Abstract
ExbB and ExbD proteins are part of the TonB-dependent energy transduction system and are encoded by the exb operon in Escherichia coli. TonB, the energy transducer, appears to go through a cycle during energy transduction, with the absence of both ExbB and ExbD creating blocks at two points: (i) in the inability of TonB to respond to the cytoplasmic membrane proton motive force and (ii) in the conversion of TonB from a high-affinity outer membrane association to a high-affinity cytoplasmic membrane association. The recent observation that ExbB exists in 3.5-fold molar excess relative to the molarity of ExbD in E. coli suggests the possibility of two types of complexes, those containing both ExbB and ExbD and those containing only ExbB. Such distinct complexes might individually manifest one of the two activities described above. In the present study this hypothesis was tested and rejected. Specifically, both ExbB and ExbD were found to be required for TonB to conformationally respond to proton motive force. Both ExbB and ExbD were also required for association of TonB with the cytoplasmic membrane. Together, these results support an alternative model where all of the ExbB in the cell occurs in complex with all of the ExbD in the cell. Based on recently determined cellular ratios of TonB system proteins, these results suggest the existence of a cytoplasmic membrane complex that may be as large as 520 kDa.
Gram-negative bacteria circumvent certain nutrient limitations imposed by the outer membrane by the use of TonB-gated transporters (also known as outer membrane receptors). These transporters actively move iron siderophores and vitamin B12 across the outer membrane and into the periplasmic space. The lack of obvious energy sources at the outer membrane is overcome by the cytoplasmic membrane energy transduction protein TonB and the proton motive force (PMF) (for a review, see reference 18). Two additional cytoplasmic membrane proteins, ExbB and ExbD, are known to be required for energy transduction (1, 7).
While TonB is clearly the energy transducer, ExbB and ExbD are each required for the transport of siderophores and sensitivities to group B colicins and bacteriophage φ80, which are indicative of a successful energy transduction event (1, 2, 7). It should also be noted that ExbB and ExbD function can be partially (∼10%) replaced by the activities of the homologous proteins TolQ and TolR in a process known as cross talk (3).
Energy transduction undoubtedly involves successive biochemical steps. Two steps dependent upon ExbB and/or ExbD have thus far been identified: the apparent coupling of TonB to the proton gradient of the cytoplasmic membrane (15) and the conversion of TonB from a (presumably) high-affinity outer membrane association to a (presumably) high-affinity cytoplasmic membrane association (16). The combined topologies of ExbB and ExbD mimic those of a signal transducer (19), with ExbD extending into the periplasmic space from its single transmembrane domain (12) and ExbB consisting of three transmembrane domains and a significant cytoplasmic domain (13, 14). The observations that each protein appears to form trimers in vivo (11) and that ExbB and ExbD interact in vitro (2) seem to be consistent with the possibilities that ExbB and ExbD occur in cells at a 1:1 ratio and that they function as a signal transducer; however, the implied hypothesis that these two proteins work in concert has never been tested. In a surprising complication, even though ExbB and ExbD are encoded on an operon, ExbB exists in the cells at a 3.5 molar excess relative to the molarity of ExbD (10). Thus, a spectrum of possibilities exists. At one extreme, all the ExbB is in complex with ExbD in an unbalanced ratio, and at the other extreme, two kinds of ExbB-containing complexes occur in the cell: simple ExbB/ExbD heterohexamers (where ExbB and ExbD are present in a 1:1 ratio) and homomultimers consisting of the majority of ExbB proteins (lacking ExbD). If indeed the majority of ExbB proteins are present as homomultimers, ExbB would almost certainly have a function different from that of ExbB/ExbD heterohexamers. The disparate topologies of the two kinds of complexes suggest that ExbB/ExbD heterohexamers might convert TonB from an outer membrane association to a cytoplasmic membrane association and that the putative ExbB homomultimers might couple TonB to the PMF. The experiments reported in this paper were initiated primarily to determine whether ExbB has an identifiable biochemical role in energy transduction in the absence of ExbD. For completeness and symmetry, the less likely converse possibility was also tested.
