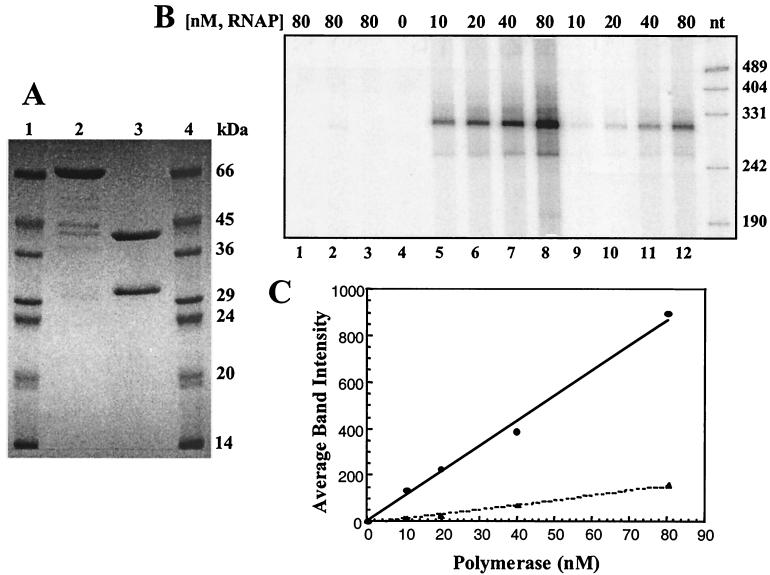
N4 RNAPII activity was purified from cells bearing either pSH+ or pSHF after induction of recombinant protein expression. The enzyme purified from cells bearing pSH+ consists of two polypeptides of sizes similar to those of the enzyme purified from infected cells (71). The enzymes from both sources have the same specific activity. A single polypeptide migrating as a species of approximately 70 kDa is obtained after purification from cells carrying pSHF (Fig. 4A). This is lower than the calculated molecular mass of 77, 726 Da but is in agreement with the anomalous migration of p4.
FIG. 4.
Relative specific activities of the heterodimeric N4 RNAPII and the p7/p4 fusion polymerase. (A) SDS-PAGE of protein samples used in transcription. Lanes 1 and 4, molecular weight markers; lane 2, purified p7/p4 fusion polymerase; lane 3, purified N4 RNAPII heterodimer. (B) Transcripts produced in vitro by the N4 RNAPII heterodimer (lanes 1, 2, and 5 to 8) and the fusion polymerase (lanes 3 and 9 to 12). Reaction mixtures contained either no template (lane 1), 75 nM tailless template (lanes 2 and 3), or 75 nM dC-tailed template (lanes 4 to 12). Polymerase was omitted from the reaction in lane 4. N4 protein p17 was present in all reaction mixtures. Sizes of DNA markers (nucleotides) are indicated to the right of the gel. (C) Quantitation of in vitro transcription products synthesized from dC-tailed templates. The amounts of 294-nt transcript synthesized by the N4 RNAPII heterodimer (solid line) and the fusion polymerase (dotted line) are plotted as a function of enzyme concentration.
The activities of the purified heterodimeric and fusion polymerases were compared in in vitro transcription reactions containing increasing amounts of the two enzymes. Reaction mixtures contained dC-tailed templates because the p7/p4 enzyme cannot utilize dsDNA templates (70; Carter, unpublished). Reaction products were analyzed by PAGE (Fig. 4B) and quantitated (Fig. 4C). In the absence of the template (Fig. 4B, lane 1), polymerase (lane 4), or dC-tailing of the template (lanes 2 and 3), no transcripts were detected. The transcripts produced in the complete reaction (Fig. 4B, lanes 5 to 12) are of the expected length (294 nt), and synthesis is linear with respect to protein concentration. The experiment demonstrates that both purified polymerases are indeed active. We estimate that the fusion polymerase possesses 20% of the activity of the heterodimeric N4 RNAPII based on quantitation data (Fig. 4C).
