Abstract
The genome of the cyanobacterium Synechocystis sp. strain PCC6803 has nine kinds of insertion sequence (IS) elements, of which ISY100 in 22 copies is the most abundant. A typical ISY100 member is 947 bp long and has imperfect terminal inverted repeat sequences. It has an open reading frame encoding a 282-amino-acid protein that appears to have partial homology with the transposase encoded by a bacterial IS, IS630, indicating that ISY100 belongs to the IS630 family. To determine whether ISY100 has transposition ability, we constructed a plasmid carrying the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible transposase gene at one site and mini-ISY100 with the chloramphenicol resistance gene, substituted for the transposase gene of ISY100, at another site and introduced the plasmid into an Escherichia coli strain already harboring a target plasmid. Mini-ISY100 transposed to the target plasmid in the presence of IPTG at a very high frequency. Mini-ISY100 was inserted into the TA sequence and duplicated it upon transposition, as do IS630 family elements. Moreover, the mini-ISY100-carrying plasmid produced linear molecules of mini-ISY100 with the exact 3′ ends of ISY100 and 5′ ends lacking two nucleotides of the ISY100 sequence. No bacterial insertion elements have been shown to generate such molecules, whereas the eukaryotic Tc1/mariner family elements, Tc1 and Tc3, which transpose to the TA sequence, have. These findings suggest that ISY100 transposes to a new site through the formation of linear molecules, such as Tc1 and Tc3, by excision. Some Tc1/mariner family elements leave a footprint with an extra sequence at the site of excision. No footprints, however, were detected in the case of ISY100, suggesting that eukaryotes have a system that repairs a double strand break at the site of excision by an end-joining reaction, in which the gap is filled with a sequence of several base pairs, whereas prokaryotes do not have such a system. ISY100 transposes in E. coli, indicating that it transposes without any host factor other than the transposase encoded by itself. Therefore, it may be able to transpose in other biological systems.
Transposable DNA elements that can move from one site to another are present in both prokaryotes and eukaryotes. These elements generally have terminal inverted repeat sequences (IRs) and a gene encoding transposase (for reviews, see references 18 and 22). Defective elements with a deletion in the transposase gene cannot transpose by themselves but can when transposase is supplied from another source. Upon transposition, they generate duplication of a sequence of several base pairs at the target site.
The insertion sequence (IS) is a small transposable element in bacteria. IS elements are classified as several families based on the homology in the amino acid sequences of the transposases encoded by the elements, IRs, and the length of the target site sequence (see reference 18). Transposases usually have a DNA-binding domain with a helix-turn-helix structure and a catalytic domain with a DDE motif. IS630 is an IS element which preferentially transposes to the TA sequence and duplicates it upon transposition (19, 36, 37). IS elements with structural features similar to those of IS630 have been found in Archaea, as well as in Eubacteria, and form a large family called the IS630 family (15, 18, 22, 34). Many IS elements belonging to other families promote the rearrangement of the genomes that carry them by deletion and inversion of a DNA segment adjacent to an element or by cointegration between an element-carrying donor and target replicons by transpositional recombination accompanied by replication of the element. Whether such genomic rearrangements are promoted by IS630 family elements, however, is not known.
In eukaryotes, the transposable element Tc1 from Caenorhabditis elegans (10) also transposes to the TA sequence as the target (41). Transposable elements similar to Tc1 have been found in other animals, ciliates, fungi, and plants, and these are collectively called the Tc1/mariner family elements (for a review, see reference 24). We have designated here the IS630 and Tc1/mariner families the IS630/Tc1 superfamily because the elements of these families encode transposases that have significant homology in the catalytic domain around a DDE motif near their C-terminal regions (7). Two Tc1/mariner family elements, Tc1 and Tc3, produce a linear DNA molecule by double-strand breaks at their ends (28, 39). The Tc3 linear molecule has a two-nucleotide (nt) overhanging sequence at each of the 3′ ends and is thought to be an intermediate in Tc3 transposition (40). Tc1/mariner family elements appear to be excised from their donor molecules, leaving an empty site with an extra sequence, called footprints (5, 9, 40), as do some other eukaryotic transposable elements belonging to different families. The transposition mechanism of IS elements belonging to the prokaryote IS630 family, however, has not been studied.
The entire genome sequence of the cyanobacterium Synechocystis sp. strain PCC6803, a photosynthetic bacterium, has been determined, and nine kinds of IS elements have been identified (14). Of these, ISY100 is present most abundantly in 22 copies per genome. ISY100 is referred to as IS1987 (6) and ISTcSa (http://www-is.biotoul.fr). In the present study, we first show that ISY100 is another prokaryotic element of the IS630 family. We then show that in an Escherichia coli K-12 strain an ISY100 member transposes to the target plasmid but does not promote cointegration between the ISY100-carrying donor and target plasmids. ISY100 is inserted specifically into the TA sequence and generates duplication of the sequence, as IS630 and Tc3 do. Furthermore, we show that ISY100 generates a linear ISY100 molecule, suggesting that ISY100 transposes via linear molecules excised from the ISY100-carrying DNA molecules, similar to the transposition of eukaryotic element Tc3. Unlike Tc3, ISY100 does not produce footprints at the site of excision. ISY100 transposes in E. coli K-12 at a very high frequency, suggesting that ISY100 transposition does not require a host protein. Therefore, ISY100 is likely to transpose in other organisms and may prove useful for genetic engineering and breeding of organisms.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used were Synechocystis sp. strain PCC6803 and E. coli K-12 strains MC1061 [F− araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL hsdR2 (rK− mK+) mcrA mcrB1] (43), NM554 [F− recA13 araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi hsdR mcrB] (26), JE6638 (F− polA1 purE trp lys proC leu thi lacZ xyl ara mtl mal man gal mel tonA tsx str rif nalA) (National Institute of Genetics collection), and BIK806 (F− thr1 leuB6 thi1 lacY1 galK2 ara14 xy15 mtl1 proA4 his4 argE31 rpsL31 tsx33 supE44 recD::Tn10) (4).
We used the following plasmids: pTrc99A (1), the pBR322-derived ampicillin resistance (Apr) plasmid with an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter in the region preceding the multiple cloning sites; pSEK80 (32), the R100-derived spectinomycin resistance (Spr) plasmid; and pHSG398 (35), the pUC18-derived chloramphenicol resistance (Cmr) plasmid. The pAU plasmids used are described below.
Media.
