Abstract
Vibrio fischeri, a luminescent marine bacterium, specifically colonizes the light organ of its symbiotic partner, the Hawaiian squid Euprymna scolopes. In a screen for V. fischeri colonization mutants, we identified a strain that exhibited on average a 10-fold decrease in colonization levels relative to that achieved by wild-type V. fischeri. Further characterization revealed that this defect did not result from reduced luminescence or motility, two processes required for normal colonization. We determined that the transposon in this mutant disrupted a gene with high sequence identity to the pgm (phosphoglucomutase) gene of Escherichia coli, which encodes an enzyme that functions in both galactose metabolism and the synthesis of UDP-glucose. The V. fischeri mutant grew poorly with galactose as a sole carbon source and was defective for phosphoglucomutase activity, suggesting functional identity between E. coli Pgm and the product of the V. fischeri gene, which was therefore designated pgm. In addition, lipopolysaccharide profiles of the mutant were distinct from that of the parent strain and the mutant exhibited increased sensitivity to various cationic agents and detergents. Chromosomal complementation with the wild-type pgm allele restored the colonization ability to the mutant and also complemented the other noted defects. Unlike the pgm mutant, a galactose-utilization mutant (galK) of V. fischeri colonized juvenile squid to wild-type levels, indicating that the symbiotic defect of the pgm mutant is not due to an inability to catabolize galactose. Thus, pgm represents a new gene required for promoting colonization of E. scolopes by V. fischeri.
One of the most intimate interactions between organisms occurs during the establishment of a long-term symbiotic relationship between a microorganism and its host, as exemplified by the Vibrio-squid symbiosis. Upon hatching, juveniles of Euprymna scolopes, a nocturnal Hawaiian squid, quickly become colonized by the luminescent marine bacterium Vibrio fischeri, establishing a symbiosis that persists throughout the lifetime of the squid (see reference 40 for a review). The bacterial symbionts colonize a specialized structure in the squid host, the light organ, located within the mantle cavity of the animal. When a sufficient cell density is reached, the bacteria luminesce, providing the counterillumination ability believed to be an antipredation mechanism for the squid host (see reference 49 for a review). Various studies characterizing the Vibrio-squid symbiosis have identified several genetic determinants involved in establishing this association (2, 14, 48, 50, 52); however, much remains to be learned about how these symbiotic partners interact.
Despite the presence of other bacteria in the seawater, the light organ of the juvenile E. scolopes is colonized primarily by V. fischeri (6); even other Vibrio species are generally unable to colonize under lab conditions (29, 36). V. fischeri cells colonize the newly hatched aposymbiotic (symbiont-free) juveniles of E. scolopes within a few hours, reaching between 105 and 106 CFU per squid within 24 h (41). The bacteria are drawn to the vicinity of the light organ by currents created by specialized ciliated appendages of the organ as seawater is vented through the mantle cavity of the juvenile squid (see reference 49 for a review). The bacterial cells embed and aggregate in a mucus-like material sloughed off the appendages and secreted from pores located at the base of the light organ (36). As they enter the pores, they encounter mucus-filled ducts (27) and an outwardly directed current created by dense cilia lining the duct walls (see reference 49 for a review); not surprisingly, nonmotile V. fischeri are symbiosis defective (14, 36). The ducts ultimately lead to the site of colonization, the internal crypts, where the microbes likely encounter a relatively hostile environment due to the presence of macrophage-like cells (35) and halide peroxidase, an enzyme implicated in oxidative stress (53). A V. fischeri mutant that cannot produce catalase does not successfully compete with a catalase-positive strain for colonization (50).
Once inside the crypts of the light organ, the bacteria multiply to a high cell density, where the microbes are exposed to amino acids and peptides which likely serve as a nutrient source (15). Inside the crypts the microbes may attach by using fimbriae to sugar residues (such as mannose [28]) on the surfaces of host epithelial cells: certain V. fischeri mutants defective in the ability to hemagglutinate animal red blood cells (a characteristic correlated with the presence of fimbriae in some bacteria) showed a decreased ability to colonize juvenile squid (B. Feliciano and E. Ruby, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. N-75, 1999). The presence of surface sugars may also serve as a nutrient source for the symbionts, a strategy employed in other host-microbe interactions. For example, Bacteroides thetaiotaomicron, a common mammalian gut commensal, induces fucosylation on the surface of ileal enterocytes in mice. The bacteria then use the fucose residues as an energy source (22).
Within hours of the onset of colonization, the squid undergoes bacterium-induced morphological changes, including increased microvillar density (25) and swelling of the epithelial lining of the crypts (30). The specialized ciliated appendages of the light organ also undergo bacterium-induced morphological changes, including lipopolysaccharide (LPS)-mediated apoptosis (12) and complete regression within 4 days of colonization (30). Thus, bacteria colonizing the light organ face continuously changing conditions, including stresses within the light organ and the dramatic morphological changes in the host, which may contribute to the specific colonization by V. fischeri. Interestingly, V. fischeri luminescence mutants, although able to establish the initial colonization, cannot induce swelling in the crypt epithelial cells and are also unable to maintain persistent symbiosis (48).
More recently identified colonization factors include RscS, a sensor kinase hypothesized to regulate symbiosis-specific genes (52), and OmpU, an outer membrane protein (2). Because colonization appears to require active participation on the part of both the bacterium and the squid host (36), it is likely that this exclusive association depends upon numerous additional host and bacterial genes. Other important bacterial factors may include those involved in chemotaxis, attachment, and protection against host stresses; however, the necessary genetic determinants for such processes have yet to be described.
In an effort to identify additional factors necessary for establishing the Vibrio-squid symbiosis, we screened a transposon mutant library and identified a colonization mutant that exhibited on average a 10-fold decrease in colonization ability relative to wild-type V. fischeri. We report here the characteristics of this symbiosis mutant of V. fischeri.