DISCUSSION
We have sequenced and cloned the two genes that encode the two subunits of N4 RNAPII, the RNAP responsible for transcription of bacteriophage N4 middle genes. Sequence comparison of the N4 RNAPII subunits to proteins in the database indicates that N4 RNAPII, a heterodimer, is homologous to the single-subunit, T7-like RNAP family and that the homology encompasses regions of both subunits p7 and p4. N4 RNAPII, with a sum of 673 amino acids, is one of the smallest members of this family described to date. An ORF (GenBank accession number BAB08093) encoding a 663-aa product which contains good matches to the DxxGR, A, and B motifs but a poor match to the consensus C motif is encoded in the Physarum polycephalum mitochondrial genome. However, at present there is no indication that this gene product is functional.
The T7-like RNAPs belong to a superfamily of single-subunit polymerases (28). The crystal structures of representative superfamily members have been determined; T7 RNAP, members of the PolI and Polα DNA polymerase families, human immunodeficiency virus type 1 (HIV-1) reverse transcriptase, and poliovirus RNA-dependent RNAP all share a common polymerase domain structure (17, 60, 61). This structure resembles a cupped hand in which thumb and fingers subdomains rise on each side of a palm subdomain (31). Sequence alignment of both RNA- and DNA-dependent RNA and DNA polymerases belonging to this superfamily identified two sequence motifs (A and C) containing highly conserved carboxylate residues. Another motif (B) is present only in DNA-dependent polymerases (13). These motifs occupy similar positions in the various structures: motifs A and C, which contain invariant aspartates, lie in the palm. These aspartates coordinate catalytically essential Mg2+ ions (43, 44, 59, 66). Motif B, which contains an invariant lysine, lies in the fingers subdomain and makes up part of the enzyme's nucleotide binding pocket (27). An additional motif common to DNA-directed polymerases [(T/D)xxGR], which lies in the palm subdomain, has also been identified (39). Recent studies indicate that this motif plays a role in stabilizing the RNA-DNA hybrid during early stages of T7 RNAP transcription initiation (25).
The locations of these four motifs and other blocks of sequence similarity in N4 RNAPII and T7 RNAP are diagrammed in Fig. 2B. Blocks of sequence similarity are colored according to their subdomain location in the T7 RNAP crystal structure (27) and are divided between the two N4 RNAPII subunits. Interestingly, the p7/p4 junction (aa 269-270) lies close to the boundary between structural subdomains. The p7 subunit contains sequences corresponding to the amino-terminal domain, the thumb, and a fragment of the palm in which the DxxGR motif is located, while p4 contains the remainder of the palm and the fingers subdomains, and all catalytically important residues within motifs A, B, and C (Fig. 2).
As a result of the high degree of sequence similarity observed, we expect that the overall tertiary structure of N4 RNAPII, as well as many secondary structural elements, will be similar to that of T7 RNAP. On this assumption, we mapped the blocks shown in Fig. 2 on the crystal structure of T7 RNAP (PDB 1ARO) (data not shown). Based on this analysis, we can make several predictions. We predict that the N4 RNAPII palm domain will possess structural elements related to those found in other single-subunit polymerases. A high degree of structural conservation has been observed for the palm domains of T7 RNAP, E. coli Klenow fragment, and HIV-1 reverse transcriptase, and the locations of the motif A and C carboxylates within the palms of these enzymes are essentially identical (29). However, in contrast to these enzymes, the N4 RNAPII sequence contains a long insertion (74 aa relative to 5 aa for T7 RNAP [Fig. 2A]) N-terminal to the sequence block (and by inference, the β hairpin [Fig. 2A]) containing motif C.