The culture media used were L broth and L-rich broth (44). L-agar plates contained 1.5% (wt/vol) agar (Wako) in L broth. When necessary, antibiotics were added to media as follows: ampicillin (Wako), 100 mg/ml; spectinomycin (Sigma), 100 mg/ml; and chloramphenicol (Sigma), 30 mg/ml (in L broth) or 90 mg/ml (in L agar).
Enzymes and synthetic oligonucleotides.
Restriction endonucleases [BamHI, BglII, BstEII, HindIII, NcoI, NdeI, NheI, SacII, and XhoI (New England Biolabs)], bacterial alkaline phosphatase, T4 DNA ligase, DNA polymerase I Klenow fragment, rTaq polymerase (Takara), RNase A (Sigma), terminal deoxynucleotidyl transferase (TdT; Boehringer Mannheim), and KOD DNA polymerase (Toyobo) were used with the buffers recommended by the suppliers.
Synthetic oligonucleotide primers (Table 1) were synthesized chemically in an OLIGO1000 M DNA synthesizer (Beckman).
TABLE 1.
Primers useda
| Primer | Sequence (5′→3′) | Position (range) |
|---|---|---|
| P1 | ggggtcgacATATGTCCGGACTTCGCTATAGTTTCTAAAG | |
| P2 | cccggatccATATGCTAGCTGTCCTCATCCGTATAATGC | |
| P3 | gggctcgagTGTTGATACCGGGAAGC | 42-58 |
| P4 | gggagatctAGGCGTAGCACCAGGCG | 900-884 |
| P5 | gggctcgagATCGATACACTCCATAATTTTAC | 51-34 |
| P6 | cccagatctCCGCGGTCCGTTCTTACTGTGGC | 899-915 |
| P7 | gggccATGGCTTACAGTTTAGA | 73-89 |
| P8 | cccggatccTAAACGCCACAGTAAGA | 921-904 |
| P9 | CAACGGTGGTATATCCAGTG | 3817-3798 |
| P10 | CAACAGTACTGCGATGAGTG | 4408-4427 |
| P11 | CTCCTTACGCATCTGTGC | 3491-3508 |
| P12 | CGGCATCAGAGCAGATTG | 4627-4610 |
| P13 | gggggatccatatgaagcttCCCCCCCCCCCC |
The positions of primers (P3, P4, and P9 to P12) are shown by coordinates given to pTrc99A (1). The positions of primers P5 to P8 are shown by coordinates given to ISY100 (see Fig. 1A). Additional nucleotides with a restriction site are indicated by lowercase letters.
PCR.
The PCR was done by the standard protocol with some modification on 50 ng of the template plasmid DNA, each primer pair, and 2.5 U of KOD DNA polymerase in a 50-μl solution. The step cycle program (total of 25 cycles) was set to denature at 94°C for 30 s, anneal at 55°C for 30 s, and extend at 74°C for 1 min. The PCR was done in a Perkin-Elmer Cetus DNA thermal cycler, and the products were separated in a 0.8% agarose gel.
Construction of pAU plasmids.
A plasmid (here called pAU1) carrying ISY100 was constructed by cloning the fragment amplified from the total DNA of Synechocystis sp. strain PCC6803 by the PCR with primers P1 and P2 (see Table 1), which hybridize to the regions flanking ISY100-a, into the NdeI site on plasmid pTrc99A after treatment of the PCR fragment with NdeI. Plasmid pAU3 carrying mini-ISY100 was constructed as follows. One fragment with the Cmr gene was amplified by PCR with primers P3 and P4 from the template pHSG398 DNA. The other pAU1 fragment without the transposase-coding region was amplified by PCR with primers P5 and P6. After XhoI and BglII treatment of the two fragments, they were mixed and ligated.
Plasmid pAU5 that carried the IPTG-inducible transposase gene and mini-ISY100 was constructed by replacing a short NcoI-BamHI segment of pAU3 in the multiple cloning sites with the transposase gene fragment which had been amplified from pAU1 by PCR with primers P7 and P8 and digested with NcoI and BamHI. Plasmid pAU7 that carried the IPTG-inducible transposase gene was constructed by religation of the pAU5 DNA that had been digested with NdeI to remove mini-ISY100.
Transposition assay.
The donor pAU plasmid and target plasmid pSEK80 were introduced in E. coli NM554 by transformation. A transformant was grown at 37°C for 18 to 22 h in L-rich broth containing chloramphenicol, ampicillin, and spectinomycin. The culture was then diluted 100-fold in the L-rich broth and, when the optical density at 600 nm (OD600) reached 0.3, IPTG was added to a final concentration of 0.5 mM. The culture was then incubated at 37°C for 18 to 22 h. Plasmid DNA was extracted from the cultured cells by the alkaline lysis method (29) and electroporated into E. coli JE6638 cells in a Gene Pulser (Bio-Rad). These cells were suspended in SOC medium and incubated at 37°C for 1 h. After appropriate dilution, cells were plated on an L plate containing spectinomycin or on a plate containing spectinomycin and chloramphenicol with or without ampicillin, followed by incubation at 37°C overnight. The transposition or cointegration frequency was calculated by dividing the number of Cmr Spr transformants by the number of Spr transformants or by dividing the number of Cmr Spr Apr transformants by the number of Spr transformants, respectively.
DNA sequencing.
DNA was sequenced by the dideoxynucleotide termination method (21, 30). The sequencing reaction was done by using a BigDye terminator cycle sequencing ready reaction kit (ABI PRISM) in a GeneAmp PCR system 9700 (Applied Biosystems). The primers used were oligodeoxyribonucleotides P9 and P10, which hybridize to the ISY100 sequence, or P11 and P12, which hybridize to the sequences flanking ISY100 in pAU3 and pAU5. Reaction products were analyzed in an ABI PRISM 377-18 sequencer (Applied Biosystems).
Southern hybridization.
E. coli NM554 cells harboring pAU3 or pAU5 were grown at 37°C overnight in L-rich broth containing chloramphenicol and ampicillin. The culture was diluted 100-fold in L-rich broth, IPTG was added to make a final concentration of 0.5 mM when the OD600 reached 0.4 to 0.6, and the culture was further incubated at 37°C for 2 h. Small DNA molecules were extracted under alkaline or neutral conditions (12, 29). The DNA samples were then electrophoresed in a 0.8% agarose gel and alkali blotted on a nylon membrane Hybond N+ (Amersham) for Southern hybridization with the oligonucleotide P9 probe that had been labeled with digoxigenin by using a DIG-oligonucleotide 3′ labeling kit (Roche). Chemiluminescence was detected with Hyperfilm (Amersham).
Determination of the ends of linear molecules.