MATERIALS AND METHODS
Strains and media.
The strains used in the present study are listed in Table 1. ESR1, the parent strain used here, was derived from V. fischeri ES114. V. fischeri cells were grown in SWT medium (6), LBS medium (9), or HEPES minimal medium (MM) (42) containing either glucose or galactose at a final concentration of 0.2%. Escherichia coli strains were grown in Luria-Bertani medium (8). Conditioned medium (CM), made from LM medium (31) as described previously (52), was utilized to determine whether the colonization mutant KV733 was defective for bioluminescence in culture. Where appropriate, antibiotics were added to the following final concentrations: chloramphenicol (CHL), 1 to 5 μg ml−1 for V. fischeri and 30 μg ml−1 for E. coli; erythromycin (ERY), 1 to 5 μg ml−1 for V. fischeri and 150 μg ml−1 for E. coli; rifampin, 100 μg ml−1; kanamycin, 100 μg ml−1 for V. fischeri and 50 μg ml−1 for E. coli. Agar was added to a final concentration of 1.5% for solid media or 0.25% for motility plates.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA1 hsdR17 (rK− mK+) glnV44 thi-1 recA1 gyrA (Nalr) relA Δ(lacIZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 54 |
| CC118λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λpir | 20 |
| S17-1λpir | thi pro hsdR hsdM+recA λpir | 43 |
| CGSC4982 | galK | CGSCb |
| V. fischeri | ||
| ES114 | WT | 6 |
| ESR1 | Rfr | 14 |
| KV733 | Rfrpgm::Tn10 | This study |
| KV828 | Rfr, carries pPS18 integrated into the chromosome | This study |
| KV1069 | Rfrpgm::erm | This study |
| KV1170 | RfrattTn7::seqA-pgm+ | This study |
| KV1171 | Rfrpgm::Tn10 attTn7::seqA-pgm+ | This study |
| KV1177 | Rfr Δpgm | This study |
| KV1306 | Rfr, carries pKV161 integrated into the chromosome | This study |
| KV1358 | Rfr, ΔgalK | This study |
| Plasmids | ||
| pBS | Blue-white screen cloning vector; Apr | Stratagene |
| pCD3 | pEVS107 (SpeI) + 3-kb NheI fragment from pPS21, contains seqA and pgm inside the Tn7 transposon | This study |
| pCNW1 | pVO8 BamHI + 5-kb galK+BglII fragment | This study |
| pEVS79 | pBC + mob; Cmr | 44 |
| pEVS104 | Conjugal helper plasmid (tra trb); Knr | 44 |
| pEVS107 | Tn7 delivery plasmid; Knr Emr | Eric Stabb |
| pKV25 | pUC19::erm | 50 |
| pKV36 | pUC19::cat | This study |
| pKV124 | pBSL181 containing promoterless lacZ and oriR6K within Tn10 ends | 52 |
| pKV161 | pEVS79 HindIII + 2.8-kb galK+HindIII fragment from pCNW1 | This study |
| pPS8 | 11-kb NheI fragment cloned from KV733, contains pgm::Tn10 and flanking DNA | This study |
| pPS12 | pBS (SacI) + 6-kb SacI fragment from pPS8, contains DNA flanking the Tn10 insertion | This study |
| pPS15 | pBS (XbaI) + 2.6-kb NheI fragment from pPS12, contains DNA upstream of pgm | This study |
| pPS18 | pPS15 BglII + 1.1-kb BamHI fragment encoding Cmr from pKV36 (results in fur::cat mutation) | This study |
| pPS19 | 14-kb NsiI fragment cloned from KV828, contains seqA+ and a portion of pgm | This study |
| pPS20 | pEVS79 (BamHI) + 2.2-kb BamHI/BglII fragment from pPS19, contains a portion of the pgm locus | This study |
| pPS21 | 20-kb XbaI fragment cloned from KV828, contains seqA+ and pgm+ | This study |
| pPS24 | pPS20 SphI/filled + 1.2-kb SmaI/EcoRV fragment encoding Emr from pKV25 (results in a pgm::erm mutation) | This study |
| pPS27 | pEVS79 (XbaI) + 3-kb Nhe fragment from pPS21 | This study |
| pPS33 | pPS20 with two MscI sites internal to pgm deleted (results in a 606-bp deletion) | This study |
| pRK2013 | Conjugal helper plasmid (tra trb); Knr | 10 |
| pTMB7 | 9.6-kb ClaI fragment cloned from KV1306, contains galK+ and flanking DNA | This study |
| pTMB9 | pTMB7 deleted between two AseI sites (results in 250-bp deletion at the 5′ end of galK) | This study |
| pUX-BF13 | Encodes Tn7 transposase (tnsABCDE); Apr | 4 |
| pVO8 | Blue-white screen cloning vector; Cmr Emr | 51 |
Nalr, nalidixic acid resistance; WT, wild type; Rfr, rifampin resistance; Apr, ampicillin resistance; Knr, kanamycin resistance; Cmr, CHL resistance; Emr, ERY resistance.
CGSC, E. coli Genetic Stock Center, Yale University, New Haven, Conn.
Molecular techniques.
All plasmid constructions were carried out by standard molecular biology techniques, with restriction and modifying enzymes obtained from New England Biolabs (Beverly, Mass.) or Promega (Madison, Wis.). Plasmids used or constructed in the present study are shown in Table 1. Cloning of DNA flanking the Tn10 transposon insertion in V. fischeri strain KV733 was accomplished by using the oriR6K (the pir-dependent origin of replication from plasmid R6K [24]) and CHL resistance elements contained within the transposon.
Genetic techniques.
Transposon mutagenesis by using the Tn10 delivery plasmid pKV124 was described previously (52). Conjugations were performed as described previously (52) by using the appropriate V. fischeri recipient strain and two E. coli strains: DH5α carrying the plasmid to be transferred and DH5α carrying a conjugal plasmid (either pRK2013 or pEVS104).
