Abstract
The nitrogen-regulated genes and operons of the Ntr regulon of Escherichia coli are activated by the enhancer-binding transcriptional activator NRI∼P (NtrC∼P). Here, we examined the activation of the glnA, glnK, and nac promoters as cells undergo the transition from growth on ammonia to nitrogen starvation and examined the amplification of NRI during this transition. The results indicate that the concentration of NRI is increased as cells become starved for ammonia, concurrent with the activation of Ntr genes that have less- efficient enhancers than does glnA. A diauxic growth pattern was obtained when E. coli was grown on a low concentration of ammonia in combination with arginine as a nitrogen source, consistent with the hypothesis that Ntr genes other than glnA become activated only upon amplification of the NRI concentration.
Escherichia coli contains six operons that are known to be part of the Ntr regulon (argThisJQMP, astCADBE, glnALG, glnHPQ, glnKamtB, and nac). These operons require σ54-RNA polymerase for expression and are activated by the phosphorylated form of enhancer-binding transcription factor NRI (NtrC) (reviewed in reference 16). A seventh operon, consisting of the ygiG gene, may also be a part of the Ntr regulon (16). E. coli also contains numerous additional genes that become activated or repressed upon nitrogen starvation (27).
The mechanism of activation by NRI∼P at σ54-dependent promoters has been studied in some detail (reviewed in reference 10). NRI∼P binds to upstream enhancer elements, oligomerizes, and displays ATPase activity. This complex interacts with σ54-RNA polymerase bound at the promoter to bring about formation of the open transcription complex. The interaction between NRI∼P and σ54-RNA polymerase requires the formation of a DNA loop, bringing the enhancer-bound activator and promoter-bound polymerase into proximity. In some cases, regulatory factors bind the intervening DNA and activate or repress transcription by topological alteration of the DNA.
The different nitrogen-regulated promoters contain distinct arrangements of NRI-binding sites that constitute their enhancer elements. The glnAp2 promoter apparently contains the most potent enhancer, consisting of two adjacent high-affinity NRI-binding sites (14, 20). The glnHp2 promoter, consisting of overlapping high-affinity sites, appears to be slightly less effective in vitro (4). The Klebsiella pneumoniae nifLA enhancer contains adjacent low-affinity NRI-binding sites and is only effective at high NRI∼P concentrations in vitro (26). Similarly, the nac promoter of Klebsiella aerogenes has a weak enhancer that is only effective at high NRI∼P concentrations in vitro; this enhancer consists of a high-affinity NRI-binding site and an adjacent site that is bound by NRI∼P only at high concentration (7). Thus, in vitro transcription studies are consistent with the hypothesis that amplitude modulation of the NRI∼P concentration results in the sequential activation of Ntr genes.
A considerable body of additional evidence also supports this hypothesis. The intracellular concentration of NRI rises dramatically in cells growing under nitrogen-limiting conditions (19), owing to the activation of the glnAp2 promoter by NRI∼P (15). Also, cells that have been genetically manipulated such that the NRI concentration is always low retain the ability to fully activate glnAp2 but are unable to grow on arginine as a nitrogen source (15) or activate the glnK promoter (2). The inability to grow on arginine probably reflects the inability to activate the astC promoter (16, 23). Similarly, the activation of the K. pneumoniae nifLA promoter requires a high concentration of NRI∼P in vivo (10). It seems reasonable that the nac, glnK, astC, and nifLA promoters should be activated by NRI∼P only at high concentrations, since the products resulting from their activation are useful under starvation conditions (16-18).
Finally, the signal transduction system that regulates the NRI phosphorylation state is able to provide rheostat-like control of NRI∼P in response to signals of nitrogen status (reviewed in reference 13). This, in combination with the observation that the NRI concentration is dramatically regulated in cells, suggests that cells may widely vary the concentration of NRI∼P in response to changes in environmental conditions.
Nevertheless, there are a few significant gaps in our knowledge. The great instability of NRI∼P has prevented its direct measurement in situ. Furthermore, most of the experiments with intact cells summarized above were conducted with log-phase cells growing under nitrogen excess or nitrogen-limiting conditions; our conclusions concerning transitions represent extrapolations from these results. Here, we focused on the growth of cells as their environment changes from nitrogen replete to nitrogen starved and measured the activation of the glnA, glnK, and nac promoters, as well as the amplification of the intracellular concentration of NRI. In addition, we examined the patterns of growth when E. coli was provided with ammonia and arginine as nitrogen sources.
