Abstract
In the methanogenic archaeon Methanococcus maripaludis, growth with ammonia results in conditions of nitrogen excess. Complete repression of nitrogen fixation (nif) gene transcription occurs, and glutamine synthetase (glnA) gene transcription falls to a basal constitutive level. In addition, ammonia completely switches off nitrogenase enzyme activity. In contrast, growth with dinitrogen as the sole nitrogen source results in nitrogen starvation, full expression of nif and glnA, and high activity of nitrogenase. Here we report that a third nitrogen source, alanine, results in an intermediate regulatory response. Growth with alanine resulted in intermediate transcription of nif and glnA, and addition of alanine to a nitrogen-fixing (diazotrophic) culture caused partial switch-off of nitrogenase. This uniformity of response occurred despite differences in regulatory mechanisms. Nitrogenase switch-off requires the nitrogen sensor homologs NifI1 and NifI2, while transcriptional regulation of nif and glnA relies on a different, unknown sensor mechanism. In addition, although nif and glnA transcription are governed by a common repressor, the numbers and arrangements of repressor binding sites differ. Thus, the nif promoter region contains two operators situated downstream of the transcription start site, while the glnA promoter region contains only one operator just upstream of two closely spaced transcription start sites. In a previous study of nif expression using ammonia, we were able to detect a role only for the first nif operator in repression. Here we show that nif repression by alanine requires the second operator as well. In contrast, in the case of glnA the single operator was sufficient for repression by ammonia or alanine. These results suggest a uniform cellular response to nitrogen that is mediated by a different mechanism in each case.
All organisms regulate nitrogen assimilation according to the nitrogen state of the cell. For example, in Escherichia coli transcription of the gene for the ammonia-assimilatory enzyme glutamine synthetase is induced when the cell is limited by nitrogen (24). Glutamine synthetase activity is also regulated by covalent modification of the enzyme (24). Other genes that may be regulated by nitrogen include those for ammonia transport and amino acid transport and utilization and other regulatory genes (31). Among free-living diazotrophs, nitrogen fixation is rigorously regulated (12), becoming active only when all nitrogen sources other than dinitrogen are exhausted.
For methanogenic archaea, an understanding of nitrogen assimilation has progressed significantly due in part to the establishment of genetic systems for Methanococcus species, including M. maripaludis (17, 26, 29). Genes for glutamine synthetase (glnA) (10, 23), nitrogen fixation (nif) (15, 18, 25), and ammonia transport (amtB) (7, 17) are homologous to those found in well-studied bacteria, indicating that nitrogen metabolism uses the same basic mechanisms. The presence of protein PII homologs suggests similarities in nitrogen sensing as well (7, 17). The PII protein of E. coli is the best-characterized member of a widespread family of nitrogen sensor proteins (1, 22).
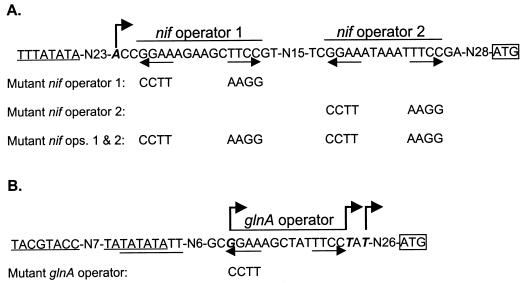
However, mechanisms of nitrogen regulation differ widely. In Proteobacteria, transcription of nitrogen-regulated genes is modulated by the two-component NtrB-NtrC activation system (20). In the gram-positive bacterium Corynebacterium glutamicum regulation occurs via the nitrogen repressor AmtR (13). In Bacillus subtilis, two homologous regulators, TnrA and GlnR, activate or repress depending on the nitrogen state of the cell (11). In contrast, we have shown recently that in M. maripaludis a novel repressor that bears no similarity to other known regulators governs a transcriptional nitrogen regulon (T. Lie, unpublished data). Previously we studied two operons, a nif operon containing the known nif genes of M. maripaludis and the glnA operon. The promoter regions of nif (9) and glnA (10) contain palindromic (inverted repeat) nitrogen operators (consensus GGAA-N6-TTCC) (Fig. 1), which we showed by mutagenesis to function in repression in vivo. Although the nif promoter region contains a second sequence that matches the nitrogen operator consensus, only the first (promoter proximal) was previously shown to be essential for repressor binding and to mediate repression with ammonia (9). Thus, the significance of the second operator remained unknown. In contrast, only one nitrogen operator exists upstream of glnA (10).
