Abstract
Virulence and immunity are poorly understood in Mycobacterium tuberculosis. We sequenced the complete genome of the M. tuberculosis clinical strain CDC1551 and performed a whole-genome comparison with the laboratory strain H37Rv in order to identify polymorphic sequences with potential relevance to disease pathogenesis, immunity, and evolution. We found large-sequence and single-nucleotide polymorphisms in numerous genes. Polymorphic loci included a phospholipase C, a membrane lipoprotein, members of an adenylate cyclase gene family, and members of the PE/PPE gene family, some of which have been implicated in virulence or the host immune response. Several gene families, including the PE/PPE gene family, also had significantly higher synonymous and nonsynonymous substitution frequencies compared to the genome as a whole. We tested a large sample of M. tuberculosis clinical isolates for a subset of the large-sequence and single-nucleotide polymorphisms and found widespread genetic variability at many of these loci. We performed phylogenetic and epidemiological analysis to investigate the evolutionary relationships among isolates and the origins of specific polymorphic loci. A number of these polymorphisms appear to have occurred multiple times as independent events, suggesting that these changes may be under selective pressure. Together, these results demonstrate that polymorphisms among M. tuberculosis strains are more extensive than initially anticipated, and genetic variation may have an important role in disease pathogenesis and immunity.
Current evidence suggests that as a species Mycobacterium tuberculosis exhibits very little genomic sequence diversity (24, 34). Most genetic variability that has been detected is associated with transposable elements and drug resistance phenotypes (5, 17, 28, 38). It follows that M. tuberculosis should exhibit very little phenotypic variation in immunologic and virulence factors. However, evidence of phenotypic diversity among clinical isolates conflicts with this hypothesis (22, 37). The presence of significant sequence diversity in M. tuberculosis would provide a basis for understanding pathogenesis, immune mechanisms, and bacterial evolution. Polymorphic genes are good candidates for virulence and immune determinants, because proteins that interact directly with the host are known to have elevated divergence. Polymorphic sequences also serve as markers for phylogenetic and evolutionary studies. Such studies are currently limited by a paucity of known genetic markers.
Recently, the genome of the M. tuberculosis laboratory strain H37Rv was completely sequenced (GenBank accession no. NC_000962) (10). This sequence provided important insights into the biology of this species but did little to address issues of sequence diversity. Furthermore, significant differences can be documented among the genomes of laboratory strains with long histories of passage and among recent clinical isolates (25). H37Rv had been passaged for many decades outside of the human host. Thus, the relevance of the H37Rv genome sequence to clinical M. tuberculosis strains has been questioned. We sequenced the genome of a clinical M. tuberculosis strain, CDC1551 (GenBank accession no. AE000516), and performed a comprehensive sequence comparison. In contrast to H37Rv, CDC1551 is a strain involved in a recent cluster of tuberculosis cases and is known to be transmissible and virulent in humans (38). The CDC1551 strain appears to be highly infectious in humans, is comparable in virulence to strain H37Rv in animal models (8), and has greater immunoreactivity than H37Rv and other clinical strains due to increased induction of tumor necrosis factor alpha, interleukin-6 (IL-6), IL-10, and IL-12 (22).
Investigators have utilized a variety of low- and high-resolution comparative genome techniques to identify differences in the genomes of Mycobacterium bovis, M. bovis BCG vaccine strains, and M. tuberculosis laboratory and clinical strains. Low-resolution analyses have included subtractive hybridizations and investigations of sequence variations associated with restriction length polymorphisms. These methods have identified a number of sequence differences between the different mycobacterial species and strains (15, 16, 21, 26). However, low resolution analysis has inherent limitations. Consequently, the polymorphisms identified have been either very large or restricted to localized areas of the M. tuberculosis genome. Comparative genomic techniques with higher resolution have also been performed recently. However, these studies have been limited by the use of H37Rv as the single reference strain (6, 7, 18). The only whole-genome comparison undertaken thus far has been an incomplete and inaccurate comparison of the H37Rv and CDC1551 strains. This study apparently used an incomplete version of the CDC1551 strain sequence and resulted, for example, in the misidentification of several comparative deletions in strain CDC1551 (7). Here we present the first comparison of the complete genomes from strains H37Rv and CDC1551, including differences resulting from large-sequence polymorphisms (LSPs) (greater than 10 bp) and single-nucleotide polymorphisms (SNPs). We discovered an unexpectedly high degree of sequence variation between the two genomes and determined that much of the variation was also present in a large panel of clinical M. tuberculosis isolates.
MATERIALS AND METHODS
Genome sequencing.
The methodologies for library construction, template preparation, sequencing, and closure of the M. tuberculosis genome were primarily those developed during the whole-genome shotgun sequencing of the 1.83-Mbp genome of Haemophilus influenzae (13). In brief, high-molecular-weight genomic DNA from strain CDC1551 was obtained from a seed lot culture maintained by William Bishai, Johns Hopkins University School of Medicine. The genomic DNA was randomly sheared by nebulization and size selected for 2.0-kb fragments. The DNA fragments were cloned into a SmaI-digested pUC18 vector. The recombinant molecules were freshly transformed by electroporation into DH10B electrocompetent cells (catalog no. 8290SA; GIBCO BRL) and transferred directly to a nutrient-rich SOB plate (13). Approximately 50,000 templates were prepared and sequenced with M13 forward and reverse primers. Sequencing reactions were analyzed on AB377 DNA sequencers. The random shotgun fragments were assembled with the TIGR assembler (35). Sequence gaps were filled by selecting a template whose forward and reverse sequence reads were in adjacent contigs. Physical gaps were ordered by combinatorial PCR based on oligonucleotide primers designed from the ends of each group of assemblies. The sequences of the physical gaps were determined by primer walking across each of the PCR products. Contigs were edited with the TIGR editor.
Annotation.
Open reading frames (ORFs) were identified with GLIMMER (30). The ORFs were searched against an in-house nonredundant amino acid database with blast_extend_repraze, which uses a BLASTP algorithm to generate pairwise amino acid alignments (4, 42). In addition to the pairwise alignments used to generate gene assignments, the ORFs were evaluated by comparison to a database of hidden Markov models generated from multiple sequence alignments for protein families and superfamilies (33). A team of annotation experts evaluated the results generated by these various tools and assigned to each ORF with a significant match an accession identification and a biological-role identification.
Suffix tree analysis.
We have developed a heuristic approach to aligning large segments (millions of base pairs in length) of closely related DNA sequence (11). The system uses a combination of three ideas: suffix trees, the longest common subsequence, and Smith-Waterman alignment. The complete nucleotide sequences of H37Rv and CDC1551 are provided as input. The alignment process then follows these steps. (i) A maximal unique match decomposition of the two genomes is performed to identify all maximal unique shared subsequences in both genomes. (ii) The matches are sorted based on a longest ascending subsequence algorithm, providing an easy and natural scan of the alignment from left to right. (iii) Gaps in the alignment are closed by performing local identification of large inserts, repeats, small mutated regions, tandem repeats, and SNPs. (iv) The alignment is outputted, including all the matches in the maximal unique match alignment and the detailed alignments of regions that do not match exactly.