The pgm and galK mutants were constructed with strains and plasmids as follows. The pgm::erm mutant was derived from pPS24 introduced into ESR1; a recombinant, KV1069, that was ERY resistant and CHL sensitive and grew poorly on MM containing galactose (MM-galactose) was subsequently identified. Similarly, the Δpgm mutant was derived by introducing pPS33 into KV1069 by conjugation. An isolate, KV1177, was sensitive to both ERY and CHL and defective for growth on MM-galactose. The galK mutant was obtained by introducing pTMB9 into ESR1. KV1358, a CHL-sensitive strain unable to grow on MM-galactose, was identified.
The pgm::Tn10 mutation was complemented with a wild-type copy of the seqA-pgm region inserted in single copy at the Tn7 insertion site (attTn7) in the V. fischeri chromosome (4; E. Stabb, unpublished data). Briefly, E. coli cells carrying the Tn7::pgm+-containing plasmid pCD3 served as a donor in a tetraparental mating, which also included E. coli cells carrying pUX-BF13 and the V. fischeri recipient (ESR1 or KV733). Because an ERY resistance cassette was included within the Tn7 ends along with pgm, ERY-resistant colonies were selected and screened for the loss of kanamycin resistance encoded by the Tn7 vector. Strains with presumptive Tn7::pgm insertions were confirmed by Southern blot analysis.
Southern blotting.
Chromosomal DNA isolated from V. fischeri strain KV733 was digested with EcoRV, which cuts once within the mini-Tn10 transposon, or BsrGI, which cuts outside the transposon. Chromosomal DNA isolated from V. fischeri strains KV733 (pgm::Tn10), KV1069 (pgm::erm), and KV1177 (Δpgm) was digested with HindIII. DNA fragments were separated by using a 0.6% agarose gel, transferred onto a nylon membrane (Hybond XL; Amersham-Pharmacia Biotech, Piscataway, N.J.), and UV-cross-linked. Detection was carried out by using the Boehringer Mannheim DIG DNA labeling kit (Roche Molecular Biochemicals, Indianapolis, Ind.). Either random priming or a PCR-based technique was used to generate digoxigenin-labeled DNA complementary to the transposon or to the pgm gene. Hybridization and detection were carried out as described previously (52).
Colonization assays.
To determine whether mutant V. fischeri strains were able to form a symbiotic association with E. scolopes, juvenile squids were placed in artificial seawater (Instant Ocean; Aquarium Systems, Mentor, Ohio) containing an inoculum of 1,000 to 5,000 cells of the desired V. fischeri strain per ml of fluid and then analyzed for the presence of bacteria in the light organ as previously described (40). The limit of detection is 14 V. fischeri cells per squid.
Luminescence assays.
KV733 and ESR1 were diluted 1:100 from an overnight culture and grown in CM in the presence or absence of synthetic V. fischeri autoinducer (600 ng of 3-oxo-hexanoyl-l-homoserine lactone [Sigma, St. Louis, Mo.] per ml). At various times after inoculation, 1-ml samples were taken for luminescence and optical density (OD) measurements. A TD-20/20 luminometer (Turner Designs, Sunnyvale, Calif.) was used to determine the level of bioluminescence of KV733 and its parent.
Phosphoglucomutase assay.
Phosphoglucomutase activity was determined by the method of Adhya and Schwartz (1).
Polymixin B, sodium dodecyl sulfate (SDS), and deoxycholate sensitivity determinations.
Polymyxin B MIC determinations were made by using a protocol modified from that of Steinberg and Lehrer (46). Briefly, polymyxin B (Sigma) was diluted to a concentration of 1,280 μg ml−1 in 0.01% acetic acid and kept frozen at −20°C. Twofold serial dilutions of the stock antibiotic ranging from 0.04 to 80 μg ml−1 were prepared in the first row of a 96-well plate in 0.01% acetic acid-0.1% bovine serum albumin. Tenfold dilutions of these concentrations (0.004 to 8 μg ml−1) were then made in SWT medium in the remaining wells. Log-phase cultures of each strain were inoculated into each dilution in triplicate to a starting concentration of 4 × 105 CFU ml−1, incubated for 24 h, and visually examined for turbidity. The lowest concentration of polymyxin B inhibiting growth was designated the MIC.
We assayed for growth of the cells in the presence of SDS by first preparing SDS to a final concentration of 10% in SWT medium. This medium was inoculated to a starting concentration of 4 × 105 CFU ml−1 with log-phase cultures in triplicate which were grown at 28°C with shaking. Then, 1-ml samples were taken for OD measurements over time.
Sensitivity to deoxycholate was determined by diluting overnight cultures 1:1,000 in SWT containing 1% deoxycholate and incubating the cells with shaking at 28°C for 6 h. Growth of the cultures was monitored hourly by diluting and plating onto SWT and then counting the resulting CFU.
LPS gel analysis.
LPS fractions were prepared by using a protocol modified from the method of B. L. Reuhs and J. S. Kim (as described in reference 39) with the following modifications: 1.5-ml samples were harvested from cultures grown to an OD at 600 nm of 0.5 to 0.7 in SWT medium, pelleted, decanted, and frozen at −80°C prior to a phenol-water extraction. The resulting LPS fractions were desalted by using MicroSpin G-25 columns (Amersham), and the volume was reduced by lyophilization in a Labconco freeze dryer 4.5 (Labconco, Kansas City, Mo.). The isolated LPS fractions were then resuspended in 20 μl of sample buffer, separated by deoxycholic acid-polyacrylamide gel electrophoresis (PAGE) on 14% acrylamide and silver stained with the Owl silver stain staining kit for electrophoresis (Owl Separation Systems, Portsmouth, N.H.) according to the manufacturer's instructions.