FIG. 1.
Promoter region sequences. Underlines indicate TATA boxes. Transcription starts are shown in boldface italics and marked with bent arrows. Horizontal arrows indicate inverted repeats. Start codons are boxed. Mutants contain the same sequences except for indicated changes in operators. (A) nif promoter region. (B) glnA promoter region; the upstream start site is constitutive while the two downstream start sites are regulated similarly by nitrogen (10).
The regulation of nitrogenase activity also varies between different microbial groups. Many diazotrophs have switch-off, the reversible down-regulation of nitrogenase activity by ammonia. In Rhodospirillum rubrum the enzymes dinitrogenase reductase ADP-ribosyl transferase and dinitrogenase reductase-activating glycohydrolase covalently modify dinitrogenase reductase and remove the modification, respectively. Their activities are regulated by the PII homologs GlnB and GlnJ (30). In contrast, switch-off in M. maripaludis occurs without detectable covalent modification of nitrogenase reductase and depends on the PII homologs NifI1 and NifI2 (16, 17). NifI1 and NifI2 diverge markedly in amino acid sequence from other members of the PII family and from each other, in a region called the T-loop that is thought to mediate interactions with other proteins (1, 22).
In the study of nitrogen regulation in many organisms, alternative nitrogen sources are used to achieve different nitrogen states in the cell (limitation versus excess). This approach allows the study of the regulatory response. Few studies have used a third nitrogen source to achieve an intermediate nitrogen state. Here we report that in M. maripaludis, alanine, an alternate nitrogen source in place of ammonia or dinitrogen, induces an intermediate nitrogen state. Two instances of transcriptional regulation (nif and glnA) and the nitrogenase switch-off mechanism are similarly tuned to the intermediate nitrogen state as evidenced by partial responses. However, these similar sensitivities are achieved by different mechanisms in each case.
MATERIALS AND METHODS
Strains, cultures, and growth conditions.
All experiments were conducted with M. maripaludis strain LL (15) (DSM stock no. 14266) and its derivatives. Strain LL was recently determined by W. Whitman to have originated from and to be identical to the wild-type strain S2 (28). Unless otherwise specified, cultures were grown in nitrogen-free liquid medium (5) under an atmosphere containing 58% H2, 20% CO2, and 22% N2 at a total pressure of 3.7 atm. In some experiments an atmosphere composed of 80% H2 and 20% CO2, or 58% H2, 21% CO2 20% Ar, was substituted for the N2-containing atmosphere. Puromycin (2.5 μg/ml), ammonium chloride (10 mM), and l-alanine (10 mM) were added from anaerobic stocks as needed. Tubes of medium (5 ml) were routinely inoculated with 0.2 ml of freshly grown culture and shaken at 37°C. After growth on ammonia, transfer to diazotrophic conditions generally required a lag period of 1 day before growth occurred, while subsequent transfers involved no noticeable lag. Therefore, cultures were routinely preadapted to diazotrophic growth and then used as inocula to initiate cultures for experiments. Cell densities were measured as optical densities at 660 nm (OD660) on a Spectronic 20 spectrophotometer.
Transcriptional fusion studies.
Strains Mm204, Mm221, and Mm222 each contain the promoter region of the nif operon (1.2 kb of DNA upstream of the nifH coding region) fused at the ATG start codon to a promoterless lacZYA fragment (9). Each construct is isolated from adjacent transcription units by transcriptional terminators and is inserted into the chromosome at the argH locus, a neutral site. Mm204 contains the wild-type nif promoter region, while Mm221 and Mm222 contain promoter regions that have been mutated in the first and second operator sequences, respectively (9) (Fig. 1A). For diazotrophic growth, tubes contained an atmosphere of H2, CO2, and N2 as specified above. For growth with ammonia or alanine, the headspace contained H2, CO2, and Ar as specified above. Cells from growing cultures (ammonia or alanine grown, OD660 of 0.3 to 0.6; dinitrogen grown, OD660 of 0.2 to 0.3) were assayed for β-galactosidase activities as described previously (21).