Polymorphism detection.
One hundred sixty-nine clinical isolates taken at random from a collection of M. tuberculosis isolates cultured at Montefiore Medical Center and 64 clinical isolates cultured at The Johns Hopkins University School of Medicine were analyzed for LSPs. Genomic DNAs from the clinical isolates and strains CDC1551 and H37Rv were bound onto Biotrans plus nylon membranes (ICN Pharmaceuticals, Costa Mesa, Calif.) in longitudinal strips using a multislot hybridization apparatus (immunoblotter; Immunetics, Cambridge, Mass.). The membrane was then turned 90° in the same slot blot apparatus and simultaneously hybridized with γ-32P-labeled probes complementary to different LSPs. Forty-four different genomic DNA samples could be slotted in an array consisting of 44 lines extending across the membrane. Hybridizing of probes for each LSP at 90° to this array permitted every probe to come into contact with every genomic DNA sample. All isolates were subjected to DNA fingerprinting using IS6110-based restriction polymorphism analysis; low-band-number isolates were also tested with a secondary fingerprinting technique (1).
SNPs.
Some apparent SNPs might represent sequencing errors rather than true SNPs. Verification of the sequence differences was accomplished by two independent methods. One hundred SNPs were chosen at random, and the base calls were independently verified by inspection of the original electropherograms at The Institute for Genomic Research (CDC1551) and The Sanger Center (H37Rv). In collaboration with Qiagen Genomics, Inc., these 100 SNPs were verified for both strains by using Masscode technology for SNP identification (19). The visual inspection of the electropherograms and the Masscode results were in good agreement and indicated that 91% (80 of 88 successful assays) of the nucleotide differences were genuine. The number of synonymous and nonsynonymous substitutions between the two strains was determined by alignment of homologous ORFs with ClustalW (36). Poor alignments were removed based on visual inspection, resulting in a comparison of 3,535 ORFs. The number of synonymous differences (Sd) and nonsynonymous differences (Sn) between homologous ORFs was calculated with the program SNAP (20).
RESULTS
Whole-genome alignment.
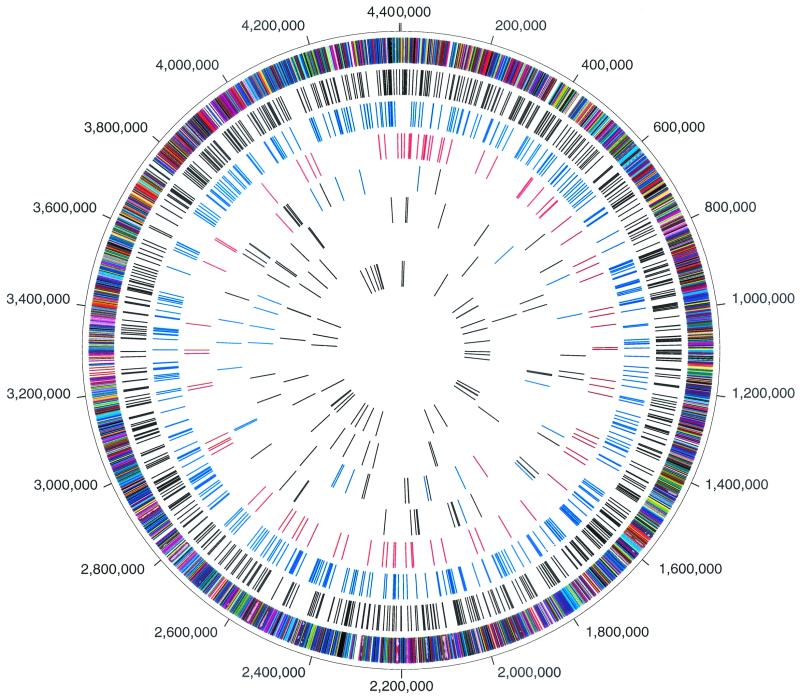
The complete genome sequence of M. tuberculosis strain CDC1551 was determined utilizing the whole-genome shotgun strategy (13). The complete annotation of the CDC1551 genome is located at The Institute for Genomic Research website at www.tigr.org/CMR and under GenBank accession no. AE000516. A whole-genome alignment was performed utilizing MUMmer software developed at The Institute for Genomic Research (11). The circular representation of the M. tuberculosis chromosome illustrated in Fig. 1 depicts the location of each predicted protein coding region as well as selected features differing between the CDC1551 and H37Rv strains, including LSPs and SNPs.
FIG.1.
Circular representation of the M. tuberculosis chromosome illustrating the location of each predicted protein-coding region as well as selected features differing between the CDC1551 and H37Rv strains. The outer concentric circle shows predicted protein-coding regions on both strands, color coded according to role category. The second concentric circle shows the location of nonsynonymous substitutions (black). The third concentric circle shows the location of synonymous substitutions (blue). The fourth concentric circle shows the location of substitutions in noncoding regions (red). The fifth concentric circle shows the location of insertions in strain CDC1551, including coding (black) and noncoding (blue) regions, and the location of phage phiRv1 (red). The sixth concentric circle shows the location of insertions in strain H37Rv, including coding (black) and noncoding (blue) regions, and the location of phage phiRv1 (red). The seventh concentric circle shows the location of IS6110 insertion elements in strains CDC1551 (blue) and H37Rv (red). The eighth (innermost) concentric circle shows the location of tRNAs (blue) and rRNA (red).
The two genomes contained notable differences. The H37Rv strain contained 37 insertions (greater than 10 bp) relative to strain CDC1551. Twenty-six insertions affected ORFs (Fig. 1; Table 1), and 11 were intergenic. The insertions in strain H37Rv included tandem repeats, additions to the 5′ or 3′ ends of ORFs, and the addition of complete ORFs. Complete ORFs included three encoding hypothetical proteins (Rv0793, Rv3427c, Rv3428c), two encoding PPE proteins (Rv3425, Rv3426), one encoding a PE_PGRS protein (Rv3519), and two encoding proteins with putative functions (Rv0794c, a dihydrolipoamide dehydrogenase, and Rv0792c, a putative transcriptional regulator). Forty-nine insertions were identified in strain CDC1551 relative to strain H37Rv. Thirty-five insertions affected ORFs (Fig. 1; Table 2), and 14 were intergenic. In addition to tandem repeats and additions to the 5′ and 3′ ends of ORFs, insertions introduced 17 complete ORFs. Eight ORFs encoded conserved hypothetical or hypothetical proteins (MT1813, MT2080, MT2080.1, MT2081, MT2420, MT2421, MT3427.1, and MT3429). The nine additional CDC1551 ORFs with functional assignments included genes encoding an adenylate cyclase (MT1360), a glycosyl-transferase (MT1800), an oxidoreductase (MT1801), a 12 transmembrane protein (MT1802), a membrane lipoprotein (MT2619), a PPE family protein (MT3248), paralogs of moaB (MT3426) and moaA (MT3427), and a gene encoding a putative transcription regulatory protein (MT3428). The changes in the 5′ end of a phospholipase C gene (MT1799) and the addition of a 12 transmembrane transport protein (MT1802) are particularly notable because of their potential role in bacterial virulence (32). It is worth noting that almost half of the insertions and deletions in both strains involved genes encoding PPE or PE_PGRS family proteins. We found only one major rearrangement of genome structure between the two strains. A prophage, initially identified in the RD3 region of M. bovis (21) and later characterized as prophage phiRv1 in strain H37Rv (10), is associated with the REP13E12 family of repeats (14). In strain H37Rv, prophage phiRv1 is integrated between coordinates 1,779,312 and 1,788,503. In strain CDC1551, phiRv1 is integrated into a second member of the REP13E12 family at CDC1551 coordinates 3,870,803 and 3,879,990.
