Abstract
Transposon Tn5-692 mutagenizes Synechococcus sp. strain PCC 7942 efficiently. The predicted product of the gene mutated in the Tn5-692-derived cell division mutant FTN2 has an N-terminal DnaJ domain, as have its cyanobacterial and plant orthologs. Anabaena sp. strain PCC 7120, when mutated in genes orthologous to ftn2 and ftn6, forms akinete-like cells.
Division in cyanobacteria, ancient phototrophic relatives of chloroplasts, may serve as a model for the study of chloroplast division. However, the genetic basis of cell division has been studied much less in cyanobacteria than in heterotrophic bacteria (5, 6, 33, 37, 45). Conditional fts mutants of Escherichia coli affected in cell division were identified by screening for the formation of nonseptate filaments at a restrictive temperature (6, 19, 27, 37). FtsZ (2), a tubulin-like GTPase, forms the basis of a cytoskeletal structure that is used by many bacteria for the mechanical constriction of the cell at the division site (34, 38, 43). Although present in vegetative cells of the cyanobacterium Anabaena sp. strain PCC 7120 (12, 50), FtsZ was not detected in the nondividing, differentiated cells called heterocysts (30).
Cyanobacterial mutants impaired in cell division were identified after chemical mutagenesis (21-26, 51). Filamentous mutants were either septate (and thus impaired in cell separation) or serpentine, dividing sporadically to produce long, multinucleoidal cells (24). The gene mutated by random cassette mutagenesis (7, 31) in a septate mutant of Synechococcus sp. strain PCC 7942 (13) was characterized; it may be involved in lipopolysaccharide assembly.
Transposon mutagenesis of Synechococcus sp. strain PCC 7942.
Mutagenesis of PCC 7942 with transposon Tn901 was used to identify a methionine-biosynthetic gene and genes involved in nitrate assimilation (35, 36, 47). The utility of Tn901 is limited by its low frequency of transposition (18). Use of Tn5 in PCC 7120 (4) was enhanced by variant Tn5-1058 and derivatives that had (i) stronger expression of the antibiotic resistance operon, (ii) enhanced transposition, and (iii) an internal origin of replication that facilitates recovery of mutated genes (for examples, see references 3, 8, 16, and 49). Tn5 derivative Tn5-692 (in plasmid pRL692; GenBank accession no. AF424805) confers resistance to erythromycin, spectinomycin, and streptomycin; contains a pMB1 oriV; and bears mutations (52) that increase its rate of transposition ca. 100-fold relative to that of Tn5-1058 (49), providing large numbers of transposon mutants of Anabaena variabilis strain ATCC 29413 (PCC 7937) (our unpublished observations) and of PCC 7942.
Wild-type PCC 7942 and its derivatives were grown in BG11 medium (42), and wild-type PCC 7120 and its derivatives were grown as described by Hu et al. (20) in 125-ml Erlenmeyer flasks at 30°C in the light (ca. 3,500 ergs cm−2 s−1) on a rotary shaker. Antibiotics were added as appropriate. E. coli was grown and transformed as described previously (44). Tn5-692 was transferred to PCC 7942 and PCC 7937 by conjugation with E. coli strain HB101 bearing pRL443, pRL528, and pRL692 (10, 14). Filters bearing exconjugants were incubated for 48 h at 30°C (light intensity, 1,500 ergs cm−1 s−1) before transfer to medium containing erythromycin and spectinomycin (10 μg of each ml−1). Colonies appeared 10 to 15 days later. The frequency of transposition was ca. 3 × 10−5 to 6 × 10−5 per recipient cell. Extensively spreading, filamentous mutants appeared at a frequency of ca. 6 × 10−7 per recipient cell.