Gel mobility shift analysis.
Mobility shift probes were obtained by PCR from plasmid templates. The probe containing the wild-type operator region was amplified from pMmp1.1 (9) with the forward primer nifwt1-1 (5′ TCTAGAATTCTATAGCATAGTTCACC 3′) and the reverse primer nifwt2 (5′ GGAATTCTATATATTGTTGACTTTCGG 3′). Plasmid templates pnifmutAG1CT1 and pnifmutAG2CT2 were generated previously (9) by cloning the EcoRI-StuI fragment (containing the promoter region of nifH) of pMmp1 (5) into the EcoRI-BamHI (filled-in) site of pGEM7 (Promega). Mutations to the operator sites were then generated by site-directed mutagenesis as described previously (9). The probe containing mutant operator 1 was amplified from pnifHpmutAG1CT1 with primers nifwt1-1 and nifwt2. The probe containing mutant operator 2 was amplified from pnifHpmutAG2CT2 with primers nifwt1-1 and nifrightag2ct2 (5′ GGAATTCTATATATTGTTGACTTTC 3′). The probe containing both mutant operators was amplified from pnifHpmutAG1CT1 with primers nifwt1-1 and mutag2ct260 (5′ GGAATTCTATATATTGTTGACTTTCCCTTATTTATAAGGGATCTTTTAGTTATTATACCC 3′). PCR was performed with Taq DNA polymerase (Roche Molecular Biochemicals) with the following conditions: 95°C for 2 min; 25 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min; and a final extension of 72°C for 10 min. Probes were purified by using the Qiagen MinElute kit, digested with EcoRI for 1 h, and filled in with Klenow fragment and [32P]dATP. Cell extracts were obtained from ammonia-grown cells by lysing cells in cold 10 mM Tris buffer (pH 7.5), sonicating them for 10 s, and centrifuging them at 10,000 × g at room temperature. Protein concentration was determined by the method of Bradford (6). Cell extracts were made from cells grown to mid- or late growth phase (OD660 of 0.6 to 0.9). Radiolabeled probe (approximately 240 fmol/ml) in buffer [10 mM Tris (pH 7.5), 300 μg of bovine serum albumin/ml, 50 μg of poly(dI-dC)/ml, 11.25% glycerol, and 10 mM dithiothreitol] was mixed with various amounts of cell extract in a total volume of 0.1 ml, incubated at 30°C for 20 min, and run on a 5% polyacrylamide gel in Tris-acetate-EDTA buffer, pH 8 (2). Radioactive bands were imaged and quantified by using a phosphorimager (Molecular Dynamics).
RNA extraction.
Diazotrophic cultures (OD660 of 0.2 to 0.3) or alanine- or ammonia-grown cultures (OD660 of 0.5 to 0.8) were transferred to screw-cap 15-ml conical tubes and centrifuged at 2,400 × g at 4°C for 15 min. Cells were resuspended in 100 μl of cold nitrogen-free medium with no sulfide added. RNA was extracted with the RNeasy kit (Qiagen) according to the manufacturer's instructions. RNA was eluted with RNase-free water followed by addition of one-half volume of super-pure-grade formamide (Sigma). Samples were stored at −20°C until use.
Northern analysis.
The EcoRI-SnaBI fragment of plasmid pJL1 was used as a probe for glnA mRNA (10). Labeling of probes was done with the Prime-It-II random primer labeling kit (Stratagene). RNA samples (approximately 3 μg) were run on an agarose-formaldehyde gel (1% [wt/vol]) (2) and transferred onto a Zeta-Probe GT blotting membrane (Bio-Rad). Hybridizations were done at 42°C with formamide buffer (50% formamide, 0.12 M Na2HPO4 [pH 7.2], 0.25 M NaCl, 7% [wt/vol] sodium dodecyl sulfate) as suggested by the Zeta-Probe GT blotting membrane instruction manual. Blots were exposed to phosphor screens, and radioactive bands were quantified with a phosphorimager.
Glutamine synthetase assay.
Cultures (ammonia and alanine grown, OD660 of 0.8 to 0.9; dinitrogen grown, OD660 of 0.2 to 0.3) were assayed for glutamine synthetase activities as described previously (10). Protein determination was done by the method of Bradford (6).