We also predict that the N4 RNAPII thumb and fingers subdomains will be shorter than the corresponding regions of T7 RNAP (Fig. 2B and Table 3). Deletions within the T7 RNAP thumb result in a loss of elongation complex stability and processivity (5). Notably, the p7/p4 heterodimer does not appear deficient in processive transcription when initiating on dC-tailed templates (data not shown). The fingers subdomain contains motif B. N4 RNAPII contains an exact match to the consensus sequence Rx3Kx7YG, as do the phage- and nuclear-encoded mitochondrial RNAPs. In contrast, the majority of linear-plasmid-encoded enzymes contain the sequence Rx3Kx7YN. The significance of this sequence change is unknown because functional data are not available for most of the plasmid-encoded enzymes. It is also noteworthy that a serine residue immediately follows motif B in the N4 RNAPII sequence; serine at this position is found only in the phage-encoded polymerases (T7, T3, SP6, and K11) (8, 38). Cross-linking studies performed using T7 RNAP suggested that the motif B lysine plays a role in nucleotide binding (34). The corresponding N4 RNAPII residue (K407) lies in the p4 subunit. Surprisingly, catalytic autolabeling of N4 RNAPII using a cross-linkable derivative of the initiating nucleotide and denatured N4 DNA as a template resulted in labeling of the p7 subunit (18). The exact site of cross-linking of the derivatized nucleotide has not been identified.
TABLE 3.
Comparison of domains between the T7 RNAP and N4 RNAPII
| Domaina | Length (aa) in:
|
|
|---|---|---|
| T7 RNAP | N4 RNAPII | |
| N terminus | 325 | 169 |
| Thumb | 86 | 57 |
| Palm | 38 | 39 |
| Palm insertion | 78 | 70 |
| Palm | 26 | 25 |
| Fingers | 186 | 117 |
| Specificity loop | 30 | 27 |
| Fingers | 15 | 10 |
| Palm | 54 | 119 |
| Foot domain | 45 | 40 |
The T7 RNAP structure contains a number of regions not found in E. coli Klenow fragment that are believed to confer RNAP-specific functions. These include the amino-terminal domain (aa 1 to 325), the palm insertion module (aa 450 to 527), and the specificity loop (aa 739 to 769) (Fig. 2B) (27). The amino-terminal 20-kDa domain of T7 RNAP (aa 1 to 172) can be cleaved at a protease-sensitive surface loop (12, 23, 41, 60). This fragment is necessary for RNA binding in vitro (41, 55), and T7 RNAP enzymes with mutations in this region display diminished binding of nascent transcripts during the abortive phase of transcription (20, 46). Amino-terminal residues are also implicated in promoter recognition and unwinding (10, 16). The remaining carboxy-terminal 80-kDa proteolytic fragment of T7 RNAP can synthesize short abortive RNA products but is deficient in processive transcription (41). The N4 RNAPII amino-terminal domain is truncated by 156 aa relative to T7 RNAP (Table 3). No sequence similarity was detected N-terminal to T7 RNAP aa 264 (Fig. 2A).
The palm insertion module closes off the back of the active site; its exact function remains to be determined. A segment of similar length and sequence is conserved in all T7-like RNAPs, including N4 RNAPII (Fig. 2B, orange blocks) (27). Residues within the specificity loop have been implicated in sequence-specific promoter recognition and interactions with the RNA product (9, 48, 52, 63). N4 RNAPII does contain an insertion corresponding to the location of the specificity loop, though we cannot predict its exact length (in Table 3, we have indicated the distance between the two flanking blocks of fingers sequence). We do not know if this insertion plays an equivalent role in promoter recognition at this time. Inherent promoter recognition is not a universal property of the single-subunit RNAPs. Yeast mitochondrial RNAP is composed of two subunits: a catalytic core (Rpo41) homologous to T7 RNAP and a specificity factor (Mtf1) required for promoter recognition (15, 37, 65). Mtf1 shares sequence similarity with eubacterial sigma factors (26). The similarity extends to its behavior: Rpo41p and Mtf1p interact in solution prior to promoter binding, and Mtf1p is released from the complex following initiation (36). Evidence exists for corresponding specificity factors in higher eukaryotes (24, 45). N4 RNAPII, although it contains the polymerase catalytic core, is unable to utilize promoter-containing DNA templates (71). N4 middle RNA synthesis is dependent on at least one additional N4 gene product, p17 (1, 70). p17 has been purified to homogeneity and characterized (Carter, unpublished). To our surprise, we found that p17 is a ssDNA binding protein that specifically recruits N4 RNAPII to ssDNA (Carter and Demidenko, unpublished). These findings suggest that N4 RNAPII must use a novel mechanism for promoter recognition.