TABLE 1.
Regions of insertions in ORFs in strain H37Rv relative to strain CDC1551
| Coordinates | Locus | Gene name or product |
|---|---|---|
| 24,721-24,737 | Rv0020c | Conserved hypothetical |
| 32,351-32,388 | Rv0029 | Conserved hypothetical |
| 206,849-206,906 | Rv0175 | Hypothetical |
| 427,358-427,373 | Rv0355c | PPE |
| 428,204-428,264 | Rv0355c | PPE |
| 840,177-840,225 | Rv0747 | PE_PGRS |
| 886,543- | Rv0792c | Transcription regulator |
| Rv0793 | Hypothetical | |
| 887,417 | Rv0794c | Dihydrolipoamide dehydrogenase |
| 1,212,121-1,212,199 | Rv1087 | PE_PGRS |
| 1,217,504-1,218,158 | Rv1091 | PE_PGRS |
| 2,062,034-2,062,124 | Rv1818c | PE_PGRS |
| 2,163,788-2,163,926 | Rv1917c | PPE |
| 2,165,426-2,165,502 | Rv1917c | PPE |
| 2,180,804-2,180,826 | Rv1928c | Short-chain dehydrogenase |
| 2,372,491-2,372,548 | Rv2112c | Conserved hypothetical |
| 2,381,412-2,383,686 | Rv2124 | Methionine synthase |
| 2,704,308- | Rv2406c | Hypothetical |
| 2,704,808 | Rv2407 | Hypothetical |
| 3,054,718-3,054,931 | Rv2741 | PE_PGRS |
| 3,171,570-3,171,624 | Rv2859c | Glutamine amidotransferase |
| 3,663,929-3,663,992 | Rv3281 | Conserved hypothetical |
| 3,730,578-3,735,861 | Rv3343c | PPE |
| 3,738,805- | Rv3345c | PE_PGRS |
| Rv3425 | PPE | |
| Rv3426 | PPE | |
| Rv3427c | Hypothetical | |
| 3,847,211 | Rv3428 | Hypothetical |
| 3,948,924-3,949,527 | Rv3514 | PE_PGRS |
| 3,949,826-3,949,943 | Rv3514 | PE_PGRS |
| 3,955,465-3,956,104 | Rv3519 | PE_PGRS |
| 4,359,144-4,359,163 | Rv3879c | Hypothetical |
TABLE 2.
Regions of insertions in ORFs in strain CDC1551 relative to strain H37Rv
| Coordinates | Locus | Gene name or product |
|---|---|---|
| 150,887-151,067 | MT0132 | PE_PGRS |
| 624,668-624,758 | MT0556 | PE_PGRS |
| 744,075-744,608 | MT0676 | Alpha-mannosidase |
| 1,121,754-1,121,769 | MT1033 | Hypothetical |
| 1,191,505-1,191,697 | MT1097 | PE_PGRS |
| 1,213,846-1,213,891 | MT1118.1 | PE_PGRS |
| 1,480,513-1,482,187 | MT1360 | Adenylate cyclase |
| 1,612,509-1,612,530 | MT1479 | Hypothetical |
| 1,632,424-1,632,451 | MT1497.1 | PE_PGRS |
| 1,633,446-1,634,201 | MT1497.1 | PE_PGRS |
| 1,885,204-1,885,214 | MT1707 | ABC transporter |
| 1,974,051-1,974,211 | MT1796 | PPE |
| 1,978,715- | MT1799 | Phospholipase C |
| MT1800 | Glycosyl transferase | |
| MT1801 | Oxidoreductase | |
| 1,985,523 | MT1802 | Membrane protein |
| 1,993,920-1,994,873 | MT1812 | Hypothetical |
| 2,130,695-2,130,710 | MT1931.1 | Hypothetical |
| 2,134,757-2,134,767 | MT1936 | Hypothetical |
| 2,143,342-2,143,387 | MT1950 | Conserved hypothetical |
| 2,160,664-2,160,941 | MT1968 | PPE |
| 2,266,057- | MT2080 | Conserved hypothetical |
| MT2080.1 | Hypothetical | |
| MT2081 | Conserved hypothetical | |
| 2,271,057 | MT2082 | Helicase |
| 2,629,977- | MT2420 | Hypothetical |
| MT2421 | Hypothetical | |
| 2,630,917 | MT2422 | PPE |
| 2,633,463-2,634,259 | MT2423 | PPE |
| 2,701,714-2,701,735 | MT2479 | Aryl sulfatase |
| 2,862,694-2,863,350 | MT2619 | Lipoprotein |
| 3,524,545-3,526,695 | MT3248 | PPE |
| 3,685,803-3,685,859 | MT3403 | Hypothetical |
| 3,705,263- | MT3426 | MoaB |
| MT3427 | MoaA | |
| MT3427.1 | Hypothetical | |
| MT3428 | Transcription regulator | |
| MT3429 | Hypothetical | |
| 3,709,688 | MT3430 | Transposase |
| 3,730,852-3,730,870 | MT3449 | PE_PGRS |
| 3,733,433-3,733,511 | MT3449 | PE_PGRS |
| 3,922,614-3,922,632 | MT3612 | PE_PGRS |
| 3,924,305-3,924,313 | MT3612 | PE_PGRS |
| 3,926,618-3,926,693 | MT3612.1 | PE_PGRS |
| 3,935,210-3,935,555 | MT3615.1 | PE_PGRS |
| 3,940,711-3,940,747 | MT3615.3 | PE_PGRS |
| 3,941,109-3,941,184 | MT3615.3 | PE_PGRS |
| 4,086,588-4,086,606 | MT3756 | PE_PGRS |
The IS3-type insertion sequence IS6110 is the principal epidemiological marker for M. tuberculosis. A number of the insertions and deletions were associated with this insertion sequence, suggesting a role for this element in genome plasticity (23, 41). Studies have shown that homologous recombination between nearby copies of IS6110 may result in genomic deletions and can be a mechanism for generating genomic diversity (12). We identified 4 copies of IS6110 in CDC1551 compared to 16 copies in H37Rv. Four of the 16 IS6110 elements present in H37Rv lacked the characteristic 3- to 4-bp direct repeat and were directly adjacent to regions that were deleted relative to strain CDC1551. Two of these deletions included the 6,807-bp region containing ORFs MT1799, MT1800, MT1801, and MT1802 and the 4,083-bp region deleted in strain H37Rv containing the molybdopterin cofactor biosynthesis gene cluster (MT3426 and MT3427) (Table 2). While strain H37Rv contained several deletions associated with a possible mechanism of homologous recombination between nearby IS6110 elements, none of the deletions in strain CDC1551 appeared to be the result of such a mechanism. The association of the IS6110 element with the deletion of important genes calls into question its utility as a neutral marker for phylogenetic analysis.