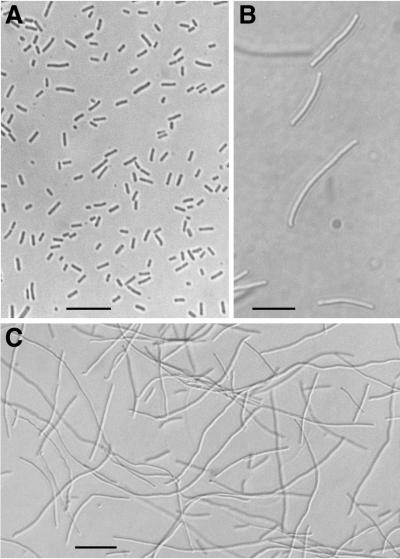
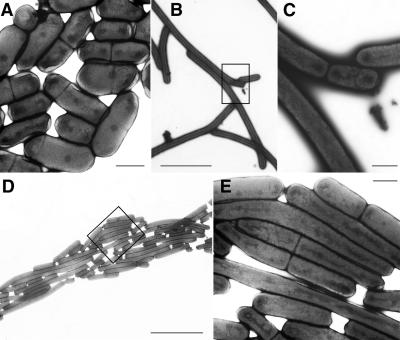
The cells of two such spreading mutants, FTN2 (Fig. 1C) and FTN6 (Fig. 1B), are up to 100-fold and 20-fold longer than wild-type cells, respectively (Fig. 1A). The growth rate of these mutants in liquid medium appeared to differ little from that of the wild type. Because their septation was not easily visualized by light microscopy, the cells were negatively stained with uranyl acetate and examined by electron microscopy. Sites of cell division in mutants FTN2 (Fig. 2B and C) and FTN6 (Fig. 2D and E) are much less frequent than in the wild-type strain (Fig. 2A). Spreading of the mutant colonies may be a consequence of the lengths of the individual cells providing a larger step size in a random-walk process of colonial growth. We use the designation “FTN” to suggest filamentation.
FIG. 1.
Morphology of wild-type PCC 7942 (A) and of mutants FTN2 (C) and FTN6 (B), grown in liquid medium and visualized by bright-field light microscopy. Scale bars represent 12.5 μm (A and B) or 25.6 μm (C).
FIG. 2.
Structures of wild-type PCC 7942 (A) and of mutants FTN2 (C; see box in panel B) and FTN6 (E; see box in panel D), negatively stained with uranyl acetate and examined by electron microscopy. The cells of both mutants divide infrequently. Scale bars represent 1 μm (A, C, and E) or 10 μm (B and D).
To clone oriV-containing Tn5-692, which lacks sites for SalI and BlnI, together with DNA contiguous with it, DNA recovered from FTN2 was cut separately with SalI and BlnI, circularized with T4 DNA ligase, and transformed to E. coli DH5α, yielding plasmids pRL2462 and pRL2733, respectively. DNA recovered from FTN6 was cut with SalI and similarly treated, yielding pRL2463. PCC 7942 was transformed with pRL2462 and pRL2463 (29). All spectinomycin- and erythromycin-resistant transformants were filamentous, establishing that the mutations were closely linked to the transposon. Mutants FTN2 and FTN6 were completely segregated, and the transposon is present in single-copy open reading frames (ORFs; data not shown) that we have provisionally designated ftn2 and ftn6.
DNA contiguous with the transposon was subcloned from pRL2462 to pBluescript SK(+) (Stratagene, La Jolla, Calif.) as XbaI-SalI and SpeI-SalI fragments, producing plasmids pRL2466 and pRL2468, respectively, and from pRL2463 to pBluescript SK(+) as XbaI-SalI and SpeI-SpeI fragments, producing plasmids pRL2465 and pRL2464, respectively. Part of plasmid pRL2733 was sequenced with primers. The expected 9-bp duplication adjacent to the site of insertion of the transposon was found in the case of FTN6 but, curiously, the two transposon-proximal 9-bp sequences recovered from FTN2 differed at one position [TGCAGGCG(C/T)]. To compare the sequences determined with the transposon-mutated genes with those from the wild-type sequences, genomic DNA from wild-type PCC 7942 was isolated (29) and the two wild-type genes were amplified piecewise by PCR and sequenced. Independent PCR amplifications confirmed the sequence TGCAGGCGC adjacent to the transposon in FTN2. Except for the final 183 bp of ftn2, which were sequenced only from pRL2733 as template, all portions of ftn2 and ftn6 were sequenced on both strands of DNA derived from a transposon recovery and on both strands of DNA PCR amplified from PCC 7942. Where there was any possible inconsistency, multiple independent PCR products were sequenced. Our sequence data include the final 282 bp of an ORF 3′ from ftn2 and the first 651 bp of an ORF 3′ from ftn6. Corresponding 3′ ORFs appear in sequence data from Synechococcus sp. strain PCC 6301 that were kindly provided by M. Sugita.