Sample DNA was prepared under the neutral condition from BIK806 cells harboring pAU5, as described in the previous section, and electrophoresed in a 0.8% agarose gel. Linear ISY100 molecules were purified from the gel by using GeneClean III (Bio 101). The 3′ ends of these molecules were determined as follows. The purified linear molecules (10 ng) were treated at 37°C for 15 min with TdT (25 U), 0.75 mM CoCl2, and 2 mM dGTP in TdT buffer (G-tailing reaction). With the reaction product as the template, PCR was done with primers P9 and P13 to amplify the fragment with the IRL end region or with primers P10 and P13 to amplify the fragment with the IRR end region. The step cycle program (total of 30 cycles) was set to denature at 94°C for 15 s, anneal at 60°C for 15 s, and extend at 74°C for 30 s. The PCR products were sequenced with primer P5 or P6 to identify, respectively, the 3′ end at IRL or IRR of the linear molecules.
To determine the 5′ end of the linear molecules, the purified molecules were treated with DNA polymerase I Klenow fragment to blunt the end, and the G-tailing reaction described above was performed.
Note that the present method can be used to determine the ends of linear molecules, even when the molecules are few or impure, because the fragment with the 3′ ends of the molecules is amplified specifically.
Detection of molecules with a circle junction and of molecules with footprints.
To detect circular ISY100 molecules, sample DNA, prepared under the alkaline condition from NM554 cells harboring pAU5, as described above, was digested with four restriction enzymes (BamHI, HindIII, NdeI, and NcoI) and used as the template for PCR with primers P9 and P10 which hybridize to the end regions of the mini-ISY100 sequence and prime DNA synthesis toward the outside of the sequence. The PCR-amplified fragments were electrophoresed in a 1.5% agarose gel. Fragments ca. 400 bp in length were purified by using GeneClean III (Bio 101), cloned, and sequenced with primer P9 or P10.
To detect molecules with footprints, part of the same sample DNA was digested with three restriction enzymes (BglII, XhoI, and SacII) and then introduced into NM554 by transformation. Structures of plasmids prepared from the Apr transformants were analyzed by restriction enzyme cleavage and DNA sequencing with primers P11 and P12.
RESULTS
Structural features of ISY100.
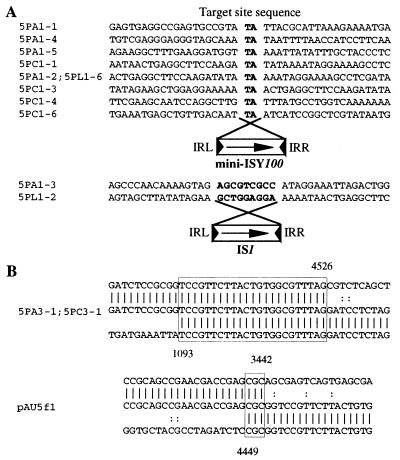
According to the Cyanobase database (http://www.kazusa.or.jp/cyano/), the Synechocystis sp. strain PCC6803 genome has 22 ISY100 members (ISY100-a, ISY100-b, etc.). Alignment of these members showed that they are highly homologous (>97.9%). Except for five members with truncation in one or two end regions, all are flanked by TA sequences. A typical ISY100 member, ISY100-a, is 947 bp in length and has imperfect terminal IRs of 24 bp (here called IRL and IRR). It has an open reading frame (ORF), which encodes a 282-amino-acid protein, probably transposase (Fig. 1A). Ten ISY100 members, however, are 1 bp shorter than ISY100-a and thus have two short ORFs because of the presence of a stretch of 9 rather than 10 adenine (A) residues within the coding region in ISY100-a (Fig. 1A).
FIG. 1.
(A) Nucleotide sequence of ISY100 (947 bp). The amino acid sequence of transposase encoded by ISY100 is shown below the nucleotide sequence. The TA sequences adjacent to both ends of the ISY100 sequence indicate the target site sequence duplicated on the transposition of ISY100. The arrows show the terminal IRs, IRL and IRR. The underlined amino acid sequence is the possible DNA-binding region with a helix-turn-helix motif. Amino acid residues of the DDE motif are circled. (B) Alignment of segments of transposases encoded by IS630/Tc1 superfamily elements. The positions of D, D, and E in the DDE motif are shown by boldface letters. Amino acid sequences were deduced from nucleotide sequences of the elements registered in the databases: D90899 (ISY100), X05955 (IS630), Z18270 (IS870), M61114 (IS1066), U67938 (IS1471), U22370 (ISRm2011-2), AP001515 (IS642), AE001010 and G69334 (ISA1083-2), S74044 (ISC1078), X01005 (Tc1), Z802210 (Tc3), AF282722 (Impala), S33560 (Bari-1), X78906 (mariner), and AF078934 (Soymar1). The numbers in parentheses show the distances (in amino acids) between amino acid sequences. Asterisks indicate the amino acid residues conserved in all of the transposase segments.
ISY100 (or ISS1987) was shown not to have homology to IS630 but to have partial homology to some IS630/Tc1 superfamily elements (6). The C-terminal half of the putative protein encoded by ISY100-a shows significant homology with the transposases encoded not only by IS630/Tc1 superfamily elements but also by IS630 and appears to have a DDE motif in the homologous regions, a finding indicative of a catalytic domain (Fig. 1B). The N-terminal region of this protein has homology with the regions encoded by IS630/Tc1 superfamily elements, i.e., IS630, Tc1, and Tc3, as well as with the IS3 family element IS981. This region appears to have a helix-turn-helix motif in the homologous regions, indicative of a DNA-binding domain (Fig. 1A). These clearly show that ISY100 belongs to the IS630/Tc1 superfamily.
Transposition of ISY100.
To see whether ISY100 has the ability to transpose, ISY100-a was examined for transposition in the E. coli K-12 background. First, an Apr plasmid pAU5 with mini-ISY100 and the IPTG-inducible transposase gene at different sites was constructed (Fig. 2A). Mini-ISY100 has an IRL and IRR of ISY100 that flank the Cmr gene instead of the transposase gene. Plasmid pAU5 then was introduced into E. coli NM554 cells that already harbored the Spr target plasmid pSEK80 (Fig. 2A). Plasmid DNA extracted from these cells, which had been cultured overnight in the presence of IPTG, was electroporated into another E. coli strain, JE6638(polA), and the cells were seeded on plates containing spectinomycin and chloramphenicol in order to select transformants carrying target plasmids with a simple insertion of mini-ISY100. Note that pAU5, a pBR322 plasmid derivative, cannot replicate in JE6638 (polA), whereas pSEK80, a derivative of plasmid R100, can. Spr Cmr transformants were obtained at the high frequency of 3.3 × 10−4 per target plasmid (Table 2). Plasmid pAU3 carries mini-ISY100 but not the IPTG-inducible transposase gene (see Fig. 2B). No Spr Cmr transformants were obtained when pAU3 was used instead of pAU5 (Table 2). This shows that the formation of Spr Cmr transformants depends on the transposase encoded by ISY100.