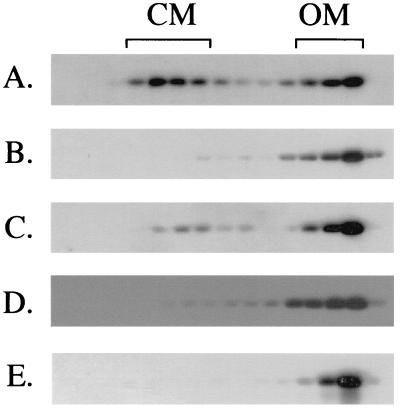
To assess whether ExbB and ExbD play individual roles in the distribution of TonB between the cytoplasmic and outer membranes, the same strain used in the earlier studies, KP1038 [exbB::Tn10 tolQ(Am)], was made recA negative by P1vir transduction of recA::cat from KP1286 (creating KP1392) and various plasmids were transformed into it. TonB has been shown to be approximately 60 and 40% distributed between the cytoplasmic and outer membranes, respectively, under normal conditions. In this study, KP1392 strains with plasmids expressing ExbB and/or ExbD or expressing neither were lysed in a French pressure cell and total lysate fractions from sucrose density gradients were analyzed for the distribution of TonB by immunoblotting (16). Strain KP1392/pKP390, expressing plasmid-encoded ExbB and ExbD at chromosomal levels, gave a wild-type level of distribution of TonB similar to that seen previously, with TonB being distributed roughly equally between the two membranes (Fig. 1A.). As expected for KP1392/pBAD24, where both ExbB and ExbD and TolQ and TolR are predicted to be absent, TonB was almost entirely associated with the outer membrane (Fig. 1B). When ExbB alone was supplied to KP1392 in trans at chromosomal levels, a small proportion of TonB occurred at the cytoplasmic membrane (Fig. 1C). When ExbD alone was supplied in trans at chromosomal levels, TonB was not detectable in the cytoplasmic membrane fraction (Fig. 1D). (ExbD was still associated with the cytoplasmic membrane under these conditions [data not shown]). The small proportion of TonB observed at the cytoplasmic membrane in KP1392/pKP392 (ExbB only) was probably due to the presence of slight amounts of ExbD and/or TolQ, resulting from the theoretical inability of the exbB::Tn10 and tolQ(Am) mutations to completely eliminate chromosomal expression of their downstream genes. Consistent with the fractionation data, KP1392/pKP392 did exhibit low-level sensitivity to B-group colicins (data not shown).
FIG. 1.
Both ExbB and ExbD are required for high-affinity association of TonB with the cytoplasmic membrane. Immunoblots of sucrose density gradient fractions with anti-TonB antibody 4F1 are shown. (A) KP1392/pKP390 (ExbB+ ExbD+); (B) KP1392/pBAD24 (ExbB− ExbD−); (C) KP1392/pKP392 (ExbB+ ExbD−); (D) KP1392/pKP378 (ExbB− ExbD+); (E) KP1456/pKP392 (ExbB+ ExbD−). CM indicates the positions of the cytoplasmic membrane fractions; OM indicates the positions of the outer membrane fractions. Strain KP1286 was constructed by P1vir transduction (17) of recA::cat from JP352 (6) into strain KP1032. Strain KP1392 was constructed by P1vir transduction of recA::cat from KP1286 into strain KP1038 (21). In each case, transductants were selected by resistance to chloramphenicol and screened for UV sensitivity. To construct plasmid pKP378, the exbD gene from pKP298 (1) was amplified by PCR and cloned into the KpnI and XbaI sites of pBAD24 (9) such that it is expressed from the initiating Met of the araB protein. To construct plasmid pKP392, the open reading frame of exbB was amplified from pLC4-14 (5) and inserted into pBAD24 at the SmaI site such that the first three codons are Met, Val, and Pro, followed by the second residue of ExbB. To construct plasmid pKP390, the entire exb operon was amplified from pLC4-14 and inserted into pBAD24 at the SmaI site with a start for ExbB identical to that of pKP392. Sequences of primers are available upon request. The sequences of all inserts derived from PCR amplification were verified to exclude the presence of unintended base changes. Cultures were grown with aeration at 37°C in M9 minimal salts medium supplemented with 0.4% glycerol (or 0.4% glucose), 0.2% Casamino Acids, 40 μg of tryptophan ml−1, 0.4 μg of thiamine ml−1, 1 mM MgSO4, 0.5 mM CaCl2, and 1.85 μM FeCl3. High-purity arabinose from Pfanstiehl (Waukegan, Ill.) was added to media at the appropriate concentrations to provide for expression of the plasmid-encoded proteins at chromosomal levels, as verified by immunoblot analysis with ExbB-specific polyclonal antibodies or ExbD-specific polyclonal antibodies (10). Arabinose concentrations in M9 medium were 0.0012% for pKP378 and 0.0003% for pKP390 and pKP392. Ampicillin was present at 100 μg ml−1 to ensure retention of the plasmids.