ftn2 and its downstream neighboring ORF are oppositely oriented and separated by a possible transcriptional terminator (CGCAaGGGGTgaaCCCCcTGCG [lowercase letters show deviations from the palindrome]), whereas ftn6 and its downstream ORF are carried on the same strand of DNA. Therefore, the phenotype of FTN2 is not, although the phenotype of FTN6 may be, attributable to a polar effect of the mutation on a downstream gene. Whereas the 152-amino-acid (aa) Ftn6 shows significant similarity (Expect value [E] [an Expect value is a BLAST indicator expressing the statistical significance of the alignment found and indicates the number of times one might expect to see such a match merely by chance] = 10−8 to 3 × 10−6) only to predicted proteins from other cyanobacteria, the predicted product of the parallel downstream ORF shows greater similarity to endopeptidase A of Arabidopsis (BLAST score, 616; E = 10−175) and of other cyanobacteria (BLAST scores, 684 to 653; E < 10−180) than to endopeptidases A from other bacteria (E ≥ 10−116).
ftn2 predicts a 631-aa protein, Ftn2, that shows greatest similarity to the predicted products of Anabaena sp. strain PCC 7120 ORF all2707 (28), which we have designated ftn2A (BLAST score, 278; E = 3 × 10−75 [1]), a Nostoc punctiforme ORF (BLAST score, 263; E = 10−70), and presumptive gene sll0169 of Synechocystis sp. strain PCC 6803 (BLAST score, 218; E = 2 × 10−55). Ftn2 also shares similarity with an Arabidopsis protein (AB016888; E = 10−11). The InterProScan program (http://www.ebi.ac.uk/interpro/scan.html) shows that Ftn2 and its cyanobacterial and plant orthologs have a DnaJ N-terminal domain (aa 6 to 70). Otherwise, they are dissimilar from other division-related proteins (6), as is the case with FTN6 and its orthologs. The presence in Ftn2 of a DnaJ domain, a single tetratricopeptide repeat (TPR) (aa 136 to 169; shown by the same program), and a leucine zipper pattern (aa 234 to 255, Prosite protein; PROSITE program at [http://ca.expasy.org/tools/scanprosite/]) suggests that Ftn2 may function as part of a complex with one or more other proteins and may be regulatory.
Proteins of the DnaJ-domain family are chaperonins that have a highly conserved J domain of approximately 70 aa, often found near the N terminus, that mediates interaction with DnaK and regulates the ATPase activity of the latter (9). dnaJ shares with fts genes the property that its inactivation leads to a filamentous phenotype (41). The dnaJ gene of Synechococcus sp. is required for growth (39).
The TPR, typically 34 aa in length and first described for the yeast cell division cycle regulator Cdc23p (46), was later found in many other proteins (11, 17, 32). Although frequently present in tandem arrays of 3 to 16 copies, single (as in FTN2) or paired TPRs are also common (32). Diverse processes involve TPR proteins, including cell-cycle control (32). No biochemical commonality connects TPR-containing proteins, although the TPR forms scaffolds that mediate protein-protein interactions and, often, the assembly of multiprotein complexes. The Arabidopsis Ftn2 ortholog is predicted to have a chloroplast transit peptide (http://HypothesisCreator.net/iPSORT/) and so may play a role in chloroplast division.
Inactivation of the ftnA genes of Anabaena sp. strain PCC 7120.