Sequencing.
Automated sequencing was carried out with forward and reverse primers complementary to vector and insert sequences. Similarity searches were performed by using the National Center for Biotechnology Information BLAST program (3, 19). Oligonucleotides were obtained from the Loyola Macromolecular Analysis Facility, Integrated DNA Technologies (Coralville, Iowa), or MWG Biotech (High Point, N.C.).
Nucleotide sequence accession number.
The following GenBank accession number was assigned to the seqA-pgm-ybgI sequence: AF474148.
RESULTS
Isolation of a symbiosis-defective V. fischeri mutant.
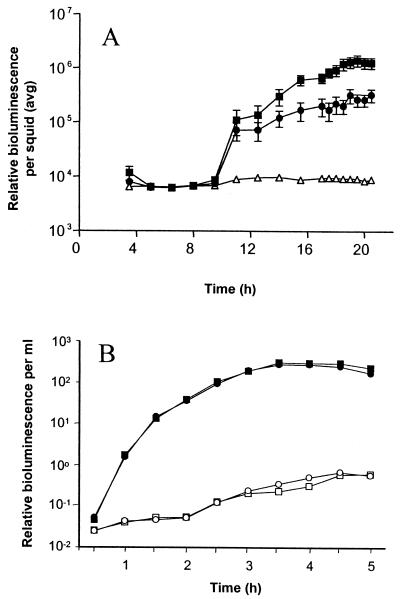
Individual transposon insertion mutants of V. fischeri ESR1 were screened for their ability to form a symbiotic association with juveniles of the squid E. scolopes as described previously (52). Briefly, individual mutants were inoculated into seawater containing newly hatched juvenile E. scolopes. Colonization was monitored noninvasively by using bioluminescence as an indirect indicator of symbiont density in the light organ of E. scolopes. One mutant, KV733, could not achieve the high levels of luminescence sustained by ESR1 in the symbiosis with E. scolopes (Fig. 1A).
FIG. 1.
Luminescence of colonization mutant KV733 and its parent. (A) Newly hatched juvenile E. scolopes squid were incubated in artificial seawater containing no V. fischeri (▵), the parent strain ESR1 (▪), or the colonization mutant KV733 (•). Bioluminescence emission was measured over time and represents an average of 11 animals for ESR1 and KV733. Error bars show the standard errors of the mean (SEM). (B) ESR1 (squares) and KV733 (circles) were grown in CM in the absence (open symbols) or presence (solid symbols) of autoinducer (3-oxo-hexanoy-l-homoserine lactone). The data shown are from a representative experiment of several replicates.
To confirm that the decreased level of symbiotic bioluminescence corresponded to decreased colonization, levels of colonization by KV733 and its parent were measured directly (Fig. 2). Juveniles inoculated with KV733 exhibited on average a 10-fold reduction in colonization levels relative to those inoculated with ESR1. This putative symbiosis-defective mutant was further characterized in culture by examining motility on soft agar plates (data not shown), growth on minimal medium (data not shown), natural bioluminescence, and response to autoinducer (Fig. 1B). In these assays, KV733 behaved like its parent, suggesting that the symbiotic defect in KV733 was not due to deficiency in motility, amino acid biosynthesis, or luminescence, all factors previously identified as important in the symbiosis. It therefore seemed likely that KV733 was defective for an as-yet-unknown symbiotic locus.
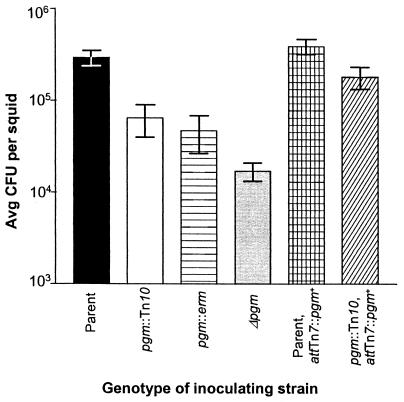
FIG. 2.
Symbiotic colonization by pgm mutant and control strains. Newly hatched juvenile E. scolopes were inoculated with the following strains: ESR1 (▪), KV733 (□), KV1069 (▤), KV1177 ( ), KV1170 (▩), or KV1171 (▨). The juveniles were exposed to ca. 1,000 cells of the parental and complemented strains or 2,000 cells of the mutant strains per ml of seawater for 3 h. The level of colonization achieved by these strains was determined by homogenization and plating 20 h after the organisms were placed together. The data shown are from a representative experiment, and the bars indicate an average of 11 squids. Error bars indicate the SEM.
Identification of the locus disrupted in KV733.
To identify the genetic defect in KV733, we determined the number and location of the transposon insertion(s) in this strain by Southern analysis. A single, large (>12-kb) BsrGI fragment that carried the transposon was identified by probing with DNA complementary to the transposon delivery plasmid, providing evidence that the strain carried a single transposon insertion (data not shown). Further analysis of KV733 chromosomal DNA revealed that the entire transposon delivery plasmid had incorporated into the chromosome at this site (data not shown).
We cloned the transposon and flanking chromosomal DNA. Sequence analysis revealed that the transposon had inserted after codon 189 of a 548-codon open reading frame (ORF). This putative protein exhibited high sequence similarity (69% identity, 83% similarity) to Pgm (phosphoglucomutase) of E. coli (26) and V. cholerae (80% identity, 89% similarity [17]). Directly upstream of the putative pgm gene in V. fischeri was an ORF that encoded a putative protein with high sequence similarity to SeqA, encoded by a gene upstream of pgm in both E. coli (51% identity, 71% similarity [5]) and V. cholerae (65% identity, 79% similarity [17]). Directly downstream of pgm in V. fischeri was an ORF encoding a predicted protein similar to proteins of unknown function in V. cholerae (74% identity, 81% similarity [17]) and Haemophilus influenzae (YbgI, 62% identity, 78% similarity [11]). Directly downstream of this gene and in the opposite orientation is an ORF encoding a product with high sequence similarity, based on partial sequence data, to GltA encoded by the gltA gene of E. coli (5).