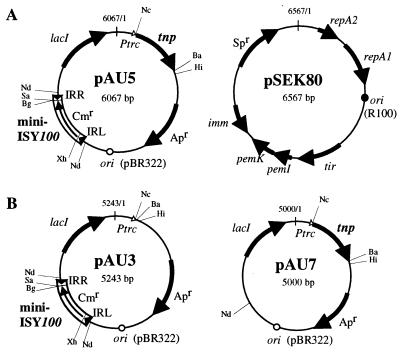
FIG. 2.
Plasmid structures. (A) pAU5 and pSEK80. (B) pAU3 and pAU7. Mini-ISY100 in pAU5 and pAU3 carries the Cmr gene, which is flanked by the IRL and IRR regions of ISY100 (51 and 53 bp, respectively). Apr, Apr gene; Ptrc, IPTG-inducible promoter; tnp, transposase gene encoded by ISY100-a; lacI, lacIq gene that represses transcription from Ptrc; ori, origin of replication of pBR322. The numbers in the pAU plasmids are coordinates given to the sequence of plasmid pTrc99A (1), from which the pAU plasmids were derived. The restriction sites shown are Ba (BamHI), Bg (BglII), Hi (HindIII), Nc (NcoI), Nd (NdeI), Sa (SacII), and Xh (XhoI). For the structural features of pSEK80, see the legend to Fig. 3.
TABLE 2.
Frequency of simple insertion or cointegration mediated by mini-ISY100
| Plasmida | tnpb | Frequency ofc:
|
|
|---|---|---|---|
| Simple insertion | Cointegration | ||
| pAU5 | + | 3.3 × 10−4 | <5.3 × 10−7 |
| pAU3 | − | <3.2 × 10−7 | <3.2 × 10−7 |
Donor plasmid carrying mini-ISY100.
Presence (+) or absence (−) of the IPTG-inducible transposase gene (tnp) in the pAU plasmid.
Frequencies of simple insertion and cointegration were calculated as described in Materials and Methods.
Structural analyses by restriction enzyme cleavage and DNA sequencing showed that plasmids in the Spr Cmr transformants carried insertion of mini-ISY100 in the TA sequence in one or the other orientations at various sites on pSEK80 (Fig. 3). The TA sequences appeared on both sides of mini-ISY100. This shows that ISY100 has the ability to transpose into the TA sequence and duplicate it.
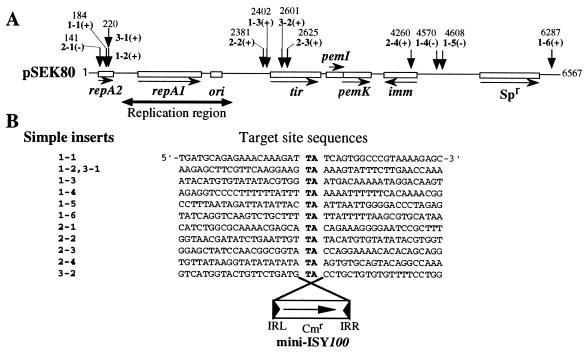
FIG. 3.
(A) Structure of plasmid pSEK80 (with coordinates 1 to 6567), showing insertion sites of mini-ISY100. The short vertical arrows indicate insertion sites. The numbers above the arrows indicate clone numbers and insertion sites. Note that mini-ISY100 is inserted such that orientation of the Cmr gene in mini-ISY100 is the same (+) as or opposite (−) to that of the Spr gene on pSEK80. repA1, the gene essential for replication initiation of plasmid R100; repA2, the gene responsible for copy number control of R100; tir, the R100 gene responsible for inhibition of plasmid RP4 transfer; pemI and pemK, genes required for stable maintenance of R100; imm, the gene responsible for immunity to superinfection of certain plasmids; Spr, Spr gene. Note that the insertion of mini-ISY100 occurred in the TA sequence at various sites on pSEK80 in regions outside of those required for plasmid replication and the Spr gene that was the selective marker. (B) Nucleotide sequences around target sites of mini-ISY100 insertion. Note that the TA sequences indicated by boldface letters appear on both sides of mini-ISY100.
Bacterial transposable elements, such as IS1 and transposon Tn3, mediate cointegration between the element-carrying donor and target plasmids, giving rise to a cointegrate with two copies of the element at the cointegration site (11, 13, 20, 23). To clarify whether mini-ISY100 can mediate cointegration, pAU5 (or pAU3) DNA was electroporated into JE6638 and then seeded onto plates containing spectinomycin, chloramphenicol, and ampicillin in order to select transformants with a cointegrate formed between the parental plasmids. No Spr Cmr Apr transformants, however, were obtained (at a frequency of less than 5.3 × 10−7 or 3.2 × 10−7; Table 2), showing that ISY100 does not have the ability to mediate cointegration.
Inhibition of cell growth by ISY100 transposase.
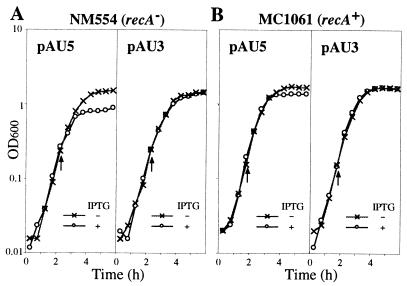
The growth of NM554 (recA) cells harboring plasmid pAU5, which carries both mini-ISY100 and the IPTG-inducible transposase gene (Fig. 2), was found to be inhibited after the addition of IPTG to the cell culture (Fig. 4A). The growth of cells harboring plasmid pAU7, which does not carry mini-ISY100 but carries the IPTG-inducible transposase gene (Fig. 2B), was also inhibited on the addition of IPTG (data not shown), whereas the growth of cells harboring pAU3, which carries mini-ISY100 but not the IPTG-inducible transposase gene (Fig. 2), was not (Fig. 4A). These findings show that transposase is responsible for cell growth inhibition. Inhibition of cell growth in another strain MC1061 (recA+) harboring pAU5 was, however, not as strong as that in NM554 (recA) cells (Fig. 4B). This finding shows that overexpression of transposase has a less deleterious effect on wild-type cells than on recA mutants.
