Abstract
When grown under K+ limitation, Escherichia coli induces the K+-translocating KdpFABC complex. The stimulation of ATPase activity by NH4+ ions was shown for the first time. Substitutions in KdpA, which is responsible for K+ binding and translocation, revealed that enzyme complexes KdpA:G232A and KdpA:G232S have completely lost their cation selectivity.
The KdpFABC complex is an inducible high-affinity K+ uptake system of Escherichia coli that belongs to the protein superfamily of P-ATPases (reviewed in references 1 and 2). The KdpFABC complex remains intact during solubilization and purification in the presence of nonionic detergents, and the solubilized enzyme retains its cation-stimulated ATPase activity (21). The catalytic subunit KdpB is homologous to the large subunit of other P-ATPases; it contains an ATP binding site and forms a phosphointermediate during the catalytic cycle (20). The KdpA subunit is involved in binding and transport of K+ (see reference 3 and references therein). The amino acid sequence reveals at least two regions with similarities to P-loop segments that form the selectivity filter of K+ channels (11, 12), from which KdpA might be evolutionarily derived (1, 7). In this study, site-directed mutagenesis was performed to investigate in detail the importance of the amino acid residues G232, G233, and G234 of KdpA, which have been aligned with the K+ channel selectivity filter motif GYG (11) for K+ affinity and cation selectivity of the KdpFABC complex.
Construction of site-directed mutants.
The E. coli strains and plasmids used in this study are listed in Table 1. A prerequisite for site-directed mutagenesis was plasmid pSM2, which was constructed from pSM1 (10) (Table 1) by introducing an additional KpnI site (silent mutation) into the kdpA gene. Further mutagenesis was performed by one-step PCR, using plasmid pSM2 as template. Mutagenized plasmids were designated according to their corresponding mutations; e.g., the pSM2 derivative carrying the mutation in kdpA that encoded the G232A change was named pSM-kdpA:G232A.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| TKW3205 | ΔkdpFABC Δatp706 thi rha lacZ nagA trkA405 trkD1 | 16 |
| DH5α | recA1 endA1 gyrA96 thi-1 relA1 hsdR17 (rK−, mK+) λ−supE44 F− φ80d lacZ ΔM15 Δ(lacZYA-argF)U169 deoR pho | GibcoBRL (Eggenstein, Germany) |
| Plasmids | ||
| pUC19 | 23 | |
| pSR3 | 3 | |
| pSR4 | kdpFABC | 17 |
| pSR5 | kdpFABC-kdpA:Q116R, derivative of pSR4 | 9 |
| pSD126 | kdpFABC-kdpA:G232D, derivative of pSR4 | 18 |
| pSM1 | kdpFABC, derivative of pSR4 | 10 |
| pSM2 | pSM1, derivative with KpnI site at position 806a | This study |
| pSM-kdpA:G232A | kdpA:G232A, derivative of pSM2 | This study |
| pSM-kdpA:G232S | kdpA:G232S, derivative of pSM2 | This study |
| pSM-kdpA:G233A | kdpA:G233A, derivative of pSM2 | This study |
| pSM-kdpA:G233S | kdpA:G233S, derivative of pSM2 | This study |
| pSM-kdpA:G233D | kdpA:G233D, derivative of pSM2 | This study |
| pSM-kdpA:G233Y | kdpA:G233Y, derivative of pSM2 | This study |
| pSM-kdpA:G234A | kdpA:G234A, derivative of pSM2 | This study |
| pSM-kdpA:G234S | kdpA:G234S, derivative of pSM2 | This study |
| pSM-kdpA:G234D | kdpA:G234D, derivative of pSM2 | This study |
Positions given are counted from the transcription start according to the study of Sugiura et al. (22).
In vivo characterization of the mutants.
In order to test the effects of the different mutations on the K+ affinity of the KdpFABC complexes in vivo, strain TKW3205, which contains no functional K+ uptake system, was transformed with the different plasmids listed in Table 1. Cells were grown as described previously (19). Ampicillin-resistant single colonies were transferred to minimal medium agar plates containing 0 to 115 mM KCl (K0 to K115). It should also be noted that nominally K+-free plates (K0) contain up to 20 μM traces of K+ due to contaminations of the chemicals used. Most of the strains carrying mutant derivatives of kdpA showed the same growth properties as TKW3205/pSM1 (wild-type kdpFABC) (data not shown). TKW3205/pSM-kdpA:G233Y did not grow on K+ at concentrations less than 0.3 mM, and TKW 3205/pSD126 and TKW3205/pSR5 need at least 0.5 mM K+ for growth, whereas TKW3205/pSM-kdpA:G232A and TKW3205/pSM-kdpA:G232S required at least 5 mM K+ for growth (data not shown), suggesting a strong effect of the corresponding substitutions on the K+ affinity of the KdpFABC complex.