In E. coli, phosphoglucomutase interconverts glucose-1-phosphate and glucose-6-phosphate and thus plays roles in both galactose metabolism and polysaccharide biosynthesis (13). The protein is part of a family of phosphoserine enzymes which contain a highly conserved serine residue within a conserved 5-amino-acid sequence critical for enzyme activity (38, 55). The deduced amino acid sequence of this region in V. fischeri, V. cholerae, and E. coli is Thr-Pro-Ser-His-Asn at amino acids 144 to 148, differing in a single Ala-Pro change from the rabbit muscle phosphoglucomutase consensus sequence Thr-Ala-Ser-His-Asn (38). On the basis of the presence of this conserved active site region in the putative V. fischeri pgm gene, as well as on the overall high sequence similarity to the pgm genes of both V. cholerae and E. coli, we conclude that the locus disrupted in KV733 is the V. fischeri pgm gene. The phenotypic and biochemical data described below further support this conclusion.
Role for pgm in the symbiosis.
Because the transposon insertion in KV733 also contains the delivery plasmid (encoding transposase), it is conceivable that, during symbiotic colonization, additional insertions that might inhibit the ability of KV733 to colonize could be generated. To determine whether the transposon insertion in KV733 was retained in the pgm gene during colonization, chromosomal DNA extracted from KV733 harvested after symbiotic colonization was subjected to Southern analysis. In each of 10 independent clones from individual animals, the transposon was present only in the pgm gene (data not shown).
Additionally, it was formally possible that KV733 carried a secondary mutation; therefore, in order to confirm that the pgm gene itself plays a role in the symbiosis, we constructed additional pgm mutants (Table 1). A pgm::erm insertion mutant (KV1069) and a Δpgm (in-frame deletion) mutant (KV1177 with a region that includes the conserved 5-amino-acid active-site sequence deleted) also exhibited defects in the ability to colonize juvenile squid similar to that of the original transposon insertion mutant, as shown in Fig. 2 (the mean colonization levels of the three pgm mutants were not statistically different [P = 0.2] as determined by one-way analysis of variance analysis). Additionally, when a pgm+ gene was inserted in single copy into the chromosome of KV733 (KV1171), the complemented strain achieved a colonization level similar to that of the wild-type parent (Fig. 2). These data indicate that the pgm gene plays a critical role in symbiotic colonization.
Phosphoglucomutase assay.
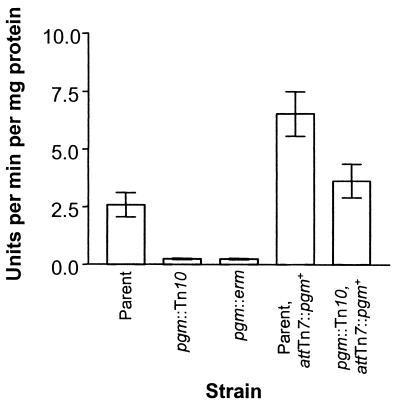
To determine whether our sequence analysis correctly predicted the function of the putative Pgm protein, we measured phosphoglucomutase activity of our wild-type and mutant strains. Whereas wild-type extracts displayed Pgm activity, the pgm mutants tested were deficient (Fig. 3). Complementation restored Pgm activity to the pgm mutant and, not surprisingly, the strain with two wild-type copies of pgm exhibited approximately twice as much Pgm activity.
FIG. 3.
Phosphoglucomutase assay. Phosphoglucomutase activity of strains ESR1 (parent), KV733 (pgm::Tn10), KV1069 (pgm::erm), KV1170 (ESR1 with chromosomal pgm+ complementation), and KV1171 (pgm::Tn10 with chromosomal pgm+ complementation). The data shown are averages from four independent experiments. Error bars show the SEM.
Growth of the pgm mutant in culture.
An E. coli pgm mutant exhibits a decreased ability to grow on minimal medium containing galactose as the sole carbon source (26). We hypothesized that if Pgm has the same function in V. fischeri, then the pgm mutants would be similarly defective. This was the case for all three pgm mutants (KV733, KV1069, and KV1177), whereas the complemented strain (KV1171) exhibited a restored ability to grow on MM-galactose as the sole carbon source (data not shown).
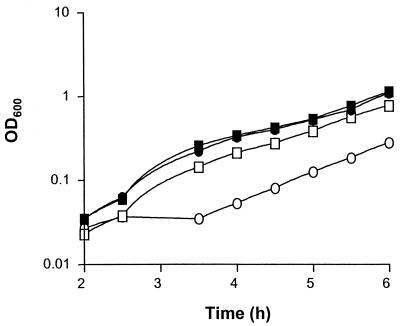
We also examined the ability of the pgm mutant KV733 to grow in different complex nutrient media. When grown in SWT (a seawater-based medium), the pgm mutant KV733 grew identically to its parent, ESR1, suggesting that KV733 does not possess a general growth defect (Fig. 4A). In contrast, when grown in LBS medium, a medium containing NaCl as the only added salt, KV733 displayed a growth defect when inoculated to a starting cell density of <106 CFU ml−1. As shown in Fig. 4B, when the pgm mutant KV733 was inoculated to a starting density of 105 CFU ml−1, its growth leveled off at an OD of ca. 0.1.
FIG. 4.