The finding that functional domains contained within one polypeptide in a given species are distributed over more than one polypeptide in another species is not unique to N4 RNAPII. A number of instances involving nucleotide polymerases have been reported in the literature. The Methanobacterium thermoautotrophicum PolB enzyme, which is a member of the single-subunit RNA/DNA polymerase superfamily, is encoded by two ORFs located ∼650 kbp apart and on opposite strands (58). The smaller PolB2 subunit contains a section of the palm and the thumb domain, while the larger PolB1 subunit contains the remainder of the palm and the fingers domain. Evidence suggests that both subunits are required for activity (30). Unlike the arrangement in N4 RNAPII, the DxxGR, A, B, and C motifs are all located in PolB1 (K. M. Kazmierczak, by inspection of GenBank accession numbers AAB85697 and AAB84714). We have searched available sequence databases for other examples of split T7-like RNAPs. A number of examples of fragmented, probably inactive enzymes in mitochondrial genomes of fungi (21, 51) and Beta vulgaris (7) have been described. Others were found by us in the Spizellomyces punctatus mitochondrial chromosome sequence. We did find one candidate pair of ORFs (ORF670 and ORF348, GenBank accession numbers NP_064021 and NP_064020) in the B. vulgaris mitochondrial genome sequence (32). These two ORFs are separated by 41 nt, in the B. vulgaris sequence. ORF670 contains motifs DxxGR and A, while ORF348 contains motifs B and C; all catalytically essential residues are present in the four motifs. Whether these ORFs encode a functional enzyme is unknown.
The conserved sequences present in the β′ subunit of E. coli RNAP are divided between two polypeptides (A and C) in the archaebacteria Sulfolobus acidocaldarius, Halobacterium halobium, and M. thermoautotrophicum (3, 47) and between polypeptides RpoC1 and RpoC2 in cyanobacteria and chloroplasts (4). In H. halobium and M. thermoautotrophicum, the β-subunit sequences are also divided between two polypeptides (B" and B′) (3, 47). In all cases, the naturally occurring breakpoints fall between blocks of conserved sequence.
Monomeric enzymes have also been split experimentally to examine how protein domains function when they are located on separate subunits. Active E. coli RNAP has been assembled from β-subunit fragments (57). In Helicobacter pylori and Wolinella succinogenes, the β and β′ subunits of RNAP are naturally fused (68, 69). Separation of the β and β′ subunits in H. pylori yields viable bacteria that are able to colonize mice (49).
In addition, examples exist of covalent linkage of genes to yield a fusion polypeptide. The second (hisD) and third (hisC) genes of the histidine operon of Salmonella enterica serovar Typhimurium have been fused into a single polypeptide (50, 67). In vitro characterization of the fused polypeptide indicated that, while the activity of the imidazolylacetol phosphate:l-glutamate aminotransferase (HisC) was normal, histidinol dehydrogenase (HisD) activity was more heat labile and had lower affinity for one of its substrates, histidinol, than the wild-type enzyme (50). The p7/p4 fused polypeptide exhibits 20% of the activity of the heterodimer in in vitro transcription elongation assays (Fig. 4), although it fully complements mutations in both polypeptides in vivo (Table 2). We have compared the sequence of the p7/p4 polypeptide to that of T7 RNAP at the N4 fusion junction. The distance between the two blocks of conserved sequence flanking the junction is longer (12 versus 9 aa) in the p7/p4 polypeptide than in T7 RNAP (Fig. 2A and Table 3). This suggests that, if the structure of the N4 RNAPII catalytic domain is similar to that of T7 RNAP, the spatial location of secondary structural elements flanking the junction should not be grossly perturbed in the fusion polymerase relative to the heterodimeric enzyme. However, we cannot rule out the possibility that the observed reduction in activity is caused by potential differences in the length or orientation of such structural elements. Additionally, the flexibility and motion of the surrounding domains, such as large protein rearrangements in the vicinity of the p7/p4 breakpoint during the transition from the initiation complex to a stable elongation complex, may be affected. We do not know if the fused N4 RNAPII polypeptide can tolerate breakpoints in other positions. Whether p7 and p4 evolved from a single polypeptide or whether, conversely, they represent the ancestral state of this class of polymerases is unknown.