To more stringently test the effect of ExbB alone, KP1456 [W3110 Δ(exb-metC) tolQ(Am)] was created in a multistep process. First, the Tn10kan marker from CAG18531 (20) was transduced into TPS13 [tolQ(Am)] (22), with screening for retention of colicin A tolerance (∼7% linkage), to create KP1451. Plasmid pTSP202 (encoding a wild-type tolQRA operon [22]) was then transformed into KP1451, to allow for production of a transducing lysate of bacteriophage P1vir. This lysate was used to transduce W3110, and the resultant Kanr transformants were screened for colicin A tolerance to create KP1434. Meanwhile, the nupG::Tn10 mutation from CAG18472 (20) was transduced into GUC41 [Δ(exb-metC)] (8) by selecting for tetracycline resistance and screening for the absence of ExbB by immunoblot analysis, to yield KP1455, where Tn10 is ∼20% linked to the Δ(exb-metC) deletion. Finally, the Tn10-linked Δ(exb-metC) deletion from KP1455 was transduced into KP1434 and the Tcr transductants were screened for the absence of ExbB as described above to create KP1456. KP1456 was completely insensitive to all TonB-dependent agents (data not shown). Fractionation of KP1456/pKP392 resulted in the absence of TonB from the cytoplasmic membrane (Fig. 1E). Thus, ExbB and ExbD were individually unable to support the association of TonB with the cytoplasmic membrane.
We had previously demonstrated the conformation of TonB to be responsive to PMF. In that assay, which is retrospective, we asked, “is TonB capable of responding to the PMF by a conformational change?” If the answer is yes, then when spheroplast membranes are subsequently de-energized by carbonyl cyanide m-chlorophenylhydrazone (CCCP) or osmotic lysis and then proteinase K is added, a proteinase K-resistant fragment is generated and detected in the immunoblots (15). If the TonB is incapable of responding to PMF (for example, TonB with a deletion of Val17 [15]), then the addition of CCCP does not convert the TonB to a proteinase K-resistant fragment. Instead, TonB is rapidly processed into undetectable fragments. This assay can thus be used to determine whether TonB could have responded to PMF (could be energized) in the first place. The PMF responsiveness of TonB requires an intact amino-terminal transmembrane domain but not the carboxy-terminal 65 amino acids (15). The absence of ExbB and/or ExbD also precludes formation of the PMF-responsive conformation (15). This assay is not sensitive to the 10% activity due to cross talk with TolQ and TolR, so strain KP1037, which carries only an exbB::Tn10 insertion and remains wild type for TolQ or TolR, sufficed for use here.
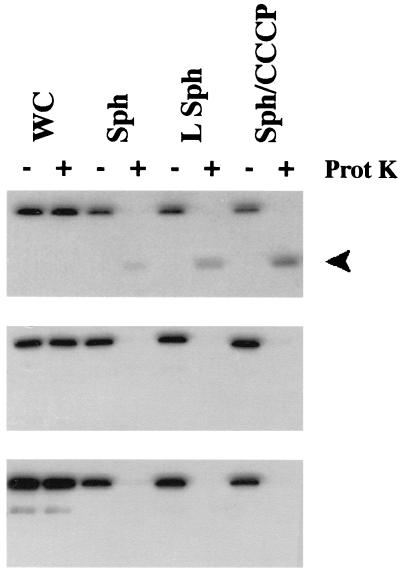
In GM1 (exbB+ exbD+), the proteinase K-resistant fragment indicative of the ability of TonB to respond to PMF is clearly visible in lysed spheroplasts and spheroplasts treated with CCCP (Fig. 2, top). The decreased amount of this fragment in the lysed spheroplasts likely represents inefficient lysis of the spheroplasts. As evident in Fig 2 (middle), strains expressing only ExbB did not support formation of the proteinase K-resistant fragment of TonB. Likewise, strains expressing ExbD alone failed to support formation of the proteinase K-resistant fragment (Fig. 2, bottom). It was in the PMF responsiveness assay that putative ExbB homomultimers might have been sufficient to convert TonB to a PMF-responsive conformation, especially since the carboxy terminus of TonB (and thus association with the outer membrane) was not required. That result would also have been most consistent with the observation that ExbB suppressors of TonB transmembrane domain mutations (which do not respond to PMF) act at the energization half of the proposed energy transduction cycle (15). Here the somewhat surprising result that both ExbB and ExbD are necessary to convert TonB to a PMF-responsive state was obtained.