MATERIALS AND METHODS
Bacteriological techniques.
Luria-Bertani broth and W salts-based defined media, preparation of plasmid DNA, preparation of competent cells, transformation of cells with DNA, sequencing of plasmid DNA, PCR amplification of DNA, preparations of P1vir phage lysates, P1-mediated transduction, recombination of DNA onto the bacterial chromosome, and long-term storage of strains were by standard techniques or were as described previously (1, 2, 5, 8, 12, 22, 24). The bacterial strains, plasmids, and oligonucleotides used in this work are described in Tables 1 to 3. Turbidity of bacterial cultures was measured with a Beckman DU65 spectrophotometer.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Construction or reference |
|---|---|---|
| YMC10 | endA1 thi-I hsdR17 supE44 ΔlacU169 hutCklebs | 3 |
| X | endA1 thi-1 hsdR17 supE44 ΔlacU169 hutCklebsamtB::Camr | 2 |
| TE2680 | recD1903::Tn10 trpDC700::putPA130 [Kans Camr] | 5 |
| YMC10Φ | endA1 thi-1 hsdR17 supE44 ΔlacU169 hutCklebs trpDC700::putPA130 [Φ(glnKp-lacZ) Kanr Cams] | 2 |
| MAAplac3 | recD1903::Tn10 trpDC700::putPA130 [Φ(glnAp2-lacZ) Kanr Cams] | TE2680 × PstI pglnAplac4 |
| YMC10ApΦ2 | endA1 thi-1 hsdR17 supE44 ΔlacU169 hutCklebs [Φ(glnAp2-lacZ) Kanr Cams] | YMC10 × MAAplac3 Plvir |
| TE2680NpΦ | recD1903::Tn10 trpDC700::putPA130 [Φ(nacp-lacZ) Kanr Cams] | TE2680 × PstI pNacLacZ |
| YMC10NpΦ | endA1 thi-1 hsdR17 supE44 ΔlacU169 hutCklebs [Φ(nacp-lacZ) Kanr Cams] | YMC10 × TE2680NpΦ P1vir |
TABLE 3.
Primers used in this study
| Primer | Sequence and description |
|---|---|
| glnApU.S.2 | CCGGAATTCATCCTCCGCAAACAAGTATTGCAGAG; upstream primer for fusing glnAp to lac in pRS551 (EcoRI) |
| glnApD.S.5 | CGCGGATCCTTACACCTGATGAGCAGGGATAGTGAC; downstream PCR primer to fuse glnAp to lac in pRS551 (BamHI) |
| 7159I | GCTGCAGGGATCCCATTGAGCGCCTGAATAGCGC; upstream PCR primer to clone glnKp into pTE103 (BamHI) |
| 4804J | GAAGCTTGAATGGTTTGATTATCACGGTCACC; downstream PCR primer to clone glnKp into pTE103 (HindIII) |
| NacpLacZus2 | CCGGAATTCGCTTTCAATCTTATTGG; upstream primer for fusing nac promoter to lac in pRS551 (EcoRI) |
| NacpLacZds2 | CGCGGATCCTGCCGCCATTACTTACA; downstream primer for fusing nac promoter to lac in pRS551 (BamHI) |
Construction of glnAp-lacZ and nacp-lacZ fusions and recombination onto the bacterial chromosome
PCR was used to amplify the glnA control region with primers glnApD.S.5 and glnApU.S.2 (Table 3). Similarly, primers NacpLacZus2 and NacpLacZds2 were used to amplify the nac control region (Table 3). The amplification products were cleaved with EcoRI and BamHI and then cloned into similarly cleaved pRS551 (25) to form a transcriptional fusion to lacZ bracketed by transcriptional terminators with flanking sequence homology to trp genes. The fusions were recombined onto the bacterial chromosome by electroporation of strain TE2680 with PstI-digested plasmid DNA and selection for kanamycin-resistant, chloramphenicol-sensitive transformants (5). The recombinants were confirmed to have a new auxotrophic requirement for tryptophan, indicating correct recombination into the trp locus. The fusions were then moved into various genetic backgrounds by P1vir-mediated generalized transduction, with selection for the nearby kanamycin resistance marker.
