Abstract
Amino acid transport by Rhizobium leguminosarum is dominated by two ABC transporters, the general amino acid permease (Aap) and the branched-chain amino acid permease (Bra). However, mutation of these transporters does not prevent this organism from utilizing alanine for growth. An R. leguminosarum permease (MctP) has been identified which is required for optimal growth on alanine as a sole carbon and nitrogen source. Characterization of MctP confirmed that it transports alanine (Km = 0.56 mM) and other monocarboxylates such as lactate and pyruvate (Km = 4.4 and 3.8 μM, respectively). Uptake inhibition studies indicate that propionate, butyrate, α-hydroxybutyrate, and acetate are also transported by MctP, with the apparent affinity for solutes demonstrating a preference for C3-monocarboxylates. MctP has significant sequence similarity to members of the sodium/solute symporter family. However, sequence comparisons suggest that it is the first characterized permease of a new subfamily of transporters. While transport via MctP was inhibited by CCCP, it was not apparently affected by the concentration of sodium. In contrast, glutamate uptake in R. leguminosarum by the Escherichia coli GltS system did require sodium, which suggests that MctP may be proton coupled. Uncharacterized members of this new subfamily have been identified in a broad taxonomic range of species, including proteobacteria of the β-subdivision, gram-positive bacteria, and archaea. A two-component sensor-regulator (MctSR), encoded by genes adjacent to mctP, is required for activation of mctP expression.
Rhizobium leguminosarum is a member of a group of α-subdivision of the proteobacteria (the rhizobia), which form a species-specific symbiotic relationship with leguminous plants in order to fix atmospheric nitrogen. The plant supplies the bacteroid (symbiotic bacteria) with a carbon source (C4-dicarboxylic acid), while the plant receives reduced nitrogen from the bacteroid. Other rhizobia include Rhizobium, Mesorhizobium, Sinorhizobium, Bradyrhizobium, and Azorhizobium species (8, 33, 53). Prior to establishing a Rhizobium-legume symbiosis, bacteria must thrive in the soil environment, competing with many organisms for nutrients. Transporters of key nutrients, such as amino acids, may give a competitive advantage to rhizobia, allowing them to better colonize roots. This may account for the large number of transporters that have been revealed by the completed genome sequences of Sinorhizobium meliloti, Mesorhizobium loti, and Agrobacterium tumefaciens (13, 26, 59).
Amino acid transport by R. leguminosarum is dominated by two permeases of the ABC transporter superfamily, the general amino acid permease (Aap) and the branched-chain amino acid permease (Bra). Aap and Bra are members of the polar amino acid transporter (transport classification [T.C.] number 3.A.1.3) and hydrophobic amino acid transporter (T.C. number 3.A.1.4) families, respectively (21, 44). However, both are atypical as they transport a broad range of amino acids (19, 56). Indeed, Bra has the broadest specificity of any characterized amino acid transporter in bacteria. Bra has a high affinity for l-alanine, l-leucine, l-histidine, and γ-aminobutyric acid (Km between 78 and 288 nM) and a lower affinity for l-glutamate (56 μM). d-α-Amino acids inhibit uptake of Bra solutes, indicating they are also transported by this permease. Conversely, Aap is specific for l-α-amino acids, with high affinity for l-glutamate, l-alanine, l-leucine, and l-histidine (Km between 200 nM and 515 nM) (19). Both Aap and Bra are required for optimal growth of free-living R. leguminosarum on a range of amino acids as a sole source of carbon and nitrogen. In fact, double aap bra mutants cannot utilize glutamate, glutamine, asparagine, proline, arginine, or γ-aminobutyrate (19). However, growth on alanine and histidine is unaffected by the loss of these transporters. Therefore, although alanine and histidine are transported by Aap and Bra, R. leguminosarum has other transporters of these amino acids.
The importance of alanine metabolism in R. leguminosarum has been highlighted by the demonstration that alanine can comprise as much as 26% of the total nitrogen secreted by bacteroids isolated from pea nodules (1). In the case of soybean bacteroids, it has been reported that alanine is the sole secretion product of nitrogen fixation (58), although this has been challenged (30). Nevertheless, these findings have questioned the established view that ammonium is the sole secretion product of N2 fixation from the bacteroid. However, it remains to be established how alanine produced by N2 fixation is secreted from the bacteroid and made available to the plant.
We report here the identification of a novel R. leguminosarum permease (MctP), which is required for optimal growth on alanine as a sole carbon and nitrogen source. Characterization of MctP confirmed that it transports alanine and other monocarboxylates such as pyruvate and lactate. However, it is distinct from characterized transporters of monocarboxylates, such as members of the lactate permease family (LctP) (T.C. number 2.A.14) and the proton-linked monocarboxylate porter family (T.C. number 2.A.1.13) (14, 44). Although MctP has significant sequence similarity to members of the sodium/solute symporter family (T.C. number 2.A.21) (42, 44), sequence comparisons indicate it is the first characterized permease of a new subfamily of bacterial transporters, with representatives present in gram-negative and gram-positive bacteria and archaea. A two-component sensor-regulator (MctSR), encoded by genes adjacent to mctP, is required for mctP expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and culture conditions.
The bacterial strains and plasmids used in this study are detailed in Table 1. R. leguminosarum strains were grown at 28°C on either tryptone yeast extract (TY) (5), acid minimal salts medium (AMS), or acid minimal salts agar (AMA) (36). The following carbon and nitrogen sources were added to minimal media at the indicated concentrations; 10 mM d-glucose, 10 mM succinate, 20 mM pyruvate, 10 mM ammonium chloride, and 20 mM alanine. The following antibiotics were used at the indicated concentrations (micrograms milliliter−1); streptomycin, 500; kanamycin, 40; tetracycline, 2 (in AMS) and 5 (in TY); gentamicin, 20; and spectinomycin, 100.