Acetylene reduction assays.
Diazotrophic cultures were grown to an OD660 of 0.2 to 0.4 and assayed for acetylene reduction as described previously (17). Strains Mm53 (nifI+) and Mm54 (ΔnifI1nifI2) (17) were used in this study.
RESULTS
Growth of M. maripaludis on alanine.
In a survey of compounds that could serve as nitrogen sources for Methanococcus species, it was found that ammonia, dinitrogen, and alanine could support the growth of M. maripaludis type strain JJ (27). We found that the same nitrogen sources served for strain LL and conducted experiments to determine the growth kinetics of this strain with each of these nitrogen sources. In batch culture, growth of hydrogenotrophic methanogens is generally hydrogen limited during most of the growth period, resulting in linear, not exponential, growth. M. maripaludis LL grew on ammonia, alanine, and dinitrogen at 0.056, 0.051, and 0.031 OD660 U/h, respectively. Growth in each case could be attributed entirely to the nitrogen source provided. Thus, in the case of ammonia or alanine, growth was similar whether the atmosphere contained N2 or entirely H2 and CO2, while no growth occurred under the latter atmosphere if no ammonia or alanine was added. These experiments show that growth with alanine may be marginally slower than that with ammonia, while growth on dinitrogen occurs at a substantially lower rate. The slower growth on dinitrogen presumably reflects the large expenditure of ATP required for nitrogen fixation (16).
Regulation of nif transcription.
The nif promoter region of M. maripaludis contains two palindromic sequences containing the nitrogen operator consensus (Fig. 1A). In a previous study (9) we constructed strains containing wild-type and mutant nif promoter regions fused to lacZ and used them to study the role of each palindrome under different nitrogen conditions. Each nif promoter-lacZ construct was inserted into the chromosome at a neutral site. The first palindrome was required for repression by ammonia, while the second palindrome played no apparent role. Here we extend these studies by investigating the possible role of the second palindrome in nif regulation during growth on alanine.
In the present study we used three strains containing nif promoter-lacZ fusion constructs. One strain contained unaltered palindromes, while the other two strains were altered in the first and second palindromes, respectively (Fig. 1A). Each strain was grown under three nitrogen conditions and assayed for β-galactosidase activity (Table 1). The wild-type promoter region mediated marked repression by ammonia but only partial repression by alanine. Altering the first palindrome eliminated all repression. Notably, altering the second palindrome eliminated repression by alanine but left repression by ammonia intact. These results show that the first palindrome (henceforth nif operator 1) is the primary cis-regulatory element and is essential for repression, while the second palindrome (nif operator 2) acts only in concert with nif operator 1 and functions only in repression by alanine.
TABLE 1.
nif promoter-lacZ expression in M. maripaludisa
| nif promoter region | Result with nitrogen source:
|
||
|---|---|---|---|
| Ammonia | Alanine | Dinitrogen | |
| Wild type | 21 ± 15 | 56 ± 10 | 129 ± 19 |
| Mutation in first operator | 452 ± 155 | 530 ± 177 | 458 ± 118 |
| Mutation in second operator | 29 ± 20 | 220 ± 27 | 227 ± 26 |
Values are in Miller units. Means and standard deviations are for triplicate cultures.