GS and β-galactosidase assays.
The γ-glutamyl transferase activity of glutamine synthetase (GS) was measured as described previously (21). Protein determinations were as described by Lowry et al. (11). Two cultures were used for each determination, and the experiments were repeated on three different occasions. Within a given experiment, values for duplicate cultures were within 10%, while the day-to-day reproducibility was ±20%. β-Galactosidase was measured by the Miller assay and expressed as Miller units, and sodium dodecyl sulfate and chloroform were used to disrupt the cells as described previously (24). In previous work, we measured the expression of the glnKp-lacZ fusion using cell sonicates (2). Comparison of the two methods indicated that the Miller assay reproducibly detects about one-fourth of the activity found in cell extracts (A. J. Ninfa and M. R. Atkinson, unpublished data).
Immunoblotting.
A crude rabbit anti-NRI antibody was kindly provided by Lawrence Reitzer. Immunoblotting was performed with the Amersham ECL Western blotting system according to the manufacturer's directions.
In vitro transcription assays.
The assay methods were identical to those described in reference 7. Transcription templates pTH8 and pLR100, containing the glnAp control region, were as described previously (7, 9, 14). The glnKp transcription template, pglnK13, was constructed by PCR amplification using primers 7159I and 4804J (Table 3), cleavage with BamHI and HindIII, and ligation into similarly cleaved pTE103, essentially as described previously (9). The plasmid was sequenced to verify that no alterations were introduced to the glnK control region during these manipulations. NRI, NRII, core RNA polymerase, and σ54 were purified as described earlier (7, 14).
RESULTS
Establishing conditions to study the transition from nitrogen excess growth to nitrogen starvation.
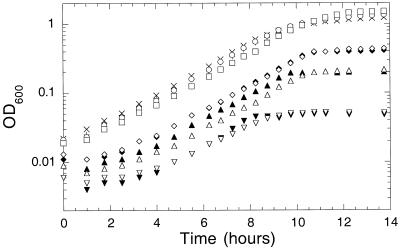
Since the preferred nitrogen source for E. coli is ammonia, we examined the effect of growth in the presence of various concentrations of ammonia. We found that while cell yield depended on the ammonia concentration, the growth rate did not (Fig. 1). Apparently, E. coli is very effective in capturing ammonia, even when it is present at low concentration. These results call into question the validity of the common practice in which medium containing limiting ammonia is used to establish low steady-state rates of growth in chemostat experiments. The amtB gene, encoding a putative ammonia transporter, had no effect on the utilization of ammonia in our experiments, even at very low ammonia concentrations (Fig. 1).
FIG. 1.
Growth of YMC10 (wild type) and X (amtB::Camr) on various concentrations of ammonium sulfate. Overnight cultures grown at 30°C in 0.4% (wt/vol) glucose and 0.2% (wt/vol) ammonium sulfate were washed and resuspended in medium containing 0.4% (wt/vol) glucose and the following concentrations of ammonium sulfate: for YMC10, 0.2 (×), 0.05 (+), 0.01 (⧫), 0.005 (▴), and 0.001% (▾); for X, 0.2 (○), 0.05(□), 0.01 (◊), 0.005 (▵), and 0.001% (▿). OD600, optical density at 600 nm.
To study the activation of the glnA, glnK, and nac promoters with high precision in dilute bacterial cultures, we engineered transcriptional fusions of these promoter regions to lacZ. These fusions were placed onto the chromosome in single copy within the trp operon (25) (see Materials and Methods). The glnKp-lacZ fusion was described previously (2); construction of the glnA and nac fusions is described in Materials and Methods. Expression of the glnAp-lacZ fusion was observed to parallel the expression of GS in adapted log-phase cells growing under nitrogen-rich and nitrogen-limiting conditions (data not shown).
Activation of glnAp, glnKp, and nacp as cells growing on ammonia became nitrogen starved.