FIG. 4.
Growth curves of cells of E. coli NM554 (A) or MC1061 (B) harboring plasmid pAU5 or pAU3. Cells were cultured overnight in L-rich broth containing chloramphenicol and ampicillin, diluted 100-fold with fresh L-rich broth containing the same drugs, and then incubated at 37°C. Arrows indicate the OD600 at which IPTG was added to make the final concentration 0.5 mM.
The relative efficiency of transformation of the Apr plasmid pAU5 in the presence of IPTG compared to that in its absence was ca. 3.1 × 10−4 in NM554. The relative efficiency of the transformation of pAU7, which carries the IPTG-inducible transposase gene but not mini-ISY100, was lower than 1.3 × 10−5, whereas that of pAU3, which carries mini-ISY100 but not the IPTG-inducible transposase gene, was 1.3. Plasmids extracted from 17 pAU5 transformants formed in the presence of IPTG consisted of 9 that were larger than pAU5, 5 that were smaller than pAU5, and 3 that were the same size as pAU5. Nucleotide sequencing analysis showed that eight of the nine larger plasmids had an insertion of mini-ISY100 in the TA sequence and one (5PA1-3) had an insertion of IS1 (which resides in the E. coli K-12 chromosome) in a 9-bp target sequence that was duplicated upon insertion (Fig. 5A and 6A). Note that all of the insertions occurred within the pAU5 transposase gene (Fig. 5A and 6A).
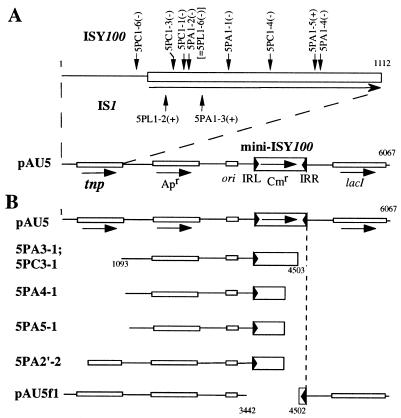
FIG. 5.
(A) Structure of pAU5, showing the insertion sites of ISY100 or IS1. The numbers indicate the insertion sites. (B) Structures of the deletion mutants from pAU5. The numbers in parentheses indicate positions at which deletions occurred. 5PC and 5PA are mutants found in colonies that were selected in the presence of IPTG on L-agar plates containing chloramphenicol and ampicillin, respectively. 5PL is a mutant found in a colony formed on an L-agar plate containing chloramphenicol and ampicillin. See Fig. 3 for nucleotide sequences around the sites of IS insertion and of deletion that occurred in pAU5.
FIG. 6.
(A) Nucleotide sequences around insertion sites of ISY100 and IS1 on pAU5. For the sites of insertion of these elements, see Fig. 5A. Note that target site sequences indicated by boldface letters appear on both sides of mini-ISY100 or IS1. (B) Nucleotide sequences around junctions where deletions occurred to generate miniplasmids. The sequence in the middle is a miniplasmid derived from pAU5, and the sequences above and below it are parental ones with a junction at which the deletion occurred. Identical bases between two sequences are indicated by vertical bars. Boxed sequences are thought to be used for deletion formation.
Of the five smaller plasmids obtained, two (5PA3-1 and 5PC3-1) were shown by nucleotide sequencing analysis to have a deletion in the transposase gene (Fig. 5B). The other three were shown by restriction enzyme cleavage analysis to also have a deletion in the transposase gene (Fig. 5B). These and the findings presented above suggest that cells harboring plasmids with a defect in the transposase gene that greatly affects cell growth are able to survive in the presence of IPTG. We therefore assume that the other three plasmids of the same size as pAU5 may bear a point mutation within the transposase gene and produce inactive transposase. Note that pAU5 derivatives with a mini-ISY100 or IS1 insertion in the transposase gene (see 5PL1-6 and 5PL1-2; Fig. 5 and 6A) also were readily obtained from cells grown in broth containing IPTG, a finding which supports this assumption. Moreover, deletions were not found at the end of ISY100 but at the short homologous sequences (Fig. 6B). This suggests that ISY100 does not promote the formation of deletion by transpositional recombination.
Generation of linear mini-ISY100 molecules.
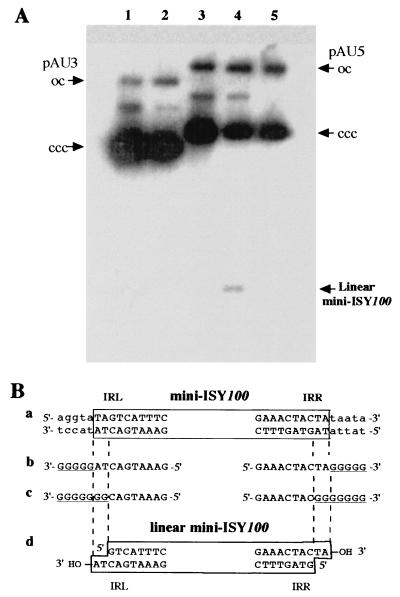
Some IS elements mediate the generation of miniplasmids from plasmids carrying them by intramolecular transposition, which is thought to be accompanied by the replication of each element. Certain IS elements also form circular and/or linear IS molecules; these are possible transposition intermediates. To examine whether pAU5-carrying mini-ISY100 generates miniplasmids or circular or linear mini-ISY100 molecules, a plasmid DNA sample was extracted under alkaline or neutral conditions from pAU5-carrying cells grown in the presence of IPTG, and the sample was electrophoresed in an agarose gel. Southern hybridization with a mini-ISY100 probe showed that only molecules of pAU5 DNA were present in the sample extracted under the alkaline condition (Fig. 7A, lane 3), but other molecules small in size were present in the sample extracted under the neutral condition (Fig. 7A, lane 4). No small molecules were detected in the DNA sample extracted under the alkaline or neutral conditions from cells harboring pAU3 with mini-ISY100 but without the transposase gene (Fig. 7A, lanes 1 and 2). These findings suggest that the small molecules generated from pAU5 are linear double-stranded DNA of mini-ISY100 produced by transposase. To confirm this, the DNA sample extracted under the neutral condition was treated with phage λ exonuclease that digests linear double-stranded DNA. The small molecules disappeared (Fig. 7A, lane 5), supporting the above suggestion that small molecules generated from pAU5 are linear double-stranded DNA.
FIG. 7.