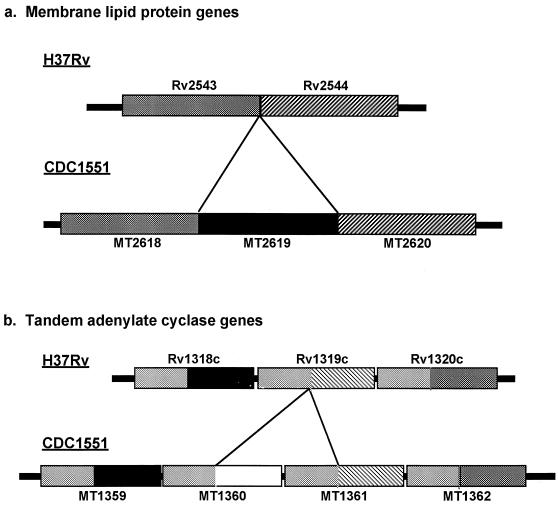
Regions differing in the copy number of several genes between strains H37Rv and CDC1551 were identified. Phylogenetic analysis of these regions revealed surprisingly contradictory evolutionary relationships among CDC1551, H37Rv, and M. bovis. Among the genes encoding membrane lipoproteins in strain H37Rv were two genes in tandem (Rv2543 and Rv2544). The homologous genome region in strain CDC1551 contained the orthologs MT2618 and MT2620, respectively. However; a third ORF, MT2619, was interspersed between the two genes (Fig. 2a). Examination of the homologous region in the M. bovis genome (www.sanger.ac.uk) revealed orthologs of the three membrane lipoprotein genes observed in the CDC1551 genome in an indistinguishable organization and nucleotide sequence. There was essentially no nucleotide diversity between orthologs. The nucleotide diversity among the paralogs was similar for all three genes, indicating equivalent evolutionary distances. Phylogenetic analysis suggested that the three paralogs arose in a common ancestor of the M. tuberculosis complex by gene duplications and that subsequent loss of MT2619 occurred in the H37Rv lineage.
FIG. 2.
(a) Schematic diagram of homologous genome region in strains H37Rv and CDC1551 encoding several membrane lipoproteins. The region in strain H37Rv contains two genes in tandem (Rv2543 and Rv2544) that are 87% identical to each other at the protein level. The homologous region in strain CDC1551 contains three genes (MT2618, MT2619, and MT2620), with the middle gene, MT2619, being unique to strain CDC1551 and 88 and 84% identical to MT2619 and MT2620, respectively. Homology between strain CDC1551 and M. bovis and equivalent evolutionary distances between paralogs suggest that the three paralogs arose in a common ancestor of the M. tuberculosis complex and subsequent loss of MT2619 occurred in the H37Rv lineage. (b) Schematic diagram of the tandem adenylate cyclase region. Two paralogous cyclases flank the region (MT1359/Rv1318c and MT1362/Rv1320c). Analysis revealed two cyclases (MT1360 and MT1361) between the two flanking genes in strain CDC1551 and only one cyclase (Rv1319c) in strain H37Rv. Rv1319c appears to be a chimera of the 5′ half of MT1361 and the 3′ half of MT1360. The 3′ halves of all orthologs share >80% nucleotide identity, while the 5′ halves appear diverse. Phylogenetic analysis indicates that the duplication events share a similar evolutionary distance. Inspection of the M. bovis sequence data reveals that this region is organized in an identical way to the H37Rv genome.
A second polymorphic region contained an unusual organization of three (Rv1318c, Rv1319c, and Rv1320c) and four (MT1359, MT1360, MT1361, and MT1362) putative adenylate cyclase genes in tandem in strains H37Rv and CDC1551, respectively (Fig. 2b). Each ORF had a strong match to the catalytic domain of guanylate or adenylate cyclase. The comparative sequence data between the two strains showed that in H37Rv, the middle gene of the cluster (Rv1319c) is actually an in-frame chimera corresponding to the 3′ and 5′ ends of genes MT1360 and MT1361, respectively, from CDC1551 (Fig. 2b). M. bovis has a structure similar to that of H37Rv. Phylogenetic analysis of this polymorphic region indicated that the in-frame chimera was generated by a deletion-fusion event. Thus, the ancestral structure was likely four tandem genes. The shared fusion in H37Rv and M. bovis could be due to a single event, indicating that these strains share a common ancestor relative to CDC1551. Notably, this hypothesis conflicts with the evolutionary scenario proposed for the membrane lipoprotein gene duplication. Together, these findings indicate that genetic variability in M. tuberculosis arises through a complex evolutionary process that involves recombination or multiple insertion-deletion events occurring independently at the same locus.
Heterogeneity of LSPs.
The comparative sequence data between CDC1551 and H37Rv provided us with a starting point for characterizing the degree and frequency of certain deletion/insertion events among clinical M. tuberculosis isolates. We tested 169 clinical M. tuberculosis isolates for the presence of a subset of seventeen CDC1551-H37Rv LSPs representing known and hypothetical genes. The seventeen probes for these LSPs, the genes involved, and their respective coordinates are listed in Table 3. The isolates included 19 restriction fragment length polymorphism (RFLP)-defined clusters and 88 unique isolates. We defined “clustered” isolates as those isolates which through DNA fingerprinting of an insertion sequence element (IS6110) shared identical banding patterns. “Unique” isolates contained unique banding patterns. Clustered isolates are thought to have a common ancestor and to be possibly linked together by recent transmission events, while unique isolates are likely to be genetically distinct strains (40). We discovered a surprisingly large degree of sequence heterogeneity among the clinical isolates. All of the 169 tested isolates lacked at least one LSP. An average of 3.7 sequences were missing for each isolate, with a range of one to seven deletions. We reproducibly demonstrated the presence or absence of specific LSPs in duplicate experiments, and we performed polymorphism specific PCR assays to reconfirm a subset of these results (data not shown).
TABLE 3.