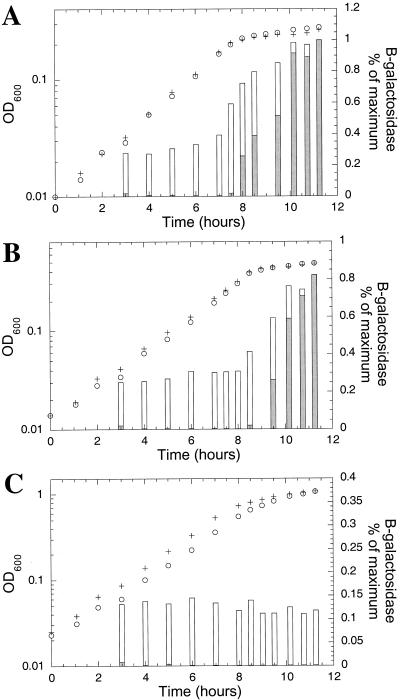
A comparison of the activation of glnAp and glnKp as cells underwent the transition from growth on ammonia to nitrogen starvation is shown in Fig. 2. A significant difference in the patterns of expression of glnKp and glnAp was observed. When grown on low concentrations of ammonium (0.005 [A] or 0.01% [B] ammonium sulfate), expression of glnAp was approximately one-third to one-fourth of the maximum level observed during exponential growth, while glnKp was essentially silent (Fig. 2A and B). In both cases, glnKp became highly expressed as the cells ran out of ammonia and growth ceased (Fig. 2A and B). In contrast, at 0.2% ammonium sulfate, where cell yield was not limited by ammonia, glnAp was expressed at approximately one-ninth of its maximum level, while glnKp was silent even after the cells stopped growing (Fig. 2C). Thus, the regulation of glnAp and glnKp was different in that under certain conditions (i.e., when cells reached stationary phase before exhaustion of the ammonia) glnAp was the only promoter expressed. Although the growth rate was not significantly altered at different concentrations of ammonia (provided as ammonium sulfate), the level of glnA transcription was clearly affected.
FIG. 2.
Induction of glnAp and glnKp in response to nitrogen starvation. Isogenic cells, wild type except that they contained either trp::Φ(glnAp-lacZ) (YMC10ApΦ2; +) or trp::Φ(glnKp-lacZ) (YMC10Φ; ○) were grown at 30°C in defined media containing 0.004% tryptophan and 0.4% glucose with either 0.005 (A), 0.01 (B), or 0.2% (C) ammonium sulfate. At the indicated times, samples were removed and assayed for β-galactosidase. Symbols: + and o, growth (optical density at 600 nm [OD600]); bars, β-galactosidase expression (white bars, YMC10ApΦ2; grey bars, YMC10Φ). Maximum expression was 2,640 Miller units for YMC10ApΦ2 and 1,680 Miller units for YMC10Φ.
In similar experiments, we observed that the nac promoter, like glnK, became activated only when growth became limited by ammonia starvation. To better focus on the relationship between the nac and glnK promoters, we compared their activation side by side in experiments where cells became limited for ammonia at two different optical densities, owing to different initial concentrations of ammonium sulfate. These experiments indicated that nac and glnK were activated at about the same point, with nac perhaps lagging slightly behind glnK (Fig. 3).
FIG. 3.
Comparison of the activation of glnKp and nacp in response to nitrogen starvation. Isogenic cells, wild type except that they contained either trp::Φ(glnKp-lacZ) (YMC10Φ; ○ and •) or trp::Φ(nacp-lacZ) (YMC10NpΦ; □ and ▪) were grown at 30°C in defined media containing 0.004% tryptophan and 0.4% glucose with either 0.005 (○ and □) or 0.01% (• and ▪) ammonium sulfate. The growth curves in both experiments for the two different strains were identical. At the indicated culture densities, samples were removed and assayed for β-galactosidase. OD600, optical density at 600 nm.
We directly examined the intracellular concentration of NRI as cells transitioned from growth on ammonia to nitrogen starvation in immunoblotting experiments (Fig. 4). Reitzer and Magasanik had previously shown that the level of NRI in log-phase cells grown on nitrogen-limiting glucose-glutamine medium was ∼10-fold higher than that in cells grown on nitrogen-rich glucose-ammonia-glutamine medium (20). We observed that a low and fairly constant level of NRI was present in cells growing on ammonia (Fig. 4). As the ammonia became depleted and cell growth stopped, the concentration of NRI increased (Fig. 4). The increase in NRI concentration occurred concurrently with the activation of glnK and nac (compare Fig. 2 and 4).
FIG. 4.