In vitro characterization of purified wild-type and mutant KdpFABC complexes.
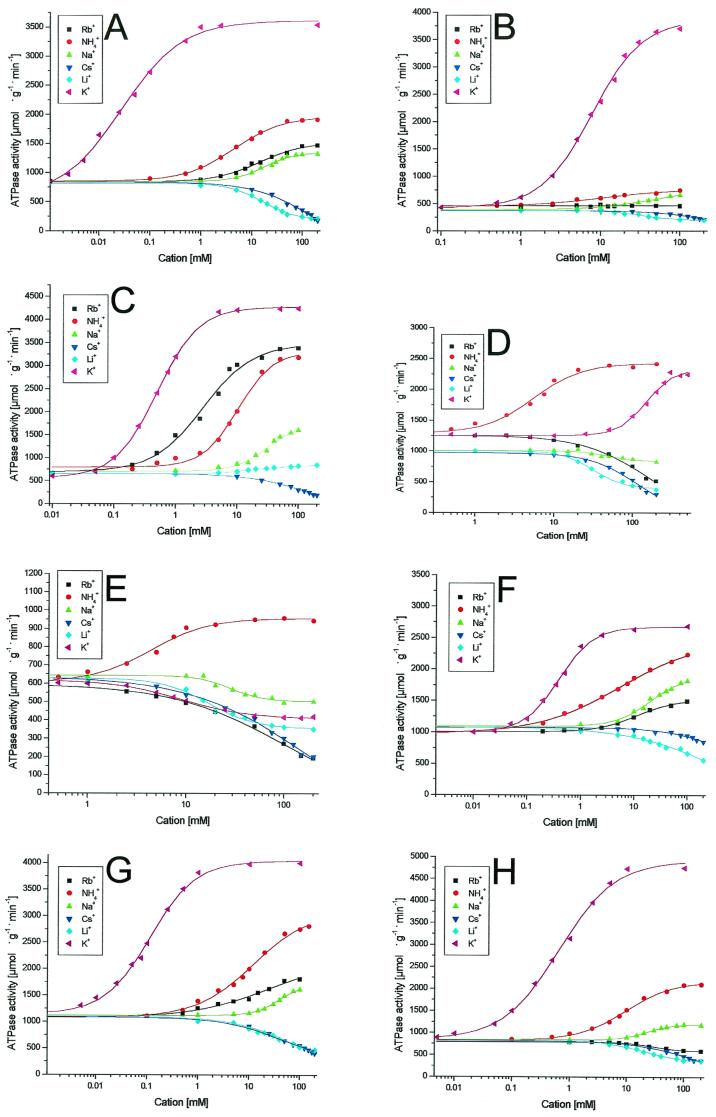
For induction of wild-type and mutant kdpFABC operons, strain TKW3205, after being transformed with one of the different plasmids, was grown in K0 medium or in medium with higher but limiting K+ concentrations, according to the influence of the particular amino acid substitution on the K+ affinity. The different KdpFABC complexes were purified as described previously (21). To remove NH4Cl, KdpFABC-containing fractions were dialyzed for 16 to 18 h at 4°C against a 200-fold volume of the buffer (50 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 10% glycerol, 0.2% Aminoxid WS 35), changed once. The cation-dependent ATPase activity, which is a common feature of P-ATPases (13, 15), was determined for the KdpFABC complexes as described previously (2). The monovalent cations tested were K+, Rb+, Na+, Li+, Cs+, and NH4+. The factors by which saturating concentrations of the different cations stimulated or inhibited the basal ATPase activities and the cation concentrations, resulting in half-maximal stimulation or inhibition (K0.5), were calculated and are summarized in Table 2. For wild-type KdpFABC (Fig. 1A), K+ causes a stimulation factor of 4 and a K0.5 of 39 μM was found. While Rb+ and Na+ were shown to slightly stimulate ATPase activity, Cs+ and Li+ inhibited ATPase activity ca. threefold. Interestingly, NH4+ stimulated the ATPase activity by a factor of 2.2 with a K0.5 of 3.9 mM, meaning that maximal activation was approximately half of that achieved with K+ but with a 100-fold lower affinity than that achieved with K+. In contrast, complex Q116R (Fig. 1B), which shows a strongly reduced K+ affinity but the same maximal rate and cation selectivity as those of the wild-type enzyme (9, 18), was hardly stimulated by NH4+. (Purified KdpFABC complexes from kdpA mutant strains are named according to the amino acid substitution, e.g., complex G232D.) Complex G232D was first described by Buurman et al. (3) and more intensively analyzed by Schrader et al. (18). The strongly reduced ability of this mutant enzyme to discriminate against Rb+ was confirmed (Fig. 1C). A maximal rate similar to those observed using K+ and Rb+ was also observed in the presence of NH4+. Figure 1D shows the cation-dependent ATPase activity of complex G232A. No significant K+ stimulation of ATPase activity was observed at concentrations up to 25 mM, and no saturation was achieved, even at 200 mM. At higher concentrations, however, the stimulating effect of K+ overlaps with the inhibiting effect of high ionic strength, a general effect observed for all enzyme complexes tested (M. van der