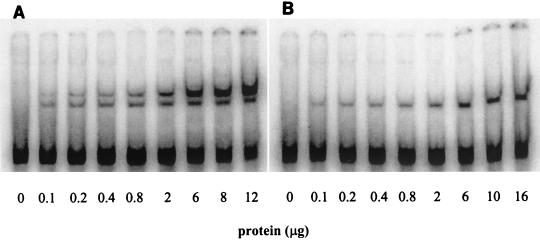
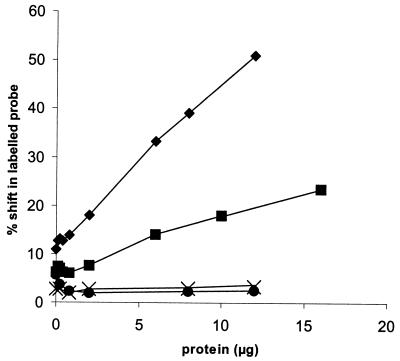
Based on these results, we hypothesized that nif operator 2 enhances binding of a repressor, thus strengthening repression at intermediate nitrogen conditions. We showed previously in gel mobility shift experiments that a repressor present in cell extracts bound specifically to the operator region containing nif operator 1 (9). Here we used various concentrations of cell extracts (of cells grown on ammonia) to make a quantitative comparison of binding to operator region DNA with and without nif operator 2. Mobility shift probes (labeled DNA fragments) were designed to correspond to the wild-type and mutant operator regions represented in Fig. 1A, beginning after the TATA box and ending before the ribosome binding site and start codon. More probe was shifted at a given concentration of cell extract when both operators were intact than when nif operator 2 was mutated (Fig. 2 and 3). Probes with mutant nif operator 1 or both mutant operators did not shift (Fig. 3). These results show that the primary and secondary binding roles of nif operators 1 and 2 observed in vitro correspond to their respective functions in regulating gene expression in vivo. The role of nif operator 1 as the primary repressor binding site explains its essential function in repression. The secondary role of nif operator 2 in strengthening repressor binding explains its function in repression by alanine, since it may be required for repressor binding at intermediate levels of active repressor.
FIG. 2.
Gel mobility shift of nif operator region DNA by cell extract. (A) Probe with both operators being wild type. (B) Probe with first operator being wild type and second operator being mutant.
FIG. 3.
Graphic representation of gel mobility shifts. ♦, from Fig. 2A, both wild-type operators; ▪, from Fig. 2B, first operator wild type, second operator mutant; •, second operator wild type, first operator mutant; ×, both operators mutant.
An additional observation emerges from these binding studies. With only nif operator 1 intact (Fig. 2B), the probe shifted to a single position. In contrast, with both operators being wild type (Fig. 2A), increasing concentrations of cell extract resulted in the appearance of a second upper band that gained intensity. At even higher concentrations of cell extract the lower band disappeared completely, and all the shifted probe occupied the upper position (data not shown). This transition may reflect a change from single-operator binding (sufficient for repression by ammonia) to a configuration where repressor binds both operators cooperatively (necessary for repression by alanine).
Regulation of glnA expression.
The glnA promoter region contains three TATA boxes corresponding to three transcription start sites (Fig. 1B) (10). The first promoter is responsible for low-level constitutive expression, while the second and third (overlapping) promoters mediate expression that is regulated via a single operator. Previously we demonstrated the role of the operator by constructing a strain (Mm312) that contained two copies of glnA (10). One copy of glnA was unaltered, while the second copy contained both an altered operator (Fig. 1B) and an in-frame deletion in the coding region. In Northern analysis, expression of the larger allele was repressed by ammonia while the expression of the shorter allele was constitutive (10).
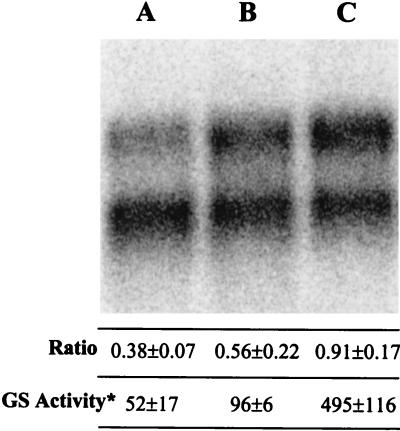
In the present study, we used Northern blots with strain Mm312 to determine the effect of alanine. In Fig. 4 the lower band represents unregulated expression due to the operator mutation while the upper band represents regulated expression. Therefore, each lane can be internally calibrated to the band representing the constitutively expressed allele, and the ratio of intensities of the upper to lower band indicates the relative degree of expression. glnA expression during growth on alanine was intermediate between that of ammonia and dinitrogen (Fig. 4). Similar trends were observed in separate experiments. Correspondingly, glutamine synthetase activity was also intermediate.
FIG. 4.
glnA mRNA levels and glutamine synthetase activities in Mm312. (A) Ammonia; (B) alanine; (C) dinitrogen. Intensity ratios of the upper and lower bands are indicated (three replicate cultures). Glutamine synthetase (GS) activities are nanomoles minute−1 milligram of protein−1 (three replicate cultures).