(A) Agarose gels (0.8%) showing generation of linear mini-ISY100 molecules. Southern hybridization was done with a mini-ISY100 probe after electrophoresis of the sample DNA prepared from cells grown in the presence of IPTG. Lanes 1 and 2, DNA samples prepared from cells harboring pAU3 under alkaline and neutral conditions, respectively; lanes 3 and 4, DNA samples prepared from cells harboring pAU5 under alkaline and neutral conditions, respectively; lane 5, the λ exonuclease-treated DNA sample of lane 4. oc, open circular DNA molecule; ccc, covalently closed circular DNA molecule. (B) Structure of linear mini-ISY100 molecules generated from pAU5. (a) Nucleotide sequences of the end regions of mini-ISY100. (b) Nucleotide sequence of the 3′ end region with oligo(G). (c) Nucleotide sequence of the 5′-end region determined by identifying the 3′-end region of the complementary strand with oligo(G). (d) Structure of linear mini-ISY100 molecules deduced from the results in sequences b and c. The sequences flanking ISY100 are indicated by lowercase letters; oligo(G) sequences are underlined.
Although no small circular molecules were detected by Southern hybridization analysis in the sample prepared from the pAU5-carrying cells (see Fig. 7A), such molecules may be present in very small numbers. In fact, Tc1, a Tc1/mariner family element, generates a small number of circular molecules with an intervening sequence, e.g., TA and TA repeats, between IRs or with a deletion in a terminal region of Tc1 (25, 27, 28). To determine whether circular molecules of mini-ISY100 are present in small numbers, the pAU5 DNA sample was digested with restriction enzymes that cleave its DNA at five sites in the region outside of mini-ISY100. A PCR then was done with relevant primers to amplify fragments with the IR-IR junction region in the circular molecules (see Materials and Methods) (Fig. 8A). Cloning and DNA sequencing of the PCR-amplified fragments showed that one clone had a 2-bp TA sequence flanked by the IRR and IRL of mini-ISY100 and that the other clones had the IRR end joined with a sequence outside or inside mini-ISY100, such that the TA sequence is always present at the joint (Fig. 8). This shows that no clones with identical structures are obtained exclusively, which suggests that the presence of any of these molecules is rare. On the basis of this and the finding that pAU5 DNA molecules with insertion of an extra mini-ISY100 copy are present in the plasmid population, we assume that the fragments are not derived from circular molecules formed by intramolecular transposition but from pAU5 molecules with two copies of mini-ISY100, in which one has been inserted from another pAU5 molecule into the target near the mini-ISY100 already present in the original pAU5 molecule.
FIG. 8.
(A) Positions of four PCR fragments derived from pAU5 deletion mutants. Small arrows indicate the positions of primers used to amplify the fragments. Note that the ends of each of the fragments shown by the solid lines actually are joined together. Inner regions of ISY100, not included in the fragments amplified by PCR, are shown by dotted lines. (B) Nucleotide sequences around the junctions, at which deletion occurred. The numbers are coordinates given to pAU5.
Structural analysis of linear DNA molecules.
To determine the 3′ ends of the linear molecules, they were purified electrophoretically and treated in the presence of dGTP with TdT that adds dGTP and forms poly(G) at the 3′ ends of linear DNA. PCR then was done with the DNA sample as the template, an oligo(C) primer, and a second primer that hybridizes to mini-ISY100 in order to amplify the 3′-end region of IRR (or IRL) of mini-ISY100 (Fig. 7). Nucleotide sequencing of the PCR-amplified fragments showed that the poly(G) sequence is attached to the 3′ end of the defined ISY100 sequence (Fig. 7B, line b). This shows that the 3′ ends of the linear molecules are the exact ends of ISY100 and that these ends have an OH group, to which TdT added dGTP (Fig. 7B, line d).
To determine the 5′ ends of the linear molecules, sample DNA purified electrophoretically was treated in the presence of deoxynucleoside triphosphates with DNA polymerase I (Klenow fragment), which blunts the ends of linear DNA with a 5′- or 3′-overhanging sequence. The DNA sample then was treated in the presence of dGTP with TdT, and the 3′ ends of the DNA were determined as described above. Nucleotide sequencing of the PCR-amplified fragments showed that the poly(G) sequence is attached to the 3′ end, 2 nt inside the defined ISY100 sequence (Fig. 7B, line c). This means that the 5′ ends of the original linear molecules are 2 nt inside of ISY100 (Fig. 7B, line d). These findings show that the linear molecules are mini-ISY100 DNA with 2-nt overhangs at its 3′ ends (Fig. 7B, line d).
Search for molecules with footprints.
Some Tc1/mariner family elements are excised for transposition and leave a footprint, such as TATGTA or TACATA at the site of excision, in which TA sequences are the target originally used for the transposition of Tc1 and Tc3 (9, 40), as well as TATGATA or TACCATA, in which the TA sequences are the target site sequence originally used for transposition of mariner (5). To examine whether mini-ISY100 leaves footprints when excised, a pAU5 DNA sample prepared from cells grown in the presence of IPTG was digested with restriction enzymes (BglII, XhoI, and SacII), each of which cleaves mini-ISY100 on pAU5 (Fig. 2), and then electroporated into E. coli NM554. Sequencing of a small plasmid (pAU5f1) from one Apr transformant obtained by this procedure showed that a DNA segment whose end is not coincident with the exact end of mini-ISY100 is deleted in plasmid pAU5f1 (Fig. 5B). Identical 3-bp sequences were present in the regions where deletion is supposed to have occurred (Fig. 6B). These findings show that plasmid pAU5f1 is not a molecule with a footprint and that when ISY100 is excised no footprints are left.
DISCUSSION
In the present study, we showed that an ISY100 member present in the genome of Synechocystis sp. strain PCC6803 encodes a protein with homology in the region containing a DDE motif to the transposases encoded by IS630/Tc1 superfamily elements and that this member has the ability to transpose into a specific sequence, TA, as do IS630/Tc1 superfamily elements. This clearly indicates that ISY100 is an element that belongs to the IS630/Tc1 superfamily. In the cyanobacterium genome, about half the ISY100 members had one ORF, as does the member analyzed here, whereas others had two short ORFs due to the presence of a stretch of 9 rather than 10 A's within the coding region for transposase. These ISY100 members with two ORFs may be nonautonomous, i.e., not able to transpose by themselves, but can transpose in the presence of members with one ORF, thereby producing transposase. Otherwise, the members are autonomous and able to produce transposase by translational frameshifting at the 9-A sequence (for a review, see reference 22), although no stem-loop or pseudoknot structures, which enhance frameshifting efficiency, are present in the region downstream of the 9-A sequence.