Laan, unpublished results); therefore, K0.5 and maximum rate of metabolism values could not be determined properly. Maximal ATPase activity was 2,280 μmol · g−1 · min−1 (measured in the presence of 300 mM KCl), which represents a 1.8-fold stimulation. Calculated from this value, the K0.5 would be ∼150 mM, which implies a 3,750-fold decrease of the K+ affinity at a minimum. In contrast, NH4+ stimulated the ATPase activity of mutant G232A with the same affinity and rate as those of the wild-type complex (Table 2). Rb+ was found to inhibit the ATPase activity of complex G232A ca. twofold. With complex G232S, comparable results were obtained (Fig. 1E). Surprisingly, K+ inhibited ATPase activity of the purified G232S complex. However, when tested in inner-membrane vesicles, high K+ concentrations moderately stimulated ATPase activity (data not shown). The wild-type enzyme, as well as all other mutant enzymes tested, did not show these discrepancies between the membrane-integrated and solubilized states. Substitutions of amino acids G233 and G234 (Table 2) only moderately affected the ion-binding properties of KdpA. The K+ affinity of complex G233Y (Fig. 1F) was reduced ∼10-fold, and the maximal activity was somewhat lower. For substitution G234D (Fig. 1H), a similar effect on affinity but an increased maximal activity were observed.
TABLE 2.
Cation-dependent ATPase activities of purified KdpFABC complexesa
| Complex | Basal ATPase activityb | Severalfold change in ATPase activity [K0.5 (mM)] withc:
|
|||||
|---|---|---|---|---|---|---|---|
| K+ | Rb+ | NH4+ | Na+ | Li+ | Cs+ | ||
| Wild type | 839 | 4.2 [0.04] | 1.7 [10] | 2.2 [3.9] | 1.4 [15] | 0.3 [15] | 0.3 [55] |
| Q116R | 406 | 9.2 [7.2] | 1.0 [−]e | 1.5 [9.9] | 1.6 [27] | 0.7 [35] | 0.7 [75] |
| G232D | 654 | 6.5 [0.50] | 5.0 [3.3] | 4.9 [9.1] | 2.4 [27] | 1.3 [18] | 0.3 [50] |
| G232A | 1,106 | 1.8 [150]d | 0.4 [78] | 1.9 [5.3] | 0.8 [40] | 0.3 [52] | 0.3 [73] |
| G232S | 618 | 0.7 [14] | 0.3 [60] | 1.6 [3.9] | 0.8 [20] | 0.6 [13] | 0.3 [45] |
| G233Y | 1,046 | 2.6 [0.40] | 1.6 [17] | 2.2 [3.6] | 1.7 [18] | 0.5 [28] | 0.8 [70] |
| G233D | 1,088 | 3.8 [0.11] | 1.7 [10] | 2.5 [8.4] | 1.5 [32] | 0.4 [31] | 0.4 [29] |
| G233A | 828 | 4.5 [0.08] | 2.0 [7.5] | 2.2 [3.7] | 1.3 [21] | 0.2 [15] | 0.3 [60] |
| G233S | 985 | 3.8 [0.12] | 2.0 [10] | 2.4 [6.2] | 1.7 [15] | 0.5 [26] | 0.6 [92] |
| G234D | 714 | 6.6 [0.53] | 0.8 [23] | 2.6 [8.8] | 1.4 [22] | 0.4 [18] | 0.4 [46] |
| G234A | 912 | 4.4 [0.10] | 1.6 [15] | 2.1 [4.2] | 1.2 [23] | 0.2 [18] | 0.2 [31] |
| G234S | 930 | 4.3 [0.17] | 1.3 [8.1] | 1.9 [7.4] | 1.0 [−]e | 0.3 [10] | 0.3 [24] |
The data presented represent average values obtained in at least three independent experiments.