Probes for LSP in strains CDC1551 and H37Rve
| Probe | Locusa | Gene name or product | Coordinatesb |
|---|---|---|---|
| 1 | MT0676 | Alpha-mannosidase | 744149-744392 |
| 2 | MT1360 | Adenylate cyclase | 1481322-1481551 |
| 3 | MT1802c | Transporter | 1982255-1982482 |
| 4 | MT1799c | Phospholipase | 1978754-1978931 |
| 5 | MT1812c | Hyp | 1994163-1994437 |
| 6 | MT2420 | Hyp | 2630855-2631147 |
| 7 | MT2619 | Membrane lipoprotein | 2862884-2863033 |
| 8 | MT3248 | PPE | 3520018-3526304 |
| 9 | MT2081/MT2082 | Hyp | 2268479-2268702 |
| 10 | MT3426d | moaB | 3705322-3705665 |
| 11 | MT3427d | moaA | 3707462-3707706 |
| 12 | MT2423d | PPE | 2633331-2633746 |
| 13 | Rv0793/Rv0794c | Hyp/dihydrolipoamide dehydrogenase | 886934-887397 |
| 14 | Rv2124c | Methionine synthase | 2381785-2383193 |
| 15 | Rv3135 | PPE | 3501335-3501499 |
| 16 | Rv3519 | Hyp | 3955704-3956104 |
| 17 | Rv3343c | PPE | 3733083-3733353 |
MT, CDC1551; Rv, H37Rv.
Probes 1 to 12 are CDC1551 coordinates, and probes 13 to 17 are H37Rv coordinates.
This locus is adjacent to an IS6110 element present in H37Rv and CDC1551.
This locus is adjacent to an IS6110 element present in H37Rv.
The probes may include the complete gene locus or only a portion of the indicated gene locus.
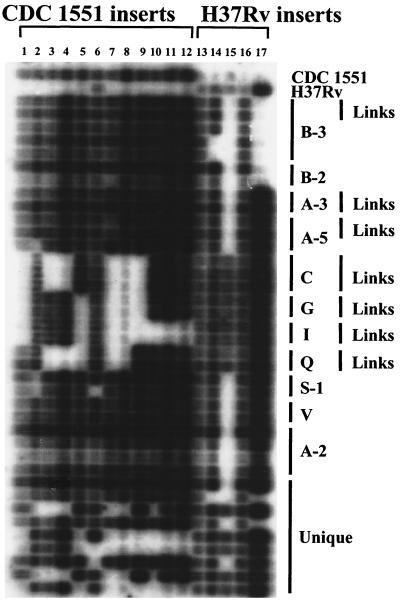
We used a second set of epidemiologically well-characterized isolates to determine whether variability of LSPs also occurred among isolates that were closely linked through epidemiological investigations (referred to herein as “epi-linked” isolates). Clusters of epi-linked isolates presumably share a recent common ancestor as part of an outbreak of disease. Thus, they represent very closely related isolates. We studied 42 isolates that included 16 RFLP-defined clusters and 15 unique isolates. Seven clusters contained a total of 17 epi-linked isolates. The pattern of LSPs was identical in all epi-linked isolates within a cluster. In contrast, four isolates within three clusters without epi-links contained deletions of at least one sequence (Fig. 3). As each cluster is likely to have arisen from a common ancestor, the additional deletions likely occurred subsequent to the common ancestor. We also found the same deletions in other clustered and unique isolates. These findings suggest that the loss of these LSPs occurred multiple times as independent events, and that new polymorphisms rarely develop within the short time frame of a clinical outbreak of disease. Alternately, the disparate clustered and unique strains containing identical deletions could have arisen from a common strain in which a unique ancestral deletion occurred. However, this second hypothesis is not supported by evidence for recurrent deletions within the phospholipase C region (16) or by the contradictory deletion-based phylogeny of the adenylate cyclase region (see above).
FIG. 3.
The distribution of CDC1551/H37Rv LSPs in clinical M. tuberculosis strains using a slot blot cross hybridization method. Strains include unique isolates and clustered strains with and without epidemiological links. Clusters are grouped by letter (and number if subtyped by a secondary fingerprinting method). Strains with epidemiological links are designated by links. Probes to LSPs are described in Table 3.
Heterogeneity of SNPs.
The comparison of the H37Rv and CDC1551 genomes identified 1,075 SNPs between the two genomes (Table 4). Approximately 85% of the substitutions occurred in coding regions (93% of the genome). We found transitions (purine to purine and pyrimidine to pyrimidine) to be more numerous than transversions (purine to pyrimidine and pyrimidine to purine), which represented 61 and 39% of the substitutions, respectively. We calculated synonymous and nonsynonymous substitutions for 3,535 pairs of homologous ORFs. There was at least one synonymous or one nonsynonymous substitution in 298 (8.4%) or 457 (12.9%) of the ORFs, respectively. In total, there were 342 and 579 synonymous (Sd) and nonsynonymous (Sn) substitutions, respectively. The proportion of synonymous differences per synonymous site corrected for the possibility that a site changed multiple times (Ds) was 3.6 × 10−4, or a 1/Ds of 1 synonymous substitution per 2,752 synonymous sites. This value for Ds was more than threefold greater than that previously described for M. tuberculosis (34), emphasizing the unexpected sequence diversity.
TABLE 4.
Nucleotide substitutions between M. tuberculosis strains CDC1551 and H37Rva
| Nucleotide in strain CDC1551 | No. of substitutions in strain H37Rv
|
|||
|---|---|---|---|---|
| A | C | G | T | |
| A | 66 | 165 | 9 | |
| C | 48 | 83 | 169 | |
| G | 165 | 89 | 44 | |
| T | 12 | 164 | 61 | |
Total synonymous substitutions, 379; total nonsynonymous substitutions, 662; total coding-to-noncoding substitutions, 40; total noncoding-to-coding substitutions, 15; total noncoding-to-noncoding substitutions, 83.
Surprisingly, the ratio of M. tuberculosis Ds to nonsynonymous substitution (Ds/Dn) was approximately 1.6, in contrast to studies of housekeeping genes in Escherichia coli and Salmonella enterica and invasion genes in S. enterica which show a Ds/Dn ratio of ranging from 4 to 17 (9, 39). The observation in E. coli and S. enterica is consistent with the generally accepted view that the many nonsynonymous substitutions are lost through purifying selection. The ratio observed in M. tuberculosis indicates that additional selective pressure is present on synonymous substitutions or there is decreased selective pressure against nonsynonymous mutations.
Patterns of synonymous and nonsynonymous substitutions can reveal information about mutations and selective pressures on genes, as well as information about population structure and recombination. We analyzed the frequency of substitution for 877 gene families to see if any contained significantly more SNPs than the genome as a whole. We calculated a P (the probability that the frequency of substitution was due to random fluctuation) for synonymous and nonsynonymous substitutions for each gene family. We used a P of <0.0001 as a cutoff to identify gene families in which the higher frequency of substitution was not likely to be due to random fluctuation (Table 5). We identified three paralogous families with significantly more synonymous substitutions and nonsynonymous substitutions (gmt [“Genome Mycobacterium tuberculosis” database] domain_PF00924, gmt domain_75, and gmt domain_337 [http://www.tigr.org/tigr-scripts/CMR2/ParalogousList.spl?db=gmt]).The gmt domain 69 also had significantly higher synonymous substitution rates, but the P for nonsynonymous substitutions was slightly above the cutoff. Interestingly, the PE and PPE paralogous family (gmt domain_PF00924) was among the three families with significantly higher synonymous and nonsynonymous substitutions. The PE and PPE genes encode a family of acidic, glycine-rich proteins that are postulated to be expressed on the extracellular surface and are considered potential antigens for host immunity (27, 31).