Growth of ESR1 and KV733 in liquid media. (A) ESR1 (squares) and KV733 (circles) were inoculated to a starting concentration of 106 CFU ml−1 in either SWT (solid symbols) or LBS (open symbols) medium and grown for 6.0 h. (B) ESR1 (▪) and KV733 (•) were inoculated to a starting concentration of 105 CFU ml−1 and grown in LBS medium for 6.5 h. Error bars indicate the SEM. (C) ESR1 (▪) was inoculated to a starting concentration of 105 CFU ml−1 and grown in LBS medium for 6 h. In parallel, KV733 was inoculated to a starting concentration of 105 CFU ml−1 and grown in LBS medium (•), LBS medium supplemented with MgSO4 (○), LBS medium supplemented with CaCl2 (▵), or LBS medium supplemented with KCl (▴) for 6 h. The data from ESR1 grown in LBS medium supplemented with individual salts were omitted for clarity.
Given the differences in the ability of the mutant to grow in the two complex media under conditions of a low inoculum, we examined the possibility that the salts present in SWT medium may be rescuing the growth defect of the pgm mutant in LBS medium. SWT medium contains NaCl, KCl, MgSO4, and CaCl2, whereas LBS medium contains NaCl as the only added salt. The growth experiments were repeated with LBS medium supplemented with the individual SWT salts (Fig. 4C). The addition of either MgSO4 or CaCl2, both salts containing divalent cations, to LBS medium allowed the pgm mutant to grow to wild-type levels when it was inoculated to a starting density of 105 CFU ml−1, whereas the addition of the monovalent salt KCl did not affect the mutant's growth. The growth of ESR1 was not affected by the addition of any of the three salts (data not shown). The consequences of this ion requirement in nature are unclear because seawater and most likely the internal crypts of the light organ contain plentiful amounts of Mg2+ and Ca2+.
Susceptibility of the pgm mutant to polymyxin B, deoxycholate, and SDS.
An E. coli pgm mutant exhibits increased susceptibility to environmental stresses (26). To investigate whether the V. fischeri pgm mutants also exhibited this phenotype, we determined the MIC of the cationic antibiotic polymyxin B for each pgm mutant and for ESR1 grown in SWT medium. All three pgm mutants had an MIC of 2 μg ml−1, a threefold increase in susceptibility relative to the MIC for ESR1 of 6 μg ml−1. Complementation with pgm+ restored the MIC to 5 to 6 μg ml−1.
We also examined the ability of the pgm mutant to grow in SWT in the presence of either 10% SDS or 1% deoxycholate. The presence of SDS interfered with the growth of KV733 to a greater extent than it did the parent strain (Fig. 5). The growth of KV733 (but not the parent strain) was similarly affected in the presence of 1% deoxycholate (data not shown). Taken together, these data indicate that the pgm mutant is more susceptible to environmental stresses.
FIG. 5.
Growth of ESR1 and KV733 in the presence of SDS. ESR1 (squares) and KV733 (circles) were inoculated in triplicate to a starting density of 4 × 105 CFU ml−1 in either SWT medium (solid symbols) or SWT medium with 10% SDS (open symbols) and grown for 6.0 h. Error bars show the SEM. The data shown are from a representative experiment.
LPS structure of the pgm mutant.
E. coli (1) and Agrobacterium tumefaciens (47) pgm mutants exhibit defects in LPS production due to an inability to produce sufficient amounts of UDP-glucose. To determine whether the V. fischeri pgm mutant had a similar defect in LPS biosynthesis, we analyzed LPS from the pgm mutant and wild-type strains by separating the LPS molecules with deoxycholic acid-PAGE and then visualizing them by silver staining (Fig. 6). The LPS extracted from the pgm mutant KV733 had a banding pattern distinct from that of ESR1, containing additional species that migrated faster than that of the smallest LPS band of ESR1 (Fig. 6). An altered banding pattern was also observed with LPS extracted from the other two pgm mutant strains (data not shown). Complementation restored the LPS banding pattern to that of the parent strain (Fig. 6 and data not shown), suggesting that the altered LPS profile of the mutant is due to the loss of pgm.
FIG. 6.
LPS profiles of the pgm mutant and wild-type strains. LPS fractions were prepared as described in Materials and Methods, separated by deoxycholic acid-PAGE, and visualized by silver staining. Lane 1, ESR1 (parent); lane 2, KV733 (pgm::Tn10); lane 3, KV1170 (ESR1 with chromosomal pgm+ complementation); and lane 4, KV1171 (pgm:: Tn10 with chromosomal pgm+ complementation). The gel is from a representative experiment.
Colonization assay of the V. fischeri galK mutant.
To test whether the inability of the pgm mutants to utilize galactose accounts for the colonizaton defect of these strains, we constructed a mutant defective for galactokinase (encoded by the galK gene) and examined the strain's ability to form a symbiotic association with E. scolopes. The galK mutant, KV1358, colonized to wild-type levels (Fig. 7). These data indicate that an inability to catabolize galactose does not account for the symbiotic defect in KV733.
FIG. 7.
Symbiotic colonization by the V. fischeri galK mutant and its parent. Newly hatched juvenile E. scolopes were exposed for 3 h at ca. 3,000 cells per ml of either ESR1 or KV1358. The level of colonization was determined by homogenization and plating 20 h after the organisms were placed together. The data shown are from a representative experiment. The bars indicate the average colonization level of 12 squids. Error bars show the SEM.
DISCUSSION
In the present study we identified a mutant of V. fischeri that failed to colonize E. scolopes to wild-type levels. This mutant, KV733, carried a transposon insertion in an ORF with high identity to the pgm (phosphoglucomutase) gene of E. coli. Our data suggest that the locus interrupted in KV733 encodes a functional homolog of the E. coli Pgm protein and that this locus, which we have designated pgm, plays a role in symbiotic colonization.
In E. coli, Pgm plays roles both in catabolizing galactose and in promoting the production of UDP-glucose, an activated form of glucose that serves as a building block for a number of polysaccharides, including LPS. Strains with a deficiency in UDP-glucose production exhibit a number of phenotypes, including an altered LPS profile (21, 32) and increased susceptibility to hydrophobic agents, bile salts, and cationic antimicrobial peptides (32). Similarly, the V. fischeri pgm mutant displayed a decrease in phosphoglucomutase activity, grew poorly on MM-galactose, exhibited an altered LPS profile, and displayed an increased susceptibility to polymyxin B, deoxycholate, and SDS. Taken together, our data indicate that KV733 exhibits phenotypes consistent with a defect in a gene with the same function as that of the E. coli pgm gene.