Averaged basal activity values for complexes without any cations are given in micromoles per gram per minute.
Values for stimulating cations are in bold letters, whereas values for inhibiting cations are in light face letters; change was measured in the presence of saturating concentrations of the different cations. Values less than 1 indicate an inhibitory effect, whereas a value of 1 represents no stimulation or inhibition. Values for cations without any effect or with only a small effect are underlines. K0.5 values [in millimolar units) for half-maximal stimulation or inhibition are in brackets.
Complex G232A showed a very low affinity for K+ such that stimulation by K+ was increasingly counteracted by the inhibitory effect of high salt concentrations (>200 mM) (Fig. 1).
[−], K0.5 value = 0.
FIG.1.
Cation-dependent ATPase activities of different KdpFABC complexes. (A) Wild-type complex. (B) Complex Q116R. (C) Complex G232D. (D) Complex G232A. (E) Complex G232S. (F) Complex G233Y. (G) Complex G233D. (H) Complex G234D. The plots of the ATPase assays of complexes G233A, G233S, G234A, and G234S did not vary significantly from those of the wild-type complex (compare panel A) and were therefore omitted. The KdpFABC complexes were purified, and ATPase activity assays were performed as described in the text.
Concluding remarks.
Based on sequence alignments with K+ channels and K+ symporters (7, 8, 11, 12) and on mutagenesis studies (3, 5), it has been suggested that the three conserved glycine residues in KdpA, G232, G233, and G234, form a selectivity filter-like structure similar to that found in the KcsA K+ channel (6). In particular, G232 appeared to be of crucial importance for K+ affinity and cation selectivity. However, the only mutation in this region that has been characterized biochemically so far is G232D (3, 18). We have systematically mutagenized this GGG motif and purified the altered KdpFABC complexes and characterized them by means of their cation-dependent ATPase activity. Using this biochemical approach, we have found that substitution G232S and even the rather conserved substitution G232A cause a dramatic decrease of K+ affinity and a complete loss of cation selectivity. This confirms and extends in vivo studies by Dorus et al. (5) and demonstrates that the growth phenotype of the corresponding mutants is indeed caused by an impaired K+ binding to KdpA. Our results stress that the K+ selectivity is mainly determined by G232, while a variety of amino acid substitutions is tolerated at positions 233 and 234. It is therefore suggested that G232 is homologous to the highly conserved N-terminal selectivity filter glycine residue in the putative P-loops of K+ symporters, which, like KdpA, might be evolutionarily derived from K+ channels (8). Furthermore, we show for the first time that NH4+ also strongly stimulates the ATPase activity of the KdpFABC complex. Surprisingly, G232 substitutions only moderately affect NH4+ affinities, indicating mechanistic differences between K+ and NH4+ binding. It remains to be established whether KdpFABC can actually transport NH4+, as has been suggested by Neijssel et al. (14) and Buurman et al. (4) on the basis of physiological studies.
Acknowledgments
We thank G. Deckers-Hebestreit for constructive criticism, E. P. Bakker, W. Epstein, and Marc Bramkamp for helpful discussion, and Heike Gerdes for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB431/P7) and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Altendorf, K., and W. Epstein. 1996. The Kdp-ATPase of Escherichia coli, p. 403-420. In A. G. Lee (ed.), Biomembranes (ATPases), vol. 5. JAI Press Inc., Greenwich, Conn.