TABLE 5.
Paralogous families with significantly higher (P < 0.0001) frequencies of substitution
| Paralogous family | Common name | Ds | Dn |
|---|---|---|---|
| Whole genome | 0.00036 | 0.00022 | |
| Domain_PF00934 | PE/PPE family | 0.001 | 0.00067 |
| Domain_337a | Conserved hypothetical | 0.027 | 0.0083 |
| Domain_69 | Conserved hypothetical | 0.004 | 0.0013 |
| Domain_75 | Conserved hypothetical | 0.032 | 0.0036 |
ORFs in paralogous family domain_337 are also found in domain_69 (http://www.tigr.org/tigrscripts/CMR2/ParalogousList.spl?db=gmt).
The fact that the H37Rv strain has been in culture for nearly a century raised the possibility that many of the nucleotide differences observed between the two strains could be a result of in vitro passage. We examined a subset of 28 of the clinical isolates for 11 nonsynonymous SNPs (4 of which resulted in conservative amino acid substitutions) initially detected through the H37Rv-CDC1551 comparison (Table 6). Seven of the eleven SNPs were polymorphic in at least 1 of the 28 isolates tested. Two of the SNPs, SNP-2904 in the gene encoding the carbon starvation A protein and SNP-4090 in a gene encoding a putative transcriptional regulator, were highly polymorphic (each allele present in ≥20% of the isolates). Thus, the SNPs discovered by the two-genome comparison appear to be broadly represented in clinical strains. The distributions of SNP-2904 and SNP-4090 are comparable to those of two SNPs previously described as highly polymorphic (gyrA-95 at codon 95 of the gyrA gene and katG-463 at codon 463 of the katG gene) (34), suggesting that they might be useful for phylogenetic analysis (Table 6).
TABLE 6.
SNPs in 28 clinical M. tuberculosis isolatesc
| Strain | SNP
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2134 | 2904 | 3135 | 3194 | 4090 | 4239 | 4246 | 4584 | 5198 | GyrA-95a | KatG-463a,b | |
| 37 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 54 | Rv | 1551 | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 98 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | ctg |
| 107 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | ctg |
| 123 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | ctg |
| 131 | Rv | Rv | NDd | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 140 | Rv | Rv | 1551 | Rv | Rv | 1551 | Rv | Rv | Rv | Rv | cgg |
| 141 | Rv | Rv | 1551 | Rv | Rv | 1551 | 1551 | Rv | 1551 | Rv | cgg |
| 155 | 1551 | 1551 | 1551 | 1551 | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 159 | Rv | ND | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 160 | Rv | 1551 | 1551 | Rv | Rv | 1551 | 1551 | Rv | ND | 1551 | cgg |
| 165 | 1551 | Rv | 1551 | Rv | Rv | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 166 | Rv | Rv | 1551 | Rv | Rv | 1551 | 1551 | Rv | 1551 | Rv | cgg |
| 174 | Rv | 1551 | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | Rv | cgg |
| 195 | Rv | Rv | 1551 | Rv | 1551 | 1551 | Rv | Rv | 1551 | Rv | cgg |
| 206 | Rv | Rv | 1551 | 1551 | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 207 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | ctg |
| 223 | Rv | 1551 | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 227 | Rv | Rv | 1551 | Rv | 1551 | 1551 | ND | Rv | 1551 | 1551 | cgg |
| 229 | ND | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | Rv | cgg |
| 251 | Rv | Rv | 1551 | Rv | Rv | 1551 | 1551 | Rv | 1551 | Rv | cgg |
| 269 | ND | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 279 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| 282 | Rv | Rv | 1551 | Rv | Rv | 1551 | 1551 | Rv | 1551 | Rv | cgg |
| 284 | Rv | Rv | 1551 | Rv | Rv | 1551 | 1551 | Rv | 1551 | 1551 | ctg |
| 304 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | ND | ND |
| 306 | Rv | Rv | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | ctg |
| 313 | Rv | 1551 | 1551 | Rv | 1551 | 1551 | 1551 | Rv | 1551 | 1551 | cgg |
| CDC1551 | 1551 | 1551 | 1551 | 1551 | 1551 | 1551 | 1551 | 1551 | 1551 | 1551 | cgg |
| H37Rv | Rv | Rv | Rv | Rv | Rv | Rv | Rv | Rv | Rv | Rv | cgg |
GyrA-95 and KatG-463 SNPs were initially identified by Sreevatsan et al. (34).
actual sequence given because SNP is identical in CDC1551 and H37Rv.
Rv, H37Rv; 1551, CDC1551.
ND, Not done.
Phylogenetic analysis using LSPs and SNPs.
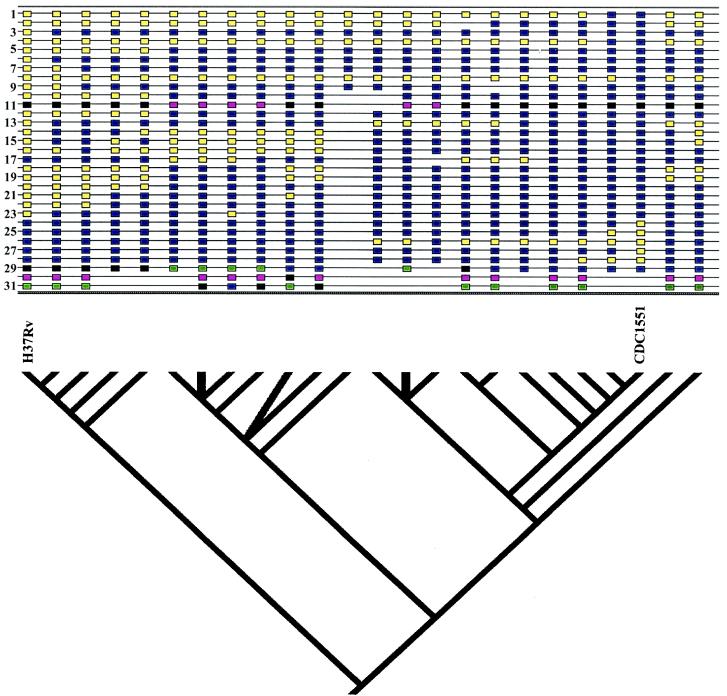
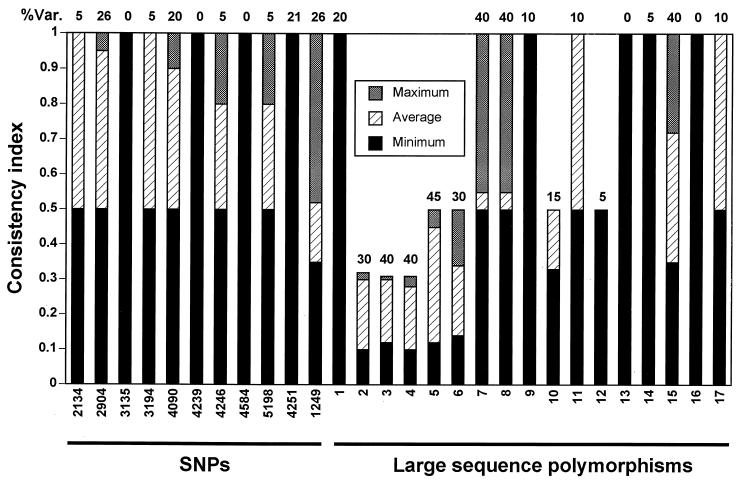
We performed a phylogenetic analysis of 21 clinical isolates, H37Rv, CDC1551, and M. bovis using a combination of data from LSPs, SNPs, and selected phenotypic traits (Fig. 4). The tree is supported by statistical analysis for most of the branching patterns. The phylogenetic analysis allows the assessment of the consistency of different markers with the consensus tree (Fig. 5). Several of the markers have a high consistency index as well as a reasonable level of variability. These markers provide information for constructing models of the phylogenetic relationships between strains. As described above, analysis of the adenylate cyclase region contradicted the model in which H37Rv, CDC1551, and M. bovis shared a common ancestor. The low consistency index of this region (marker 2) confirms on a population level that the cyclase region would be expected to be a poor phylogenetic marker, explaining this apparent contradiction. Markers with low consistency indices represent regions of the genome which may have evolved by mechanisms of convergence, recombination, and mutational frequencies outside the average for the species. Such sites can be useful in discriminating among strains linked by other markers, such as insertion sequence markers.