Interestingly, in the seawater-based SWT medium, wild-type V. fischeri grew to high cell densities despite the presence of 10% SDS. This is in contrast to a recent report indicating that wild-type V. fischeri cells grown in LBS medium were susceptible to 0.009% SDS (2). It is possible that the three additional salts found in SWT medium (MgSO4, CaCl2, and KCl) protect the cells against lysis by the detergent. In support of this hypothesis, the addition of Mg2+ to growth medium has been shown to stabilize the outer membrane of a Salmonella enterica serovar Typhimurium LPS mutant (45). Similarly, we saw that divalent cations were required for optimal growth of the pgm mutant in LBS medium.
In addition, our data suggested that the pgm gene itself is required for symbiotic colonization: the original transposon insertion mutant and two other pgm mutants that we constructed exhibited similar defects in colonization, and complementation restored the ability of KV733 to colonize. The colonization defect conferred by the mutation in pgm is similar to that caused by some amino acid auxotrophies (15). It is more severe than either a mutation in the luxA gene, which causes a fourfold decrease only after the first day of colonization (48), or a mutation in catalase, which results in a competitive defect (50). We have observed that the pgm mutant is at least 10-fold reduced in colonization as early as 12 h postinoculation, a time at which the wild-type strain has not yet achieved its population maximum (C. R. DeLoney and K. L. Visick, unpublished observations). These data suggest that the pgm mutant is not simply growing normally until it reaches a population maximum that is 10-fold reduced from that which can be achieved by the wild-type strain. Rather, it appears that this mutant is deficient at even earlier stages of colonization. This result is consistent with our hypothesis that the pgm mutant fails to resist host-imposed stresses (see below).
What role does pgm play in symbiotic colonization? Given the pleiotrophic effects of pgm mutants, several explanations could be proposed to explain the symbiotic defect. One possibility was that the symbiotic defect resulted from the inability of the pgm mutant to use galactose as a carbon source. We speculated that galactose moieties on the surfaces of cells in the light organ might serve as a carbon source for the bacteria. A similar mechanism seems to be employed during colonization of the mouse gut by B. thetaiotaomicron, which induces the presentation of the sugar fucose on host cells and then uses it as an energy source (22). However, our construction of a second galactose utilization mutant, one not defective for LPS biosynthesis (data not shown) that colonized normally suggested that this hypothesis was incorrect.
A second possible explanation is that alterations in LPS, correlated with susceptibility to stresses, prevent the mutant from colonizing to wild-type levels because it cannot withstand environmental stresses in the squid. Although relatively little is known about the environment of the light organ, the presence of defensive host cells (primitive macrophages [35]) and proteins (halide peroxidase [53]), as well as the exclusivity of the symbiosis, suggests that the bacteria likely encounter host-derived stresses during colonization.
Comprising the vast majority of the outer leaflet of the outer membrane of gram-negative bacteria, LPS plays a crucial role in maintaining membrane integrity (see reference 34 for a review). Divalent cations such as magnesium form cross-bridges between neighboring LPS molecules by binding to phosphate residues on inner core heptose components (which are often derived from UDP-glucose [see reference 37 for a review]). These interactions give strength and rigidity to the otherwise fluid outer membrane. H. influenzae pgm mutants, presumably defective for UDP-glucose synthesis, contain LPS species with a reduced number of sugar residues in the inner core (21). If the pgm mutation caused a parallel defect on the inner core of V. fischeri LPS, which would not be surprising given the conserved nature of LPS structure among gram-negative bacteria (16), then the strength of the outer membrane likely would be reduced. Our sensitivity assays suggest that this is indeed the case. However, it is not clear whether the moderate increase in susceptibility to agents such as polymyxin B that the pgm defect confers in culture can account for the 10-fold decrease in colonization ability.
A third explanation for the symbiotic defect in KV733 could be the loss or reduction of outer membrane proteins necessary for colonization. Some LPS mutants (“deep rough”) that are devoid of part of the core oligosaccharide (for reviews, see references 33 and 37) show significant decreases in the amounts of the major outer membrane proteins. The V. fischeri outer membrane protein OmpU plays a role in symbiotic colonization (2) and may be affected by the LPS defect. Our preliminary data suggest that this is not the case; we have detected no distinct differences in the banding patterns of proteins extracted from wild-type and pgm mutant cells (K. Visick, unpublished observations).
In E. coli, UDP-glucose plays a number of roles other than LPS synthesis, including (i) protecting against thermal and osmotic stresses through the production of trehalose (7); (ii) serving as a building block for periplasmic β-d-glucans, which are implicated both in osmotic adaptation and in cell signaling (23); and (iii) regulating (negatively) σS, the stationary-phase sigma factor (18). Defects in any of these cellular processes could potentially result in the observed symbiotic phenotype in V. fischeri, and future research will have to address these possibilities.
The role of Pgm in colonization is not limited to the Vibrio-squid symbiosis. V. cholerae (32) and H. influenzae (21) pgm mutants similarly display defects in pathogenic colonization. The requirement of a metabolic enzyme in establishment of the symbiosis between V. fischeri and E. scolopes demonstrates the complexity of the association, as well as how little is known about the necessary genetic determinants. We anticipate that our characterization of the role of Pgm in this symbiotic association will provide opportunities for the comparison of symbiotic and pathogenic colonization. As we continue to search for insights on how these mutualistic partners interact, studies such as these will add to our knowledge of how symbiotic relationships—both beneficial and detrimental—are established between bacteria and animals.