- 2.Altendorf, K., M. Gaßel, W. Puppe, T. Möllenkamp, A. Zeeck, C. Boddien, K. Fendler, E. Bamberg, and S. Dröse. 1998. Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. 163(Suppl.):137-146. [PubMed]
- 3.Buurman, E. T., K.-T. Kim, and W. Epstein. 1995. Genetic evidence of two sequentially occupied K+ binding sites in the Kdp transport ATPase. J. Biol. Chem. 270:6678-6685. [DOI] [PubMed] [Google Scholar]
- 4.Buurman, E. T., M. J. Teixera de Mattos, and O. M. Neijssel. 1991. Futile cycling of ammonium ions via the high affinity potassium uptake system (Kdp) of Escherichia coli. Arch. Microbiol. 155:391-395. [DOI] [PubMed] [Google Scholar]
- 5.Dorus, S., H. Mimaru, and W. Epstein. 2001. Substrate-binding clusters of the K+-transporting Kdp ATPase of Escherichia coli investigated by amber suppression scanning mutagenesis. J. Biol. Chem. 276:9590-9598. [DOI] [PubMed] [Google Scholar]
- 6.Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and cation selectivity. Science 280:69-77. [DOI] [PubMed] [Google Scholar]
- 7.Durell, S. R., E. P. Bakker, and H. R. Guy. 2000. Does the KdpA subunit from the high affinity K+-translocating P-type Kdp-ATPase have a structure similar to that of K+ channels? Biophys. J. 78:188-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durell, S. R., Y. Hao, T. Nakamura, E. P. Bakker, and H. R. Guy. 1999. Evolutionary relationship between K+ channels and symporters. Biophys. J. 77:775-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fendler, K., S. Dröse, W. Epstein, E. Bamberg, and K. Altendorf. 1999. The Kdp-ATPase of Escherichia coli mediates an ATP-dependent, K+-independent electrogenic partial reaction. Biochemistry 38:1850-1856. [DOI] [PubMed] [Google Scholar]
- 10.Gaßel, M. 1999. Charakterisierung, Reinigung und Rekonstitution des Kdp-Komplexes aus Escherichia coli unter besonderer Berücksichtigung der Untereinheiten KdpC und KdpF sowie Untersuchungen zur Identifikation der Plekomakrolidbindestelle von P- und V-ATPasen. Ph.D. thesis. University of Osnabrück, Osnabrück, Germany.
- 11.Jan, L. Y., and Y. N. Jan. 1994. Potassium channels and their evolving gates. Nature 371:119-122. [DOI] [PubMed] [Google Scholar]
- 12.Jan, L. Y., and Y. N. Jan. 1997. Cloned potassium channels from eukaryotes and prokaryotes. Annu. Rev. Neurosci. 20:91-123. [DOI] [PubMed] [Google Scholar]
- 13.Møller, J. V., B. Juul, and M. le Maire. 1996. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1286:1-51. [DOI] [PubMed] [Google Scholar]
- 14.Neijssel, O. M., E. T. Buurman, and M. J. Teixera de Mattos. 1990. The role of the futile cycles in the energetics of bacterial growth. Biochim. Biophys. Acta 1018:252-255. [DOI] [PubMed] [Google Scholar]
- 15.Pedersen, P. L., and E. Carafoli. 1987. Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem. Sci. 12:146-150. [Google Scholar]
- 16.Puppe, W. 1991. Kalium-Transport bei Escherichia coli: Molekulargenetische und biochemische Untersuchungen zu funktionellen Domänen der Kdp-ATPase. Ph.D. thesis. University of Osnabrück, Osnabrück, Germany.
- 17.Puppe, W., A. Siebers, and K. Altendorf. 1992. The phosphorylation site of the Kdp-ATPase of Escherichia coli: site-directed mutagenesis of the aspartic acid residues 300 and 307 of the KdpB subunit. Mol. Microbiol. 6:3511-3520. [DOI] [PubMed] [Google Scholar]
- 18.Schrader, M., K. Fendler, E. Bamberg, M. Gassel, W. Epstein, K. Altendorf, and S. Dröse. 2000. Replacement of glycine 232 by aspartic acid in the KdpA subunit broadens the ion specificity of the K+-translocating KdpFABC complex. Biophys. J. 79:802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siebers, A., and K. Altendorf. 1988. The K+-translocating Kdp-ATPase from Escherichia coli. Purification, enzymatic properties and production of complex- and subunit-specific antisera. Eur. J. Biochem. 178:131-140. [DOI] [PubMed] [Google Scholar]
- 20.Siebers, A., and K. Altendorf. 1989. Characterization of the phosphorylated intermediate of the K+-translocating Kdp-ATPase from Escherichia coli. J. Biol. Chem. 264:5831-5838. [PubMed] [Google Scholar]
- 21.Siebers, A., R. Kollmann, G. Dirkes, and K. Altendorf. 1992. Rapid, high-yield purification and characterization of the K+-translocating Kdp-ATPase from Escherichia coli. J. Biol. Chem. 267:12717-12721. [PubMed] [Google Scholar]
- 22.Sugiura, A., K. Nakashima, K. Tanaka, and T. Mizuno. 1992. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 6:1769-1776. [DOI] [PubMed] [Google Scholar]
- 23.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]