ADDENDUM IN PROOF
Cyanophage P60 RNAP is 574 aa, making it the shortest known member of the T7 RNAP family (GenBank accession no. AAL73254).
Acknowledgments
S. H. Willis and K. M. Kazmierczak contributed equally to this work.
We thank Roger Hendrix, Graham Hatfull, and the members of the Pittsburgh Bacteriophage Institute for confirmation of and corrections to the sequence of the N4 genome spanning positions 960 to 7997, and we thank Bill McAllister for many discussions and for providing helpful comments on the manuscript. S.H.W. performed some of the work described in this report while at the laboratory of F. Stahl (University of Oregon, Eugene). We thank him for his hospitality.
This work was supported by NIH grant AI 12575 to L.B.R.-D. K.M.K. and R.H.C. were partially supported by NIH grants T32 GM 07197 and T32 GM 07183, respectively.
REFERENCES
- 1.Abravaya, K., and L. B. Rothman-Denes. 1989. In vitro requirements for N4 RNA polymerase II-specific initiation. J. Biol. Chem. 264:12695-12699. [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berghofer, B., L. Krockel, C. Kortner, M. Truss, J. Schallenberg, and A. Klein. 1988. Relatedness of the archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 16:8113-8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergsland, K. J., and R. Haselkorn. 1991. Evolutionary relationships among eubacteria, cyanobacteria, and chloroplasts: evidence from the rpoC1 gene of Anabaena sp strain 7120. J. Bacteriol. 173:3446-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonner, G., E. M. Lafer, and R. Sousa. 1994. Characterization of a set of T7 RNA polymerase active site mutants. J. Biol. Chem. 269:25120-25128. [PubMed] [Google Scholar]
- 6.Bonner, G., D. Patra, E. Lafer, and R. Sousa. 1992. Mutations in T7 RNA polymerase that support the proposal for a common polymerase active site structure. EMBO J. 11:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahoon, A. B., and D. B. Stern. 2001. Plastid transcription: a menage a trois? Trends Plant Sci. 6:45-46. [DOI] [PubMed] [Google Scholar]
- 8.Cermakian, N., T. M. Ikeda, P. Miramontes, B. F. Lang, M. W. Gray, and R. Cedergren. 1997. On the evolution of the single-subunit RNA polymerases. J. Mol. Evol. 45:671-681. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham, G. M. T., D. Jeruzalmi, and T. A. Steitz. 1999. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature 399:80-83. [DOI] [PubMed] [Google Scholar]
- 10.Cheetham, G. M. T., and T. A. Steitz. 2000. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10:117-123. [DOI] [PubMed] [Google Scholar]
- 11.Cho, N.-Y., M. Choi, and L. B. Rothman-Denes. 1995. The bacteriophage N4 single-stranded DNA binding protein (N4SSB) is the transcriptional activator of E. coli RNA polymerase at N4 late promoters. J. Mol. Biol. 246:461-471. [DOI] [PubMed] [Google Scholar]
- 12.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 81:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delarue, M., O. Poch, N. Tordo, D. Moras, and P. Argos. 1990. An attempt to unify the structure of polymerases. Protein Eng. 3:461-467. [DOI] [PubMed] [Google Scholar]