Switch-off of nitrogenase activity.
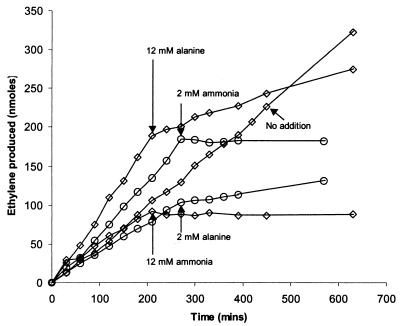
We used the acetylene reduction assay to monitor nitrogenase activity. As before (16, 17), addition of ammonia to diazotrophic cultures completely stopped accumulation of ethylene, indicating a cessation of nitrogenase activity (Fig. 5). Ammonia had the same effect whether added to a 12 mM or a 2 mM concentration, and in a previous study complete switch-off occurred at an ammonia concentration as low as 25 μM (17). Alanine at either 12 or 2 mM had only a partial effect (i.e., incomplete switch-off). No switch-off occurred when either ammonia or alanine was added to the mutant Mm54, which lacks the nifI1 and nifI2 genes previously shown to be required for switch-off (16, 17) (data not shown).
FIG. 5.
Switch-off of nitrogenase activity in Mm53. Acetylene was added at time zero. Ammonia or l-alanine was added at the times and final concentrations indicated. Additions at 2 mM concentrations were performed in a separate experiment.
DISCUSSION
In M. maripaludis, use of alanine as a nitrogen source consistently elicited an intermediate regulatory response compared to those with ammonia and dinitrogen. The effect did not appear to rely on a limiting concentration, since at least in the case of switch-off, 2 mM alanine had as great an effect as a 12 mM concentration. The cell may perceive alanine as a poorer nitrogen source than ammonia. The mechanism for alanine import or for conversion of alanine into usable nitrogen for the cell (perhaps alanine dehydrogenase-catalyzed ammonia production or amine transfer to glutamate to form glutamine) may be relatively inefficient.
The intermediate response to alanine occurred at the transcriptional as well as posttranscriptional levels. Furthermore, the two systems seem similarly tuned to the degree of nitrogen starvation versus excess. This is the case even though transcriptional regulation and switch-off appear to use different sensory mechanisms to determine the nitrogen state of the cell. Thus, switch-off, but not transcriptional regulation, relies on NifI1 and NifI2 (16, 17), both paralogs of the PII family of proteins shown to sense nitrogen in Proteobacteria (1, 22). The sensor for transcriptional regulation is unknown but does not depend on the NifI proteins (16).
While the sensor for transcriptional regulation by nitrogen remains unknown, we have recently identified the gene encoding the repressor protein (Lie, unpublished). As expected, a mutation in this gene derepressed both nif and glnA. The same repressor apparently regulates both operons, as predicted from their similar operator sequences. Growth on alanine apparently results in an intermediate repressor activity, the amount of repressor present in the cell combined with factors that affect its tendency to bind to operator DNA. In this light, it is perhaps surprising that the nif operon and glnA respond similarly to intermediate nitrogen, since repressor interaction with the nif operon requires two operators, while glnA uses only one. However, the operator for glnA falls between the TATA boxes and the transcription starts, whereas the operators in the nif promoter region lie downstream from the transcription start site (Fig. 1). This more-downstream position may necessitate tighter binding of the repressor (via cooperative binding to two operators) for interference with transcription initiation. In addition, nucleotides flanking the consensus part of the operators differ, possibly affecting binding affinities. A more detailed analysis of repressor-operator interactions awaits the purification of the repressor protein.
Cooperative binding to multiple operators is common in the bacteria. In the classical example, two secondary operators are found in the lac promoter region of E. coli (8). This theme extends to nitrogen regulation in the gram-positive bacterium C. glutamicum, where a repressor binds to a single, a double, or even a truncated single operator in the promoter regions of different nitrogen-regulated genes (3, 13, 14). For the archaea, repression has been demonstrated in several other instances (4, 19). A repression mechanism analogous to the bacterial repression paradigm apparently occurs in Archaea in spite of a basal transcription apparatus that resembles a simplified eukaryal system (4, 19). The existence of multiple operators adds another facet to this analogy.