Immunoblotting analysis of the NRI concentration during growth on ammonia and the transition to nitrogen starvation. Strain YMC10 (wild type) was grown at 30°C on defined medium containing 0.4% glucose and 0.005% ammonium sulfate. Samples were harvested for Western blot analysis at the indicated times. The standard lane (std) contains 6 ng of purified NRI. Each sample lane contains 5 μg of crude protein extract. OD600, optical density at 600 nm.
Activation of glnAp, glnKp, and nacp as cells grow on glutamine as the sole nitrogen source.
Most studies of Ntr gene activation are focused on the expression of genes in log-phase cultures growing on glutamine as the sole nitrogen source. Here, we examined lac fusion expression as adapted cells grow on glutamine, providing a view of glnA, glnK, and nac regulation under these conditions (Fig. 5). When cells grew on glutamine, a reduction in the rate of growth occurred in mid-log phase, at an optical density at 600 nm (OD600) of ∼0.5 (Fig. 5B). Prior to this reduction in growth, fusion expression was similar to that in cells growing on ammonia, namely, glnA was partially activated and glnK and nac were not activated (Fig. 5A). At the point where the growth rate was reduced, fusion expression was similar to that seen in cells that depleted a limiting ammonia concentration, namely, all three promoters were sharply activated. The sharp transition between these two states occurred in mid-log phase; thus, use of mid-log phase glutamine-adapted cultures for assessment of Ntr gene expression levels is somewhat risky.
FIG. 5.
Activation of glnA, glnK, and nac promoter fusions in cells growing on glutamine as the sole nitrogen source. Overnight cultures were grown in 0.4% glucose-0.2% glutamine to stationary phase. Cells were diluted into similar medium, except that it contained 0.1% (wt/vol) glutamine, and incubated at 30°C. (A) Expression of β-galactosidase. Symbols: □, glnA-lacZYA; ◊, glnKp-lacZYA; ○, nacp-lacZYA. (B) Growth of the three cultures. Symbols are as in panel A.
In vitro transcription from the glnK promoter.
Since the glnAp2 and nac promoters had been examined in vitro but the glnK promoter had not, we examined the NRI∼P dependence of transcription from this promoter by using purified components. NRI∼P stimulates the isomerization of the closed promoter-polymerase complex to the open complex, competent for transcription initiation. The formation of the open complex may be assayed by examining the rate at which uninitiated complexes are formed in the presence of ATP alone or by measuring the formation of short initiated complexes in the absence of a single nucleotide. Because complexes of the latter type are very stable, their formation permits assessment of activation at promoters where the open complex is unstable (7). In our transcription assays, as before (7), we controlled the concentration of NRI∼P by adding various concentrations of NRI in the presence of excess NRII (NtrB). As templates we used supercoiled plasmids containing a strong transcriptional terminator positioned downstream from the promoter of interest (7, 9, 14).
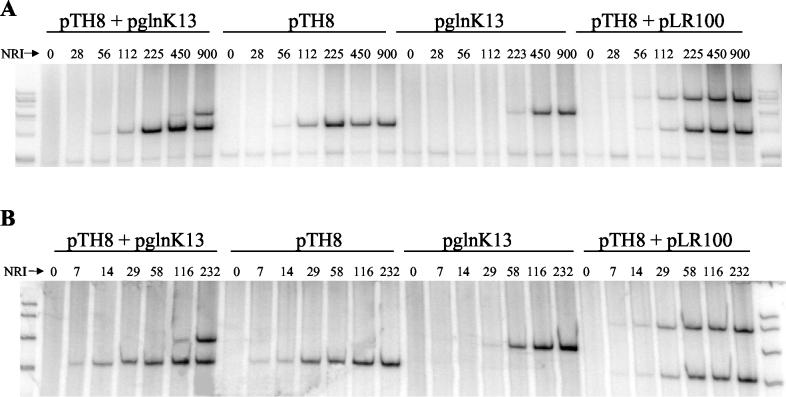
As expected, activation of glnAp2 required a lower concentration of NRI∼P than did activation of glnKp when single promoters were examined, as well as when both promoters were present in the same transcription reaction mixture (Fig. 6). In contrast, two templates containing the glnAp2 promoter positioned different distances from the transcriptional terminator were simultaneously activated as the concentration of NRI∼P was increased (Fig. 6). The relative behaviors of the promoters in the in vitro transcription system were the same regardless of whether open complexes (Fig. 6A) or initiated complexes (Fig. 6B) were assayed; thus, the open complexes formed at these two promoters may have similar stabilities. The open complex at the glnK promoter seemed to be considerably more stable than the corresponding open complex at the nac promoter since we could see evidence of their formation in experiments where heparin challenge preceded initiation (7) (Fig. 6B).