Orthologs ftn2A (see above) of ftn2 and ftn6A (all1616) (BLAST score, 76; E = 10−13) of ftn6 were identified in the genome of PCC 7120 (http://www.kazusa.or.jp/cyano/Anabaena). ftn2A is transcribed on the same strand of DNA as its downstream gene, whereas ftn6A is oriented opposite to its downstream gene. To inactivate ftn2A and ftn6A, a copy of each, truncated at both ends, was prepared by PCR with PCC 7120 DNA as template and primer pair 5′-CCGAATTCGTGGCAGTGGAAAATCGTGGG-3′ and 5′-CCGAATTCCACTTGCACGATTGGGATC-3′ and primer pair 5′-CCGAATTCGCCCTACTCATTAACTATAG-3′ and 5′-CCGAATTCCGGAGCGATCGCTTGTTTG-3′, respectively. The copies were cloned in the EcoRI site of pRL498 (15), producing plasmids pRL2471 and pRL2474, respectively. The clones were transferred by conjugation (14) to wild-type PCC 7120 with selection on AA plus nitrate agar medium containing 25 μg of neomycin ml−1. Homologous recombination was confirmed by Southern blotting (data not shown).
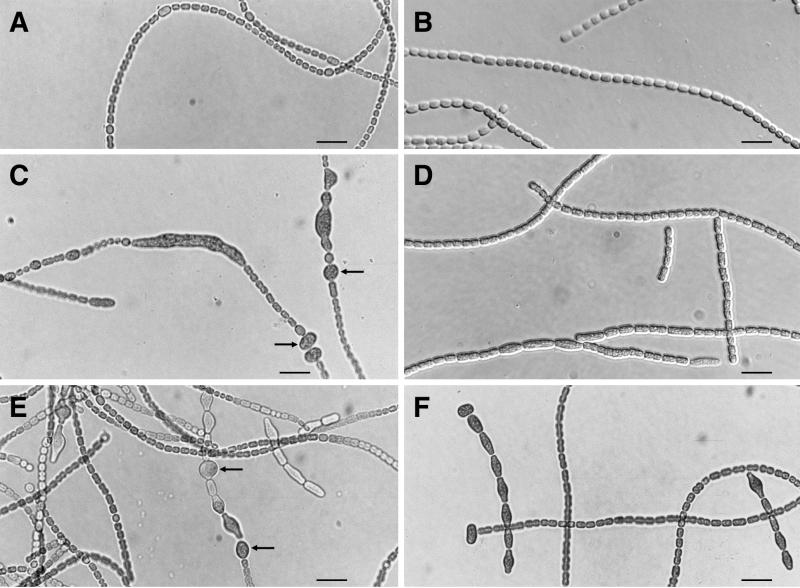
Anabaena sp. strain PCC 7120, a filamentous cyanobacterium, is capable of cellular differentiation (48). Cells of ftn2A (Fig. 3D) and ftn6A (Fig. 3F) Anabaena sp. strains, i.e., of PCC 7120::pRL2471 and PCC 7120::pRL2474, grown in the presence of nitrate were often up to twice as long as cells of the wild-type strain (Fig. 3B). In medium free of combined nitrogen, ftn2A (Fig. 3C) and ftn6A (Fig. 3E) mutants formed very elongated vegetative cells (those of ftn2A were up to 60-fold longer than those of the wild-type strain [Fig. 3A]), heterocysts of nearly normal size (but also sometimes up to fourfold larger, with an increase in both length and width), and enlarged akinete-like cells. Unlike mutant FTN6A, mutant FTN2A failed to segregate completely (data not shown). Wild-type PCC 7120 bears many copies of insertion sequences, including those in several ORFs whose putative products resemble protein kinases (40). Therefore, PCC 7120, which does not normally form akinetes in the laboratory, may itself exhibit a mutant phenotype. The presence of the greatly enlarged cells, which by their shape and frequent contiguity to heterocysts somewhat resemble akinetes, suggests that Ftn2A and Ftn6A may be involved in cellular differentiation as well as in division. The functions of ftn2, ftn2A, ftn6, and ftn6A remain to be elucidated.
FIG. 3.
Morphology of Anabaena sp. strain PCC 7120 wild type (A and B) and mutants FTN2A (C and D) and FTN6A (E and F) grown in liquid medium free of combined nitrogen (AA/8 [20]) (A, C, and E) or supplemented with 5 mM nitrate (B, D, and F). Scale bars represent 12.5 μm. Akinete-like cells are indicated by arrows.
Nucleotide sequence accession number.
The sequences of ftn2 and ftn6 have been submitted to GenBank under accession no. AF421196 and AF421197.