- 14.Falco, S. C., and L. B. Rothman-Denes. 1979. Bacteriophage N4-induced transcribing activities in E. coli. I. Detection and characterization in cell extracts. Virology 95:454-465. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, R. P., and D. A. Clayton. 1988. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8:3496-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross, L., W. J. Chen, and W. T. McAllister. 1992. Characterization of bacteriophage T7 RNA polymerase by linker insertion mutagenesis. J. Mol. Biol. 228:488-505. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann, G. R., C. Biebricher, S. J. Glaser, F. Grosse, M. J. Katzameyer, A. J. Lindner, H. Mosig, H.-P. Nasheuer, L. B. Rothman-Denes, A. R. Schaffner, G. J. Schneider, K.-O. Stetter, and M. Thomm. 1988. Initiation of transcription- a general tool for affinity labeling of RNA polymerases by autocatalysis. Biol. Chem. Hoppe-Seyler 369:775-788. [PubMed] [Google Scholar]
- 19.Haynes, L. L., and L. B. Rothman-Denes. 1985. N4 virion RNA polymerase sites of transcription initiation. Cell 41:597-605. [DOI] [PubMed] [Google Scholar]
- 20.He, B., M. Rong, R. K. Durbin, and W. T. McAllister. 1997. A mutant T7 RNA polymerase that is defective in RNA binding and blocked in the early stages of transcription. J. Mol. Biol. 265:275-288. [DOI] [PubMed] [Google Scholar]
- 21.Hermanns, J., and H. D. Osiewacz. 1994. Three mitochondrial unassigned open reading frames of Podospora anserina represent remnants of a viral-type RNA polymerase gene. Curr. Genet. 25:150-157. [DOI] [PubMed] [Google Scholar]
- 22.Higgins, D. G., J. D. Thompson,, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, R. A., and C. C. Richardson. 1987. Enzymatic properties of a proteolytically nicked RNA polymerase of bacteriophage T7. J. Biol. Chem. 262:3790-3799. [PubMed] [Google Scholar]
- 24.Ikeda, T. M., and M. W. Gray. 1999. Characterization of a DNA binding protein implicated in transcription in wheat mitochondria. Mol. Cell. Biol. 19:8113-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imburgio, D., M. Anikin, and W. T. McAllister. 2002. A conserved DX2GR motif in T7 RNA polymerase stabilizes the RNA:DNA hybrid during transcription initiation. J. Mol. Biol. 319:37-51. [DOI] [PubMed] [Google Scholar]
- 26.Jang, S. H., and J. A. Jaehning. 1991. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial σ factors. J. Biol. Chem. 266:22671-22677. [PubMed] [Google Scholar]
- 27.Jeruzalmi, D., and T. A. Steitz. 1998. Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J. 17:4101-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyce, C., and T. Steitz. 1994. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 63:777-822. [DOI] [PubMed] [Google Scholar]
- 29.Joyce, C. M., and T. A. Steitz. 1995. Polymerase structures and function: variations on a theme. J. Bacteriol. 177:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelman, Z., S. Pietrokowski, and J. Hurwitz. 1999. Isolation and characterization of a split B-type DNA polymerase from the archaeon Methanobacterium thermoautotrophicum ΔH. J. Biol. Chem. 247:28751-28761. [DOI] [PubMed] [Google Scholar]
- 31.Kohlstaedt, L., J. Wang, J. Friedman, P. Rice, and T. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 32.Kubo, T., S. Nishizawa, A. Sugarawa, N. Itchoda, A. Estiati, and T. Mikami. 2000. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNAcys (GCA). Nucleic Acids Res. 28:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, J., J. A. Maga, N. Cermakian, R. Cedergren, and J. E. Feagin. 2001. Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol. Biochem. Parasitol. 113:261-269. [DOI] [PubMed] [Google Scholar]
- 34.Maksimova, T. G., A. A. Mustayev, E. F. Zaychikov, D. L. Lyakhov, V. L. Tunitskaya, A. K. Akbarov, S. V. Luchin, V. O. Rechinsky, B. K. Chernov, and S. N. Kochetkov. 1991. Lys631 residue in the active site of the bacteriophage T7 RNA polymerase. Affinity labelling and site-directed mutagenesis. Eur. J. Biochem. 195:841-847. [DOI] [PubMed] [Google Scholar]