FIG. 4.
M. tuberculosis strain phylogeny based on a combination of LSPs, SNPs, and selected phenotypic traits. The tree shown is a consensus of the most parsimonious trees found using the heuristic search algorithm. Character state boxes are shown at the top (for LSPs, a blue box indicates presence and a yellow box indicates absence; for SNPs and other characters, colors correspond to different character states). Characters 1 to 12 are SNPs, characters 13 to 28 are LSPs, character 29 belongs to the Musser group, character 30 is Smear positive or negative, and character 31 is Site (pulmonary or extrapulmonary). Unresolved branching patterns are collapsed.
FIG. 5.
Consistency index for characters in the phylogenetic tree based on the LSPs and SNPs. Each column corresponds to the consistency index for a particular character (SNPs and LSPs). Each bar shows the minimum (black), average (striped), and maximum (grey) value of the consistency index over a large number of different trees (including multiple equally parsimonious trees and multiple distance trees). The percent variability indicates the variability of each marker in the 28 isolates tested. Calculations of the consistency indices were made using the MacClade program.
DISCUSSION
Several studies have compared closely related strains of bacteria, including Helicobacter pylori, several Chlamydia species, and pathogenic and nonpathogenic E. coli (2, 3, 25, 29). In general, these studies were limited to the sequencing of a comparative strain or species and identification of polymorphisms between the two but failed to extend the findings beyond the two strains being compared.
Several studies of the Mycobacterium genus describe LSPs among the M. bovis BCG vaccine strains and virulent M. bovis as well as among other tubercle bacilli (6, 15, 21, 18). While these studies describe several regions of LSP, the methodologies of subtractive hybridization and RFLP analysis limit the resolution and therefore the utility of the markers described. Betts et al. (7) in their analysis of the M. tuberculosis proteome attempt a comparative analysis of the genome contents of the H37Rv and CDC1551 strains. Several conclusions drawn from this analysis appear to be incorrect based on our present analysis. In particular, they describe eight ORFs completely unique to H37Rv, including Rv0278c, Rv0279c, Rv0746c, Rv0747, and Rv1087, all of the PE_PGRS family. The identification of a 5,742-bp deletion associated with ORFs Rv0278c and Rv0279c and a deletion of 4,910 bp associated with ORFs Rv0746c, Rv0747, and Rv0748 is incorrect and may be the result of analyzing an incomplete version of the sequence of strain CDC1551. Two other regions of strain H37Rv, bp 2,714,308 to 2,714,808 and bp 3,933,523 to 3,936,659, are incorrectly identified as being deleted in strain CDC1551. Their analysis of insertions in the strain CDC1551 genome relative to strain H37Rv also appears to contain several errors. They failed to identify approximately 15 regions of insertion in strain CDC1551 relative to H37Rv. Among these regions was a 4,425-bp region containing ORFs MT3426 and MT3427 and moaB and moaA paralogs, respectively. Most of the coordinates reported by Betts et al. (7) are incorrect with respect to strain CDC1551. This is again likely due to the analysis of an incomplete genome sequence.
We initially undertook the sequencing and annotation of a second M. tuberculosis strain, CDC1551, in an attempt to correlate genotypic changes with strain phenotype. Prior to our study it was generally accepted that little sequence variation existed. Our study demonstrates that a much higher level of polymorphism is present among the M. tuberculosis species. This discovery complicates attempts to associate specific genotypic changes with phenotypic differences. However, it also provides several important opportunities.
First, it is likely that a subset of the polymorphic sequences code for genes involved in host-pathogen interactions. A statistical analysis of the single-base substitution frequency in 877 gene families identified several families in which the frequency of substitution was significantly greater than that observed for the genome as a whole. This included the PE/PPE gene family, which encodes acidic, glycine-rich proteins that are postulated to be expressed on the extracellular surface and are considered potential antigens for host immunity (27, 31). The higher substitution frequency in this family might be the result of antigenic variation or may be due to other interactions with the host. Three other gene families representing conserved hypothetical proteins also had a higher substitution frequency. These genes warrant further investigation as potential candidates for virulence or immunogenicity. The LSPs that we discovered may also encode proteins involved in disease pathogenesis as demonstrated by the polymorphisms in one of the phospholipase C genes and members of the PE/PPE gene family.
Second, the level of LSPs and SNPs that we discovered among the M. tuberculosis isolates suggested that we could also develop a set of markers that would be valuable in studying the phylogenetics of the M. tuberculosis species and other tubercle bacilli. Analysis of two LSPs, an ORF encoding a membrane lipoprotein and an ORF encoding a putative adenylate cyclase, suggested conflicting scenarios of the evolutionary relationship of CDC1551, H37Rv, and M. bovis. However, phylogenetic analysis of 21 clinical isolates along with strains CDC1551, H37Rv, and M. bovis demonstrated that the adenylate cyclase region had a low consistency index and would be a poor phylogenetic marker, explaining its apparent contradiction with the membrane lipoprotein region. Other polymorphisms with high consistency indexes were discovered that are likely to be excellent markers for investigating the evolution and phylogenetics of this species.