FIG. 2.
Both ExbB and ExbD are required for the PMF responsiveness of TonB. (Top) KP1037/pKP390 (ExbB+ ExbD+); (middle) KP1037/pKP392 (ExbB+ ExbD−); (bottom) KP1037/pKP378 (ExbB− ExbD+). Strains were grown as described in the legend to Fig. 1. Samples from whole cells (WC), spheroplasts (Sph), lysed spheroplasts (LSph), and spheroplasts treated with CCCP (Sph/CCCP) as described previously (15) are shown. Exposure to proteinase K (Prot K) is indicated by the plus signs. The position of the fragment indicative of TonB PMF responsiveness is indicated by the arrow. The immunoblot was developed with anti-TonB antibody 4F1 as described previously (15).
The results of this study support the idea that all of the ExbB in the cell will occur in complex with all of the ExbD in the cell. If that is the case, the recently determined ratios of TonB system proteins (10), in concert with cross-linking and crystallization results, lead to an interesting prediction about what the complex may look like. The extreme carboxy terminus of TonB has been shown to crystallize as a dimer, although in that study it was not clear whether the dimer represented a functional state (4). If there are indeed two TonB proteins per complex, then based on the ratio determinations, there will be 4 to 5 ExbD and 14 to 15 ExbB proteins per complex. Based on molecular masses of 26.1 kDa for TonB, 15.5 kDa for ExbD, and 26.1 kDa for ExbB, the cytoplasmic membrane energy transduction complex may be as large as 520 kDa.
Acknowledgments
We thank R. Larsen for construction of pKP390, pKP392, KP1434, and KP1451 and the demonstration that GUC41 is missing both exbB and exbD. We thank Q. Zhang for construction of pKP378, T. Nevinski for construction of strain KP1286, and C. Bulathsinghala for construction of strain KP1456.
This work was supported by grant GM42146 to K.P. from the NIGMS.
REFERENCES
- 1.Ahmer, B. M. M., M. G. Thomas, R. A. Larsen, and K. Postle. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177:4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun, V., S. Gaisser, C. Herrman, K. Kampfenkel, H. Killman, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C., A. Mooser, A. Pluckthun, and A. Wlodawer. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535-27540. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, L., and J. Carbon. 1975. Biochemical construction and selection of hybrid plasmids containing specific segments of the E. coli genome. Proc. Natl. Acad. Sci. USA 72:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Economou, A., J. A. Pogliano, J. Beckwith, D. B. Oliver, and W. Wickner. 1995. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171-1181. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, E., K. Günter, and V. Braun. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutermann, S. K., and L. Dann. 1973. Excretion of enterochelin by exbA and exbB mutants of Escherichia coli. J. Bacteriol. 114:1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44:271-281. [DOI] [PubMed] [Google Scholar]
- 11.Higgs, P. I., P. S. Myers, and K. Postle. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kampfenkel, K., and V. Braun. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174:5485-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kampfenkel, K., and V. Braun. 1993. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 268:6050-6057. [PubMed] [Google Scholar]
- 14.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389-396. [DOI] [PubMed] [Google Scholar]
- 15.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 16.Letain, T. E., and K. Postle. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Gram-negative bacteria. Mol. Microbiol. 24:271-283. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 19.Postle, K. 1993. TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 25:591-601. [DOI] [PubMed] [Google Scholar]
- 20.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skare, J. T., and K. Postle. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 5:2883-2890. [DOI] [PubMed] [Google Scholar]
- 22.Sun, T. P., and R. E. Webster. 1986. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J. Bacteriol. 165:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]