Tc1, an IS630/Tc1 superfamily element, generates circular molecules with a ≥2-bp sequence intervening between the two IR ends of Tc1 (25, 27, 28). Although some elements belonging to other families generate circular molecules as transposition intermediates, the molecules from Tc1 are thought to be by-products not intermediates of transposition. We detected no circular molecules by Southern hybridization, but fragments with an intervening TA sequence between IRR and IRL by PCR (Fig. 8). These molecules are, however, thought not to be transposition intermediates but to be derived from pAU5 DNA molecules with two copies of ISY100, based on our finding that pAU5 DNA molecules with an insertion of an extra copy of ISY100 are present in the pAU5 DNA population.
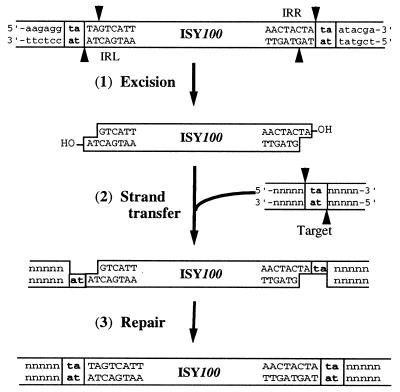
We showed that transposase promotes the production of linear mini-ISY100 molecules with 2-nt overhangs at the 3′ ends. Tc3, an eukaryotic element, generates a linear molecule with a 2-nt overhanging sequence at its 3′ ends, and this molecule is thought to be excised from the original site by double strand breaks promoted by Tc3 transposase, becoming a transposition intermediate (40). The linear molecule generated from the prokaryotic ISY100, therefore, may also act as a transposition intermediate, being inserted into the target by a cut-and-paste mechanism (Fig. 9), as proposed for Tc3. We also showed that ISY100 mediates neither cointegration nor generates miniplasmids. This finding supports the idea that ISY100 transposes by a cut-and-paste mechanism without accompanying replication of itself. In this mechanism, the transposases encoded by IS630/Tc1 superfamily elements may recognize terminal-end regions, as well as the target TA sequence, resulting in site-specific transposition of these elements. IS630, a prokaryotic element, tends to be inserted into palindromic sequences with the TA sequence as the core (37). ISY100, however, does not appear to be inserted into the palindromic sequences. Note that, unlike ISY100, some other prokaryotic transposable elements generate a linear DNA molecule with the 5′ overhang or blunt ends as a transposition intermediate (2, 3, 31). Linear molecules generated from all of the elements, including ISY100, however, have the 3′ ends coincident with the exact ends of the elements. This supports the idea that the hydroxyl group at the 3′ end functions in the reaction of strand transfer to the target DNA (2, 3, 31).
FIG. 9.
Model of ISY100 transposition. (Step 1) Transposase binds to the end regions of the element and stagger cuts the double-stranded DNA, generating the linear ISY100 molecule with 2-nt overhangs. (Step 2) The hydroxyl group (OH) at the 3′ end of a strand attacks the 5′ end of the TA target sequence to transfer that strand to the sequence. (Step 3) Host enzymes repair gaps of 4 nt, resulting in generation of a complete copy of ISY100 flanked by TA sequences at a new position.
Some Tc1/mariner family elements are excised for transposition and leave a footprint at the site of excision (5, 9, 40). These footprints are thought to be generated by repair of a gap with 2-nt overhangs generated upon excision, such that the gap is filled with a sequence of several base pairs. No footprints, however, were detected in the case of ISY100, even though excision of this element is supposed to generate the same gap in the ISY100-carrying plasmid DNA molecule as that generated by Tc1/mariner family elements. This suggests that eukaryotes have a repair system to leave footprints, whereas prokaryotes do not.
In the present study, we showed that ISY100 transposase inhibits the growth of E. coli cells. This deleterious effect is supported by the finding that transposase induction causes accumulation of the transposase-producing plasmid with an ISY100 (or IS1) insertion within the transposase gene. Transposases have the nonspecific DNA-binding ability, as well as the IR-specific DNA-binding ability (17, 33). Because ISY100 is not present in E. coli, its transposase may bind to DNA nonspecifically or specifically to sequences with partial homology to the IRs of ISY100, having caused inhibition of chromosome replication or the expression of some genes essential for cell growth.
Some transposable elements require a host factor for transposition. We showed that cyanobacterium ISY100 can transpose in E. coli. This suggests that ISY100 does not require a host factor but does transposase encoded by itself or that ISY100 does require a cyanobacterium factor but can transpose in E. coli because E. coli may have a homolog to the cyanobacterium factor. Some Tc1/mariner family elements transpose in vivo in species that differ from the original ones (for a review, see reference 24), and three of them—Tc1, Himar1, and Mos1—transpose in vitro only with their transposases (16, 38, 42). These results support the suggestion that ISY100 does not require a host factor.
In addition to ISY100, we examined the transposition of ISY523, the second most abundant IS element, which is present in 16 copies in the genome of Synechocystis sp. strain PCC6803. This element (871 bp in length) seems to belong to one of several subfamilies of the IS5 family, whose members transpose to the target sequence TTA (or TAA). It has imperfect terminal IRs of 17 bp and one ORF encoding transposase with a DDE motif characteristic of the members of the subfamily. We constructed a plasmid (pAU6) with mini-ISY523 and the IPTG-inducible transposase gene, as we did pAU5 with mini-ISY100. This mini-ISY523 on pAU6 does not transpose to the target plasmid pSEK80 in E. coli cells grown in the presence of IPTG (data not shown). Induction of transposase by IPTG, however, inhibits the growth of cells carrying pAU6 (data not shown), as in the case of ISY100, indicating that “active” transposase is produced. These findings suggest that ISY523, unlike ISY100, requires a host factor(s) in the cyanobacterium.
ISY100, a very small element, transposes at a high frequency and does not appear to require a host factor; therefore, it may be used as a tool for genetic engineering and breeding of various kinds of bacteria and even plants and animals. Considering that cyanobacteria are the original source of chloroplasts in plants (for a review, see reference 8) and that Tc1/mariner family elements have not been shown to transpose in plants, we are currently engaged in an examination of the transposition of ISY100 in plants.
Acknowledgments
We thank T. Tabata for the information on IS elements present in Synechocystis sp. strain PCC6803. We also thank K. Tanaka for providing the cyanobacterium strain.