We demonstrated that the Ds is more than threefold greater than estimated previously (24, 34). Interestingly, the Ds-to-Dn ratio was close to 1, unlike the much higher level expected if selection was against nonsynonymous but not synonymous substitutions. This could be due to decreased selective pressure against nonsynonymous mutations. For example, the prolonged passage of strain H37Rv in culture may have permitted the accumulation of nonsynonymous mutations in many genes that would otherwise be under strict selection in vivo. Alternatively, selective pressure may exist for certain synonymous substitutions. This may be due to codon bias aimed at maintaining a high G+C content (65.6%) and thus limiting the number of synonymous substitutions. Other explanations consistent with M. tuberculosis biology include a low recombination frequency, a small population size, or a recent bottleneck in M. tuberculosis evolution.
The sequencing of approximately 8.8 Mbp of M. tuberculosis (the combined complete sequences of strains CDC1551 and H37Rv) provided approximately 74 LSPs and more than 1,000 SNPs as potentially informative markers. Based on the consistency indices of the 17 LSPs and 10 SNPs evaluated, 5 and 2, respectively, would be good phylogenetic markers. This compares favorably with the effort by Sreevatsan et al. (34) in sequencing 2 Mbp of 26 selected genes and the identification of 32 SNPs, of which 2 were present at high frequency. The comprehensive comparison based on a genome-wide scan between just two isolates suggests that polymorphisms between M. tuberculosis strains may be more extensive than initially anticipated and that such polymorphisms may have great value in providing comparative information for the basis of human colonization, infectivity, and virulence and informative loci for the analysis of evolutionary and phylogenetic relationships within the Mycobacterium genus and among clinical isolates.
Acknowledgments
We thank J. Parkhill, The Sanger Center, for review of the H37Rv electropherograms; A. Duesterhoeft and K. Dix, Qiagen Genomic, Inc., for Masscode verification of SNPS; and The Sanger Center for the availability of the M. bovis sequence data.
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, grants RO1-AI40125 and AI46669.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City: an analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834-846. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Sague, C. S., W. Dooley, M. D. Hutton, J. Otten, A. Breeden, J. T. Crawford, A. E. Pitchenik, C. Woodley, G. Cauthen, and W. R. Jarvis. 1992. Hospital outbreak of multidrug-resistant Mycobacterium tuberculosis infections. Factors in transmission to staff and HIV-infected patients. JAMA 268:1280-1286. [DOI] [PubMed] [Google Scholar]
- 6.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 7.Betts, J. C., P. Dodson, S. Quan, A. P. Lewis, P. J. Thomas, K. Duncan, and R. A. McAdam. 2000. Comparison of the proteome of Mycobacterium tuberculosis strain H37Rv with clinical isolate CDC 1551. Microbiology 146:3205-3216. [DOI] [PubMed] [Google Scholar]
- 8.Bishai, W. R., A. M. Dannenberg, Jr., N. Parrish, R. Ruiz, P. Chen, B. C. Zook, W. Johnson, J. W. Boles, M. L. Pitt. 1999. Virulence of Mycobacterium tuberculosis CDC1551 and H37Rv in rabbits evaluated by Lurie's pulmonary tubercle count method. Infect. Immun. 67:4931-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, E. F., J. Li, H. Ochman, and R. K. Selander. 1997. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J. Bacteriol. 179:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 11.Delcher, A. L., S. Kasif, R. D. Fleischmann, J. Peterson, O. White, and S. L. Salzberg. 1999. Alignment of whole genomes. Nucleic Acids Res. 27:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, Z., C. Doig, D. T. Kenna, N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:449-512. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, S. V., B. Heym, J. Parkhill, B. Barrell, and S. T. Cole. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881-892. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 16.Ho, T. B. L., B. D. Robertson, G. M. Taylor, R. J. Shaw, and D. B. Young. 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jereb, J. A., D. R. Burwen, S. W. Dooley, W. H. Haas, J. T. Crawford, L. J. Geiter, M. B. Edmond, J. N. Dowling, R. Shapiro, A. W. Pasculle, et al. 1993. Nosocomial outbreak of tuberculosis in a renal transplant unit: application of a new technique for restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates. J. Infect. Dis. 168:1219-1224. [DOI] [PubMed] [Google Scholar]
- 18.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokoris, M., K. Dix, K. Moynihan, J. Mathis, B. Erwin, P. Grass, B. Hines, and A. Duesterhoeft. 2000. High-throughput SNP genotyping with the masscode system. Mol. Diagn. 5:329-340. [DOI] [PubMed] [Google Scholar]
- 20.Korber, B. 2000. HIV signature and sequence variation analysis, p. 55-72. In R. G. Allen and G. H. Learn (ed.), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 21.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manca, C., L. Tsenova, C. E. Barry III, A. Bergtold, S. Freeman, P. A. Haslett, J. M. Musser, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162:6740-6746. [PubMed] [Google Scholar]
- 23.McHugh, T. D., and S. H. Gillespie. 1998. Nonrandom association of the IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J. Clin. Microbiol. 36:1410-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musser, J. M., A. Amin, and S. Ramaswamy. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 26.Philipp, W. J., S. Nair, G. Guglielmi, M. Lagranderie, B. Gicquel, and S. T. Cole. 1996. Physical mapping of Mycobacterium bovis BCG Pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology 142:3135-3145. [DOI] [PubMed] [Google Scholar]
- 27.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 28.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resitance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 29.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skeiky, Y. A., P. J. Ovendale, S. Jen, M. R. Alderson, D. C. Dillon, S. Smith, C. B. Wilson, I. M. Orme, S. G. Reed, and A. Campos-Neto. 2000. T cell expression cloning of a Mycobacterium tuberculosis gene encoding a protective antigen associated with the early control of infection. J. Immunol. 165:7140-7149. [DOI] [PubMed] [Google Scholar]
- 32.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnhammer, E. L., S. R. Eddy, and R. Durbin. 1997. Pfam; a comprehensive database of protein domain families based on seed alignments. Proteins 3:405-420. [DOI] [PubMed] [Google Scholar]
- 34.Sreevatsanm, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutton, G. G., O. White, M. D. Adams, and A. R. Kerlavage. 1995. TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci. Tech. 1:9-19. [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valway, S. E., M. P. Sanchez, T. F. Shinnick, I. Orme, T. Agerton, D. Hoy, J. S. Jones, H. Westmoreland, and I. M. Onorato. 1998. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 338:633-639. [DOI] [PubMed] [Google Scholar]
- 38.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, F. S., T. S. Whittam, and R. K. Selander. 1997. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J. Bacteriol. 179:6551-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren, R. M., S. S. Shah, and D. Alland. 1997. Multiple drug resistance: a world wide threat, p. 77-96. In A. Malin and K. P. W. J. McAdam (ed.), Bailliere's clinical infectious diseases, vol. 4. Mycobacterial diseases. Part I: clinical frontiers. Bailliere Tindall, London, United Kingdom.
- 41.Warren, R. M., S. L. Sampson, M. Richardson, G. D. Van Der Spuy, C. J. Lombard, T. C. Victor, and P. D. van Helden. 2000. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol. Microbiol. 37:1405-1416. [DOI] [PubMed] [Google Scholar]
- 42.Waterman, M. S. 1988. Computer analysis of nucleic acid sequences. Methods Enzymol. 164:765-793. [DOI] [PubMed] [Google Scholar]