Acknowledgments
Samples were prepared for electron microscopy and micrographed by Sally Burns, Michigan State University Center for Electron Optics. We thank Mimoru Sugita, Nagoya University, for the sequences of ftn2- and ftn6-containing contigs from Synechococcus sp. strain PCC 6301.
This work was supported by the U.S. Department of Energy under grant DOE-FG02-91ER20021.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 3.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 4.Borthakur, D., and R. Haselkorn. 1989. Tn5 mutagenesis of Anabaena sp. strain PCC 7120: isolation of a new mutant unable to grow without combined nitrogen. J. Bacteriol. 171:5759-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouche, J. P., and S. Pichoff. 1998. On the birth and fate of bacterial division sites. Mol. Microbiol. 29:19-26. [DOI] [PubMed] [Google Scholar]
- 6.Bramhill, D. 1997. Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13:395-424. [DOI] [PubMed] [Google Scholar]
- 7.Broedel, S. E., and R. E. Wolf. 1990. Genetic tagging, cloning, and DNA sequence of the Synechococcus sp. strain PCC 7942 gene (gnd) encoding 6-phosphogluconate dehydrogenase. J. Bacteriol. 172:4023-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, Y., and C. P. Wolk. 1997. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J. Bacteriol. 179:258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheetham, M. E., and A. J. Caplan. 1998. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, M. F., J. C. Meeks, Y. A. Cai, and C. P. Wolk. 1998. Transposon mutagenesis of heterocyst-forming filamentous cyanobacteria. Methods Enzymol. 297:3-17. [Google Scholar]
- 11.Das, A. K., P. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty, H. M., and D. G. Adams. 1995. Cloning and sequence of ftsZ and flanking regions from the cyanobacterium Anabaena PCC 7120. Gene 163:93-99. [DOI] [PubMed] [Google Scholar]
- 13.Dolganov, N., and A. R. Grossman. 1993. Insertional inactivation of genes to isolate mutants of Synechococcus sp. strain PCC 7942: isolation of filamentous strains. J. Bacteriol. 175:7644-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed]
- 15.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119-138. [DOI] [PubMed]
- 16.Ernst, A., T. Black, Y. Cai, J.-M. Panoff, D. N. Tiwari, and C. P. Wolk. 1992. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J. Bacteriol. 174:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goebl, M., and M. Yanagida. 1991. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16:173-177. [DOI] [PubMed] [Google Scholar]
- 18.Golden, S. S. 1988. Mutagenesis of cyanobacteria by classical and gene-transfer-based methods. Methods Enzymol. 167:714-727. [DOI] [PubMed] [Google Scholar]
- 19.Hirota, Y., A. Ryter, and F. Jacob. 1968. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harbor Symp. Quant. Biol. 33:677-693. [DOI] [PubMed] [Google Scholar]
- 20.Hu, N.-T., T. Thiel, T. H. Giddings, and C. P. Wolk. 1982. Anabaena and Nostoc cyanophages from sewage settling ponds. Virology 114:236-246. [DOI] [PubMed] [Google Scholar]
- 21.Ingram, L. O., and E. L. Thurston. 1970. Cell division in morphological mutants of Agmenellum quadruplicatum, strain BG-1. Protoplasma 71:55-75. [Google Scholar]
- 22.Ingram, L. O., and C. Van Baalen. 1970. Characteristics of a stable, filamentous mutant of a coccoid blue-green alga. J. Bacteriol. 102:784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingram, L. O., C. Van Baalen, and W. D. Fisher. 1972. Cell division mutations in the blue-green bacterium Agmenellum quadruplicatum strain BG1: a comparison of the cell wall. J. Bacteriol. 11:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingram, L. O., and W. D. Fisher. 1973. Novel mutant impaired in cell division: evidence for a positive regulating factor. J. Bacteriol. 113:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingram, L. O., and W. D. Fisher. 1973. Mechanism for the regulation of cell division in Agmenellum. J. Bacteriol. 113:1006-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingram, L. O., G. J. Olson, and M. M. Blackwell. 1975. Isolation of a small-cell mutant in the blue-green bacterium Agmenellum quadruplicatum. J. Bacteriol. 123:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, R. B., and L. Shapiro. 2000. Proteins on the move: dynamic protein localization in prokaryotes. Trends Cell Biol. 10:483-488. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8(Suppl.):205-213, 227-253. [DOI] [PubMed] [Google Scholar]