FIG. 6.
Transcription from glnAp2 and glnKp by purified components of E. coli. Transcription reaction mixtures contained the indicated supercoiled templates (10 nM each), core RNA polymerase (100 nM), σ54 (200 nM), NRII (100 nM), and the indicated concentration of NRI (nanomolar units). Reaction mixtures were incubated in the presence of ATP for the formation of open complexes (A) or in the presence of ATP, CTP, and GTP for the formation of short initiated complexes (B). Complex formation was stopped by addition of heparin, complexes were extended by addition of the missing nucleotide(s), and transcripts were recovered by phenol extraction and ethanol precipitation, subjected to electrophoresis on sequencing gels, and detected by autoradiography as described previously (7). Transcripts were labeled by use of [α-32P]UTP. Templates pTH8 and pLR100 contained the glnA promoter positioned different distances upstream from the phage T7 terminator in the pTE103 vector. Template pglnK13 contained the glnK promoter positioned upstream from the phage T7 terminator in the pTE103 vector.
Patterns of growth on the combination of ammonia and arginine.
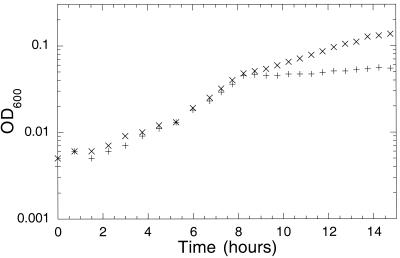
Arginine can serve as the sole nitrogen source for E. coli, supporting a doubling time of ∼4 h in defined medium with excess glucose. Growth on arginine requires the presence of NRI, as the astCADBE operon is part of the Ntr regulon (16, 23). We examined the growth of E. coli when both ammonia at low concentration and arginine at high concentration were provided. The rate of growth on ammonia plus arginine was indistinguishable from that observed with ammonia alone until all the ammonia was consumed. At that point, the cells began growing at the rate characteristic of cells using only arginine. In some cases, a short lag was detected. Thus, the pattern of growth on the mixture of arginine and ammonia was diauxic (Fig. 7).
FIG. 7.
Growth of E. coli on limiting ammonium sulfate and excess arginine is diauxic. Growth of YMC10 (wild type) at 30°C on defined minimal medium containing 0.4% glucose and 0.001% ammonium sulfate (+) or 0.001% ammonium sulfate and 0.2% arginine (×). OD600, optical density at 600 nm.
DISCUSSION
The Ntr regulon has some resemblance to a developmental gene cascade, in that genes are sequentially activated in response to an environmental stimulus. Our results support the hypothesis that, in the Ntr system, temporal staging of gene expression results from amplitude modulation of the phosphorylated form of the enhancer-binding activator protein. This amplitude modulation gives rise to the sequential activation of transcription because of the distinct features of the enhancers and promoter architecture at the various regulated promoters.
In cells growing on ammonia, the glnAp promoter was partially activated, while the glnK and nac promoters were not. This suggests that, when cells grew on defined glucose-ammonia medium, the level of NRI∼P was sufficient to permit significant expression of glnA while glnK and nac remained silent. The modest (approximately threefold) regulation of glnA expression by ammonia concentration that we observed probably reflects the fine regulation of the NRI∼P level when it is at the low end of its physiological range. Our immunoblotting analysis of NRI did not detect a significant increase in NRI as the ammonia concentration of the medium was reduced by consumption. The fairly uniform growth rate of bacteria irrespective of ammonia concentration suggests that this fine control of NRI∼P at the low end of its physiological range and the attendant fine control of GS expression and activity enable cells to grow optimally without recourse to activation of the other Ntr genes.