- 35.Malone, C., S. Spellman, D. Hyman, and L. B. Rothman-Denes. 1988. Cloning and generation of a genetic map of bacteriophage N4 DNA. Virology 162:328-336. [DOI] [PubMed] [Google Scholar]
- 36.Mangus, D. A., S. H. Jang, and J. Jaehning. 1994. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J. Biol. Chem. 269:26568-26574. [PubMed] [Google Scholar]
- 37.Masters, B. S., L. L. Stohl, and D. A. Clayton. 1987. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 51:89-99. [DOI] [PubMed] [Google Scholar]
- 38.McAllister, W. T., and C. A. Raskin. 1993. The phage RNA polymerases are related to DNA polymerases and reverse transcriptases. Mol. Microbiol. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 39.Mendez, J., L. Blanco, J. M. Lazaro, and M. Salas. 1994. Primer-terminus stabilization at the φ29 DNA polymerase active site. J. Biol. Chem. 269:30030-30038. [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Muller, D. K., C. T. Martin, and J. E. Coleman. 1988. Processivity of proteolytically modified forms of T7 RNA polymerase. Biochemistry 27:5763-5771. [DOI] [PubMed] [Google Scholar]
- 42.Ohmori, H., L. L. Haynes, and L. B. Rothman-Denes. 1988. Structure of the ends of the coliphage N4 genome. J. Mol. Biol. 202:1-10. [DOI] [PubMed] [Google Scholar]
- 43.Osumi-Davis, P., N. Sreerama, D. Volkin, C. Middaugh, R. Woody, and A. Woody. 1994. Bacteriophage T7 RNA polymerase and its active-site mutants. Kinetic, spectroscopic and calorimetric characterization. J. Mol. Biol. 237:5-19. [DOI] [PubMed] [Google Scholar]
- 44.Osumi-Davis, P. A., M. C. de Aguilera, R. W. Woody, and A. Y. Woody. 1992. Asp537, Asp812 are essential and Lys631, His811 are catalytically significant in bacteriophage T7 RNA polymerase activity. J. Mol. Biol. 226:37-45. [DOI] [PubMed] [Google Scholar]
- 45.Parisi, M. A., and D. A. Clayton. 1991. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252:965-969. [DOI] [PubMed] [Google Scholar]
- 46.Patra, D., E. M. Lafer, and R. Sousa. 1992. Isolation and characterization of mutant bacteriophage T7 RNA polymerases. J. Mol. Biol. 224:307-318. [DOI] [PubMed] [Google Scholar]
- 47.Puhler, G., H. Leffers, F. Gropp, P. Palm, H.-P. Klenk, F. Lottspeich, R. A. Garrett, and W. Zillig. 1989. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc. Natl. Acad. Sci. USA 86:4569-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raskin, C. A., G. A. Diaz, and W. T. McAllister. 1993. T7 RNA polymerase mutants with altered promoter specificities. Proc. Natl. Acad. Sci. USA 90:3147-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raudonikiene, A., N. Zakharova, W. W. Su, J. Y. Jeong, L. Bryden, P. S. Hoffman, D. E. Berg, and K. Severinov. 1999. Helicobacter pylori with separate β- and β′-subunits of RNA polymerase is viable and can colonize conventional mice. Mol. Microbiol. 32:131-138. [DOI] [PubMed] [Google Scholar]
- 50.Rechler, M. M., and C. B. Bruni. 1971. Properties of a fused protein formed by genetic manipulation. Histidinol dehydrogenase-imidazolylacetol phosphate: l-glutamate aminotransferase. J. Biol. Chem. 246:1806-1813. [PubMed] [Google Scholar]
- 51.Robison, M. M., J. C. Royer, and P. A. Horgen. 1991. Homology between mitochondrial DNA of Agaricus bisporus and an internal portion of a linear mitochondrial plasmid of Agaricus bitorquis. Curr. Genet. 19:495-502. [DOI] [PubMed] [Google Scholar]