Acknowledgments
We thank Paul Smith for his assistance in the characterization of the pgm gene and many plasmid constructions, Erika Enk for the construction of KV1069, Cionne Wolfe for isolating the V. fischeri galK gene, and Eric Stabb for plasmids and advice in advance of publication. We also thank David Keating, Ned Ruby, Eric Stabb, Jon Visick, and Alan Wolfe for their insightful comments and critical review of the manuscript.
This work was supported by NIH grant GM59690 awarded to K.L.V. and by the National Science Foundation Research Fellowship in Microbial Biology awarded in 2001 to C.R.D.
REFERENCES
- 1.Adhya, S., and M. Schwartz. 1971. Phosphoglucomutase mutants of Escherichia coli K-12. J. Bacteriol. 108:621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aeckersberg, F., C. Lupp, B. Feliciano, and E. G. Ruby. 2001. Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J. Bacteriol. 183:6590-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao, Y., D. P. Lies, F. Haian, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 8.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.Dunlap, P. V. 1989. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J. Bacteriol. 171:1199-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. Fitzhugh, C. A. Fields, J. D. Gocayne, J. D. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 12.Foster, J. S., M. A. Apicella, and M. J. McFall-Ngai. 2000. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Biol. 226:242-254. [DOI] [PubMed] [Google Scholar]
- 13.Fraenkel, D. G. 1996. Glycolysis, p. 189-198. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 14.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf, J., and E. G. Ruby. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 95:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronow, S., and H. Brade. 2001. Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? J. Endotoxin Res. 7:3-23. [PubMed] [Google Scholar]
- 17.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. A. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleischmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 19.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero, M., V. DeLorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hood, D. W., M. E. Deadman, T. Allen, H. Masoud, A. Martin, J. R. Brisson, R. Fleischmann, J. C. Venter, J. C. Richards, and E. R. Moxon. 1996. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol. Microbiol. 22:951-965. [DOI] [PubMed] [Google Scholar]
- 22.Hooper, L. V., J. Xu, P. G. Falk, T. Midtvedt, and J. I. Gordon. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy, E. P. 1996. Membrane-derived oligosaccharides (periplasmic β-d-glucans) of Escherichia coli, p. 1064-1071. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 24.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 25.Lamarcq, L. H., and M. J. McFall-Ngai. 1998. Induction of a gradual, reversible morphogenesis of its hosts epithelial brush border by Vibrio fischeri. Infect. Immun. 66:777-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, M., and N. Kleckner. 1994. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J. Bacteriol. 176:5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFall-Ngai, M. J. 1999. Consequences of evolving with bacterial symbionts: insights from the squid-vibrio associations. Annu. Rev. Ecol. Syst. 30:235-256. [Google Scholar]
- 28.McFall-Ngai, M., C. Brennan, V. Weis, and L. Lamarcq. 1998. Mannose adhesin-glycan interactions in the Euprymna scolopes-Vibrio fischeri symbiosis, p. 273-276. In Y. L. Gal and H. O. Halvorson (ed.), New developments in marine biothechnology. Plenum Press, New York, N.Y.
- 29.McFall-Ngai, M. J., and E. G. Ruby. 1991. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491-1494. [DOI] [PubMed] [Google Scholar]
- 30.Montgomery, M. K., and M. McFall-Ngai. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719-1729. [DOI] [PubMed] [Google Scholar]
- 31.Nealson, K. H. 1978. Isolation, identification, and manipulation of luminous bacteria. Methods Enzymol. 57:153-166. [Google Scholar]
- 32.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 34.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyholm, S. V., and M. J. McFall-Ngai. 1998. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol. Bull. 195:89-97. [DOI] [PubMed] [Google Scholar]
- 36.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raetz, C. R. H. 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles, p. 1035-1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 38.Ray, W. J., Jr., M. A. Hermodson, J. M. Puvathingal, and W. C. Mahoney. 1983. The complete amino acid sequence of rabbit muscle phosphoglucomutase. J. Biol. Chem. 258:9166-9174. [PubMed] [Google Scholar]
- 39.Reus, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. Kumar Kolli, and S. G. Pueppke. 1998. Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64:4930-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruby, E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591-624. [DOI] [PubMed] [Google Scholar]
- 41.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 42.Ruby, E. G., and K. H. Nealson. 1977. Pyruvate production and excretion by the luminous marine bacteria. Appl. Environ. Microbiol. 34:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 44.Stabb, E. V., and E. G. Ruby. New RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol., in press. [DOI] [PubMed]
- 45.Stan-Lotter, H., M. Gupta, and K. E. Sanderson. 1979. The influence of cations on the permeability of the outer membrane of Salmonella typhimurium and other gram-negative bacteria. Can. J. Microbiol. 25:475-485. [DOI] [PubMed] [Google Scholar]
- 46.Steinberg, D. A., and R. L. Lehrer. 1997. Designer assays for antimicrobial peptides: disputing the “one size fits all” theory. Methods Mol. Biol. 78:169-186. [DOI] [PubMed] [Google Scholar]
- 47.Uttaro, A. D., G. A. Cangelosi, R. A. Geremia, E. W. Nester, and R. A. Ugalde. 1990. Biochemical characterization of avirulent exoC mutants of Agrobacterium tumefaciens. J. Bacteriol. 172:1640-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 182:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visick, K. L., and E. G. Ruby. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 180:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence, John Wiley & Sons, Ltd., Chichester, England.
- 52.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weis, V. M., A. L. Small, and M. J. McFall-Ngai. 1996. A peroxidase related to the mammalian antimicrobial protein myleperoxidase in the Euprymna-Vibrio mutualism. Proc. Natl. Acad. Sci. USA 93:13683-13688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recominants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zielinski, N. A., A. M. Chakrabarty, and A. Berry. 1991. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266:9754-9763. [PubMed] [Google Scholar]