Acknowledgments
We thank Gwen Wood for a critical reading of the manuscript.
This work was supported by grant GM-55255 from the National Institutes of Health and grant 35319-09927 from the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture.
REFERENCES
- 1.Arcondeguy, T., R. Jack, and M. Merrick. 2001. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65:80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. Current protocols in molecular biology. J. Wiley and Sons, Inc., New York, N.Y.
- 3.Beckers, G., L. Nolden, and A. Burkovski. 2001. Glutamate synthase of Corynebacterium glutamicum is not essential for glutamate synthesis and is regulated by the nitrogen status. Microbiology 147:2961-2970. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in Archaea. Curr. Opin. Microbiol. 4:208-213. [DOI] [PubMed] [Google Scholar]
- 5.Blank, C. E., P. S. Kessler, and J. A. Leigh. 1995. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J. Bacteriol. 177:5773-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 8.Choy, H., and S. Adhya. 1996. Negative control, p. 1287-1299. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 9.Cohen-Kupiec, R., C. Blank, and J. A. Leigh. 1997. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl. Acad. Sci. USA 94:1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen-Kupiec, R., C. J. Marx, and J. A. Leigh. 1999. Function and regulation of glnA in the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 181:256-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 12.Halbleib, C. M., and P. W. Ludden. 2000. Regulation of biological nitrogen fixation. J. Nutr. 130:1081-1084. [DOI] [PubMed] [Google Scholar]
- 13.Jakoby, M., L. Nolden, J. Meier-Wagner, R. Kramer, and A. Burkovski. 2000. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol. Microbiol. 37:964-977. [DOI] [PubMed] [Google Scholar]
- 14.Jakoby, M., M. Tesch, H. Sahm, R. Kramer, and A. Burkovski. 1997. Isolation of the Corynebacterium glutamicum glnA gene encoding glutamine synthetase I. FEMS Microbiol. Lett. 154:81-88. [DOI] [PubMed] [Google Scholar]
- 15.Kessler, P. S., C. Blank, and J. A. Leigh. 1998. The nif gene operon of the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 180:1504-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler, P. S., C. Daniel, and J. A. Leigh. 2001. Ammonia switch-off of nitrogen fixation in the methanogenic archaeon Methanococcus maripaludis: mechanistic features and requirement for the novel GlnB homologues NifI1 and NifI2. J. Bacteriol. 183:882-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kessler, P. S., and J. A. Leigh. 1999. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152:1343-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leigh, J. A. 2000. Nitrogen fixation in methanogens: the archaeal perspective. Curr. Issues Mol. Biol. 2:125-131. [PubMed] [Google Scholar]
- 19.Leigh, J. A. 1999. Transcriptional regulation in Archaea. Curr. Opin. Microbiol. 2:131-134. [DOI] [PubMed] [Google Scholar]
- 20.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Ninfa, A. J., and M. R. Atkinson. 2000. PII signal transduction proteins. Trends Microbiol. 8:172-179. [DOI] [PubMed] [Google Scholar]
- 23.Possot, O., L. Sibold, and J. P. Aubert. 1989. Nucleotide sequence and expression of the glutamine synthetase structural gene, glnA, of the archaebacterium Methanococcus voltae. Res. Microbiol. 140:355-371. [DOI] [PubMed] [Google Scholar]
- 24.Reitzer, L. J. 1996. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine, p. 391-407. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 25.Souillard, N., and L. Sibold. 1989. Primary structure, functional organization and expression of nitrogenase structural genes of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Mol. Microbiol. 3:541-551. [DOI] [PubMed] [Google Scholar]
- 26.Tumbula, D. L., and W. B. Whitman. 1999. Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol. Microbiol. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Whitman, W. B. 1989. Order II. Methanococcales Balch and Wolfe 1981, 216VP, p. 2185-2190. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 3. The Williams & Wilkins Co., Baltimore, Md.
- 28.Whitman, W. B., J. Shieh, S. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 29.Whitman, W. B., D. L. Tumbula, J. P. Yu, and W. Kim. 1997. Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors 6:37-46. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2001. Functional characterization of three GlnB homologs in the photosynthetic bacterium Rhodospirillum rubrum: roles in sensing ammonium and energy status. J. Bacteriol. 183:6159-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]