- 52.Rong, M., B. He, W. T. McAllister, and R. D. Durbin. 1998. Promoter specificity determinants of T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 95:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothman-Denes, L. B., and G. C. Schito. 1974. Novel transcribing activities in N4-infected Escherichia coli. Virology 60:65-72. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Sastry, S., and B. M. Ross. 1998. RNA-binding site in T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 95:9111-9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Severinov, K. 2000. RNA polymerase structure-function: insights into points of transcriptional regulation. Curr. Opin. Microbiol. 3:118-125. [DOI] [PubMed] [Google Scholar]
- 57.Severinov, K., A. Mustaev, M. Kashlev, S. Borukhov, V. Nikiforov, and A. Goldfarb. 1992. Dissection of the β subunit in the E. coli RNA polymerase into domains by proteolytic cleavage. J. Biol. Chem. 267:12813-12819. [PubMed] [Google Scholar]
- 58.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Chuch, C. J. Daniels, J.-I. Mao, P. Rice, J. Nolling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sousa, R. 1996. Structural and mechanistic relationships between nucleic acid polymerases. Trends Biochem. Sci. 21:186-190. [PubMed] [Google Scholar]
- 60.Sousa, R., Y. J. Chung, J. P. Rose, and B.-C. Wang. 1993. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 Å resolution. Nature 364:593-599. [DOI] [PubMed] [Google Scholar]
- 61.Steitz, T. A. 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274:17395-17398. [DOI] [PubMed] [Google Scholar]
- 62.Studier, W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 63.Temiakov, D., P. E. Mentesana, K. Ma, A. Mustaev, S. Borukhov, and W. T. McAllister. 2000. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl. Acad. Sci. USA 97:14109-14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.VanderLaan, K., S. C. Falco, and L. B. Rothman-Denes. 1977. The program of RNA synthesis in N4-infected Escherichia coli. Virology 76:596-601. [DOI] [PubMed] [Google Scholar]
- 65.Winkley, C. S., M. J. Keller, and J. A. Jaehning. 1985. A multicomponent mitochondrial RNA polymerase from Saccharomyces cerevisiae. J. Biol. Chem. 260:14214-14223. [PubMed] [Google Scholar]
- 66.Woody, A., S. Eaton, P. Osumi-Davis, and R. Woody. 1996. Asp537 and Asp812 in bacteriophage T7 RNA polymerase as metal ion-binding sites studied by EPR, flow-dialysis, and transcription. Biochemistry 35:144-152. [DOI] [PubMed] [Google Scholar]
- 67.Yourno, J., T. Kohno, and J. R. Roth. 1970. Enzyme evolution: generation of a bifunctional enzyme by fusion of adjacent genes. Nature 228:820-824. [DOI] [PubMed] [Google Scholar]
- 68.Zakharova, N., P. S. Hoffman, D. E. Berg, and K. Severinov. 1998. The largest subunits of RNA polymerase from gastric helicobacters are tethered. J. Biol. Chem. 273:19371-19374. [DOI] [PubMed] [Google Scholar]
- 69.Zakharova, N., B. J. Paster, I. Wesley, F. E. Dewhirst, D. E. Berg, and K. V. Severinov. 1999. Fused and overlapping rpoB and rpoC genes in helicobacters, campylobacters, and related bacteria. J. Bacteriol. 181:3857-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zehring, W. A., S. C. Falco, C. Malone, and L. B. Rothman-Denes. 1983. Bacteriophage N4-induced transcribing activities in E. coli. III. A third cistron required for N4 RNA polymerase II activity. Virology 126:678-687. [DOI] [PubMed] [Google Scholar]
- 71.Zehring, W. A., and L. B. Rothman-Denes. 1983. Purification and characterization of coliphage N4 RNA polymerase II activity from infected cell extracts. J. Biol. Chem. 258:8074-8080. [PubMed] [Google Scholar]
- 72.Zivin, R., W. Zehring, and L. B. Rothman-Denes. 1981. Transcriptional map of bacteriophage N4. Location and polarity of N4 RNAs. J. Mol. Biol. 152:335-356. [DOI] [PubMed] [Google Scholar]