This research was supported by a Grant-in-Aid of Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 2.Bainton, R., P. Gamas, and N. L. Craig. 1991. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell 65:805-816. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin, H. W., and N. Kleckner. 1992. Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. Proc. Natl. Acad. Sci. USA 89:4648-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biek, D. P., and S. N. Cohen. 1986. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J. Bacteriol. 167:594-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan, G., D. Garza, and D. Hartl. 1990. Insertion and excision of the transposable element mariner in Drosophila. Genetics 125:103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassier-Chauvat, C., M. Poncelet, and F. Chauvat. 1997. Three insertion sequences from the cyanobacterium Synechocystis PCC6803 support the occurrence of horizontal DNA transfer among bacteria. Gene 195:257-266. [DOI] [PubMed] [Google Scholar]
- 7.Doak, T. G., F. P. Doerder, C. L. Jahn, and G. Herrick. 1994. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci. USA 91:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas, S. E. 1998. Plastid evolution: origins, diversity, trends. Curr. Opin. Genet. Dev. 8:655-661. [DOI] [PubMed] [Google Scholar]
- 9.Eide, D., and P. Anderson. 1988. Insertion and excision of Caenorhabditis elegans transposable element Tc1. Mol. Cell. Biol. 8:737-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emmons, S. W., L. Yesner, K. S. Ruan, and D. Katzenberg. 1983. Evidence for a transposon in Caenorhabditis elegans. Cell 32:55-65. [DOI] [PubMed] [Google Scholar]
- 11.Galas, D. J., and M. Chandler. 1982. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J. Mol. Biol. 154:245-272. [DOI] [PubMed] [Google Scholar]
- 12.He, M., A. Wilde, and M. A. Kaderbhai. 1990. A simple single-step procedure for small-scale preparation of Escherichia coli plasmids. Nucleic Acids Res. 18:1660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heffron, F., B. J. McCarthy, H. Ohtsubo, and E. Ohtsubo. 1979. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell 18:1153-1163. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 15.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, J. C. Venter, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 16.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 17.Maekawa, T., J. Amemura-Maekawa, and E. Ohtsubo. 1993. DNA binding domains in Tn3 transposase. Mol. Gen. Genet. 236:267-274. [DOI] [PubMed] [Google Scholar]
- 18.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsutani, S., H. Ohtsubo, Y. Maeda, and E. Ohtsubo. 1987. Isolation and characterization of IS elements repeated in the bacterial chromosome. J. Mol. Biol. 196:445-455. [DOI] [PubMed] [Google Scholar]
- 20.McCormick, M., W. Wishart, H. Ohtsubo, F. Heffron, and E. Ohtsubo. 1981. Plasmid cointegrates and their resolution mediated by transposon Tn3 mutants. Gene 15:103-118. [DOI] [PubMed] [Google Scholar]
- 21.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsubo, E., and Y. Sekine. 1996. Bacterial insertion sequences. Curr. Top. Microbiol. Immunol. 204:1-26. [DOI] [PubMed] [Google Scholar]
- 23.Ohtsubo, E., M. Zenilman, and H. Ohtsubo. 1980. Plasmids containing IS elements are potential transposons. Proc. Natl. Acad. Sci. USA 77:750-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plasterk, R. H., Z. Izsvak, and Z. Ivics. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15:326-332. [DOI] [PubMed] [Google Scholar]
- 25.Radice, A. D., and S. W. Emmons. 1993. Extrachromosomal circular copies of the transposon Tc1. Nucleic Acids Res. 21:2663-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raleigh, E. A., N. E. Murray, H. Revel, R. M. Blumenthal, D. Westaway, A. D. Reith, P. W. Rigby, J. Elhai, and D. Hanahan. 1988. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 16:1563-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, A. M., and T. P. Snutch. 1984. Isolation of the closed circular form of the transposable element Tc1 in Caenorhabditis elegans. Nature 311:485-486. [DOI] [PubMed] [Google Scholar]
- 28.Ruan, K., and S. W. Emmons. 1984. Extrachromosomal copies of transposon Tc1 in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 81:4018-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekine, Y., N. Eisaki, and E. Ohtsubo. 1996. Identification and characterization of the linear IS3 molecules generated by staggered breaks. J. Biol. Chem. 271:197-202. [DOI] [PubMed] [Google Scholar]
- 32.Sekine, Y., K. Izumi, T. Mizuno, and E. Ohtsubo. 1997. Inhibition of transpositional recombination by OrfA and OrfB proteins encoded by insertion sequence IS3. Genes Cells 2:547-557. [DOI] [PubMed] [Google Scholar]
- 33.Sekino, N., Y. Sekine, and E. Ohtsubo. 1995. IS1-encoded proteins, InsA and the InsA-B′-InsB transframe protein (transposase): functions deduced from their DNA-binding ability. Adv. Biophys. 31:209-222. [DOI] [PubMed] [Google Scholar]
- 34.Sensen, C. W., H. P. Klenk, R. K. Singh, G. Allard, C. C. Chan, Q. Y. Liu, S. L. Penny, F. Young, M. E. Schenk, T. Gaasterland, W. F. Doolittle, M. A. Ragan, and R. L. Charlebois. 1996. Organizational characteristics and information content of an archaeal genome: 156 kb of sequence from Sulfolobus solfataricus P2. Mol. Microbiol. 22:175-191. [DOI] [PubMed] [Google Scholar]
- 35.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 36.Tenzen, T., S. Matsutani, and E. Ohtsubo. 1990. Site-specific transposition of insertion sequence IS630. J. Bacteriol. 172:3830-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenzen, T., and E. Ohtsubo. 1991. Preferential transposition of an IS630-associated composite transposon to TA in the 5′-CTAG-3′ sequence. J. Bacteriol. 173:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tosi, L. R., and S. M. Beverley. 2000. cis and trans factors affecting Mos1 mariner evolution and transposition in vitro, and its potential for functional genomics. Nucleic Acids Res. 28:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Luenen, H. G., S. D. Colloms, and R. H. Plasterk. 1993. Mobilization of quiet, endogenous Tc3 transposons of Caenorhabditis elegans by forced expression of Tc3 transposase. EMBO J. 12:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Luenen, H. G., S. D. Colloms, and R. H. Plasterk. 1994. The mechanism of transposition of Tc3 in C. elegans. Cell 79:293-301. [DOI] [PubMed] [Google Scholar]
- 41.van Luenen, H. G., and R. H. Plasterk. 1994. Target site choice of the related transposable elements Tc1 and Tc3 of Caenorhabditis elegans. Nucleic Acids Res. 22:262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vos, J. C., I. De Baere, and R. H. Plasterk. 1996. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 10:755-761. [DOI] [PubMed] [Google Scholar]
- 43.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 44.Yoshioka, Y., H. Ohtsubo, and E. Ohtsubo. 1987. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J. Bacteriol. 169:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]