- 29.Koksharova, O., M. Schubert, S. Shestakov, and R. Cerff. 1998. Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 36:183-194. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn, I., L. Peng, S. Bedu, and C.-C. Zhang. 2000. Developmental regulation of the cell division protein FtsZ in Anabaena sp. strain PCC 7120, a cyanobacterium capable of terminal differentiation. J. Bacteriol. 182:4640-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labarre, J., F. Chauvat, and P. Thuriaux. 1989. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 171:3449-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamb, J. R., S. Tugendreich, and P. Hieter. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20:257-259. [DOI] [PubMed] [Google Scholar]
- 33.Levin, P. A., and R. Losick. 2000. Asymmetric division and cell fate during sporulation in Bacillus subtilis, p. 167-189. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 34.Lowe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 35.Luque, I., A. Herrero, E. Flores, and F. Madueño. 1992. Clustering of genes involved in nitrate assimilation in the cyanobacterium Synechococcus. Mol. Gen. Genet. 232:7-11. [DOI] [PubMed] [Google Scholar]
- 36.Madueño, F., W. E. Borrias, G. A. Van Arkel, and M. G. Guerrero. 1988. Isolation and characterization of Anacystis nidulans R2 mutants affected in nitrate assimilation: establishment of two new mutant types. Mol. Gen. Genet. 213:223-228. [Google Scholar]
- 37.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee, A., and J. Lutkenhaus. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17:462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oguchi, K., K. Nimura, H. Yoshikawa, and H. Takahashi. 1997. Sequence and analysis of a dnaJ homologue gene in cyanobacterium Synechococcus sp. PCC7942. Biochem. Biophys. Res. Commun. 236:461-466. [DOI] [PubMed] [Google Scholar]
- 40.Ohmori, M., M. Ikeuchi, N. Sato, P. Wolk, T. Kaneko, T. Ogawa, M. Kanehisa, S. Goto, S. Kawashima, S. Okamoto, H. Yoshimura, H. Katoh, T. Fujisawa, S. Ehira, A. Kamei, S. Yoshihara, R. Narikawa, and S. Tabata. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:271-284. [DOI] [PubMed] [Google Scholar]
- 41.Paciorek, J., K. Kardys, B. Lobacz, and K. I. Wolska. 1997. Escherichia coli defects caused by null mutations in dnaK and dnaJ genes. Acta Microbiol. Pol. 46: 7-17. [PubMed] [Google Scholar]
- 42.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 43.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Shapiro, L., and R. Losick. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89-98. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., M. S. Boguski, M. Goebl, and P. Hieter. 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307-317. [DOI] [PubMed] [Google Scholar]
- 47.Tandeau de Marsac, N., W. E. Borrias, C. J. Kuhlemeier, A. M. Castets, G. A. van Arkel, and C. A. M. J. J. van den Hondel. 1982. A new approach for molecular cloning in cyanobacteria: cloning of an Anacystis nidulans met gene using a Tn901-induced mutant. Gene 20:111-119. [DOI] [PubMed] [Google Scholar]
- 48.Wolk, C. P. 2000. Heterocyst formation in Anabaena, p. 83-104. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 49.Wolk, C. P., Y. Cai, and J.-M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, C.-C., S. Huguenin, and A. Friry. 1995. Analysis of genes encoding the cell division protein FtsZ and a glutathione synthetase homologue in the cyanobacterium Anabaena sp. PCC 7120. Res. Microbiol. 146:445-455. [DOI] [PubMed] [Google Scholar]
- 51.Zhevner, V. D., V. M. Glazer, and S. V. Shestakov. 1973. Mutants of Anacystis nidulans with modified process of cell division. Mikrobiologiya 42:290-297. (In Russian.) [PubMed]
- 52.Zhou, M., A. Bhasin, and W. S. Reznikoff. 1998. Molecular genetic analysis of transposase-end DNA sequence recognition: cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. J. Mol. Biol. 276:913-925. [DOI] [PubMed] [Google Scholar]