The glnK and nac promoters became strongly activated when cells stopped growing due to ammonia starvation, suggesting that at this point the NRI∼P concentration was significantly increased. Our immunoblotting analysis confirmed that NRI concentration was increased as cells became starved. Experiments examining the use of arginine as a nitrogen source suggested that the astCADBE operon (16, 23) was not expressed when cells had ammonia available but rather was only activated as ammonia became depleted. Thus, the glnK, nac, and astC promoters define a group of promoters that are regulated differently from glnAp.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant features and construction | Reference |
|---|---|---|
| pRS551 | lac fusion vector | 25 |
| pglnAplac4 | glnA promoter fused at +165 to lac operon in pRS551 | |
| pTH8 | glnA promoter cloned into pTE103 | 9 |
| pLR100 | glnA promoter cloned into pTE103 | 14 |
| pTE103 | pUC8 multicloning site with phage T7 terminator | 6 |
| pglnK13 | glnK promoter cloned into pTE103 | |
| pNacLacZ | nac promoter fused to lac operon in pRS551 |
Acknowledgments
This work was supported by grant GM57393 from the NIH-NIGMS to A.J.N.
REFERENCES
- 1.Atkinson, M. R., and A. J. Ninfa. 1993. Mutational analysis of the bacterial signal-transducing protein kinase/phosphatase nitrogen regulator II (NRII or NtrB). J. Bacteriol. 175:7016-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., and A. J. Ninfa. 1998. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 29:431-447. [DOI] [PubMed] [Google Scholar]
- 3.Backman, K., Y.-M. Chen, and B. Magasanik. 1981. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 78:3743-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmona, M., F. Claverie-Martin, and B. Magasanik. 1997. DNA bending and the initiation of transcription at σ54-dependent bacterial promoters. Proc. Natl. Acad. Sci. USA 94:9568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliot, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-pfrA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliot, T., and E. P. Geiduschek. 1984. Defining a bacteriophage T4 late promoter: absence of a “−35” region. Cell 36:211-219. [DOI] [PubMed] [Google Scholar]
- 7.Feng, J., T. J. Goss, R. A. Bender, and A. J. Ninfa. 1995. Activation of transcription initiation from the nac promoter of Klebsiella aerogenes. J. Bacteriol. 177:5523-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hattori, M., and Y. Sakaki. 1986. Dideoxy sequencing using denatured plasmid templates. Anal. Biochem. 152:232-238. [DOI] [PubMed] [Google Scholar]
- 9.Hunt, T. P., and B. Magasanik. 1985. Transcription of glnA by purified bacterial components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc. Natl. Acad. Sci. USA 82:8453-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kustu, S., E. Santero, J. Keener, D. Popham, and D. Weiss. 1989. Expression of σ54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 12.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Ninfa, A. J., P. Jiang, M. R. Atkinson, and J. A. Peliska. 2000. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr. Top. Cell. Regul. 36:31-75. [DOI] [PubMed] [Google Scholar]
- 14.Ninfa, A. J., L. J. Reitzer, and B. Magasanik. 1987. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell 50:1039-1046. [DOI] [PubMed] [Google Scholar]
- 15.Pahel, G., D. M. Rothstein, and B. Magasanik. 1982. Complex glnA-glnL-glnG operon of Escherichia coli. J. Bacteriol. 150:202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reitzer, L. J. 1996. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine, p. 391-407. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 18.Reitzer, L. J. 1996. Sources of nitrogen and their utilization, p. 380-390. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 19.Reitzer, L. J., and B. Magasanik. 1983. Isolation of the nitrogen assimilation regulator, NRI, the product of the glnG gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:5554-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reitzer, L. J., and B. Magasanik. 1985. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc. Natl. Acad. Sci. USA 82:1979-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee, S. G., P. B. Chock, and E. R. Stadtman. 1985. Glutamine synthetase from Escherichia coli. Methods Enzymol. 113:213-241. [DOI] [PubMed] [Google Scholar]
- 22.Saiki, R. K. 1990. Amplification of genomic DNA, p. 13-20. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols, a guide to methods and applications. Academic Press, San Diego, Calif.
- 23.Schneider, B. L., A. K. Kiupakis, and L. J. Reitzer. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180:4278-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions, p. 107-111. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single copy and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 26.Wong, P.-K., D. Popham, J. Keener, and S. Kustu. 1987. In vitro transcription of the nitrogen fixation regulatory operon nifLA of Klebsiella pneumoniae. J. Bacteriol. 169:2876-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]