TABLE 1.
Bacterial strains, cosmids, and plasmids used in this study
| Strain, cosmid, or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| 3841 | Rhizobium leguminosarum bv. viciae Smr derivative of strain 300 | 24 |
| A34 | Rhizobium leguminosarum bv. viciae (formerly known as 8401 pR11JI) | 10 |
| VF39SM | Rhizobium leguminosarum bv. viciae Smr | 38 |
| RU1357 | A34 braE::TnphoA ΔaapJQM::ΩSp | 20 |
| RU1180 | 3841 mctP::TnlacZ | This study |
| RU1580 | pRU895 homogenote in 3841; mctS::GeneJumper Kmr transposon | This study |
| RU1581 | pRU897 homogenote in 3841; mctP::GeneJumper Kmr transposon | This study |
| RU1582 | pRU898 homogenote in 3841; mctR::GeneJumper Kmr transposon | This study |
| RU1583 | pRU899 homogenote in 3841; mctR::GeneJumper Kmr transposon | This study |
| RU1584 | pRU900 homogenote in 3841; mctS::GeneJumper Kmr transposon | This study |
| RU1585 | pRU901 homogenote in 3841; mctR::GeneJumper Kmr transposon | This study |
| RU1586 | pRU902 homogenote in 3841; mctS::GeneJumper Kmr transposon | This study |
| RU1587 | pRU903 homogenote in 3841; mctS::GeneJumper Kmr transposon | This study |
| RU1601 | 3841 pRU923 | This study |
| RU1602 | RU1580 pRU923 | This study |
| RU1603 | RU1581 pRU923 | This study |
| RU1604 | RU1582 pRU923 | This study |
| RU1632 | RU1357 pRU976 | This study |
| RU1633 | RU1357 pRU986 | This study |
| Cosmid | ||
| pRU3147 | pLAFR1 cosmid containing mct region from strain 3841 | This study |
| Plasmids | ||
| pCR2.1 TOPO | Cloning vector for PCR products; Kmr, Ampr | Invitrogen Life Technologies |
| pCR-Blunt-TOPO | Cloning vector for PCR products; Kmr | Invitrogen Life Technologies |
| pDONR201 | PCR cloning vector for Gateway Technology | Invitrogen Life Technologies |
| pJQ200SK | pACYC derivative, P15A origin of replication; Gmr | 39 |
| pLAFR1 | Wide-host-range mobilizable P-group cloning vector; Tcr | 11 |
| pPHJI1 | P-group chaser plasmid | 16 |
| pRK415-1 | Broad-host-range P-group cloning vector; Tcr | 27 |
| pJP2 | pTR102 GUS (mini-RK2 derivative) with artificial MCS; Ampr Tcr | 37 |
| pMN402 | GFP+ expression vector; hygromycin resistance | 47 |
| pOT2 | Promoter probe vector with promoterless gfpUV; Gmr | 2 |
| pOT2GFP+ | Promoter probe vector with promoterless gfp+; Gmr | This study |
| pOT3GFPuv | pOT2 with Gateway vector conversion cassette (reading frame A; Invitrogen Life Technologies) cloned into SmaI site | This study |
| pOT3GFP+ | pOT2GFP+ with Gateway vector conversion cassette (reading frame A; Invitrogen Life Technologies) cloned into SmaI site | This study |
| pRU738 | 2-kb PCR product (primers P237 and P238; mctP) from pRU3147 cloned into pCR2.1-TOPO | This study |
| pRU739 | 2-kb EcoRI fragment from pRU738 in pRK415-1 with mctP under the control of the lac promoter | This study |
| pRU747 | 2-kb EcoRI fragment from pRU738 in pRK415-1 with mctP not under the control of the lac promoter | This study |
| pRU785 | 3.6-kb SalI fragment from pRU3147 in pRK415-1 with mctRP not under the control of the lac promoter | This study |
| pRU788 | 3.6-kb SalI fragment from pRU3147 in pRK415-1 with mctRP under the control of the lac promoter | This study |
| pRU887 | 4.8-kb PCR product (primers P238 and P285; mctSRP) from pRU3147 cloned into pCR2.1-TOPO | This study |
| pRU890 | pJQ200SK containing a 4.4-kb BamHI fragment from pRU887 | This study |
| pRU895 | pRU890 mctS::GeneJumper Kmr transposon | This study |
| pRU897 | pRU890 mctP::GeneJumper Kmr transposon | This study |
| pRU898 | pRU890 mctR::GeneJumper Kmr transposon | This study |
| pRU899 | pRU890 mctR::GeneJumper Kmr transposon | This study |
| pRU900 | pRU890 mctS::GeneJumper Kmr transposon | This study |
| pRU901 | pRU890 mctR::GeneJumper Kmr transposon | This study |
| pRU902 | pRU890 mctS::GeneJumper Kmr transposon | This study |
| pRU903 | pRU890 mctS::GeneJumper Kmr transposon | This study |
| pRU917 | PCR product (primers P316 and P317) containing the putative mctP promoter region from pRU3147 in pDONR201 | This study |
| pRU922 | mctP promoter region from pRU917 in pOT3GFPuv | This study |
| pRU923 | mctP promoter region from pRU917 in pOT3GFP+ | This study |
| pRU935 | 1.8-kb PCR product (primers P345 and P343; mctS) from pRU3147 cloned into pCR-Blunt II-TOPO | This study |
| pRU936 | 2.7-kb PCR product (primers P342 and P343; mctSR) from pRU3147 cloned into pCR-Blunt II-TOPO | This study |
| Strain, cosmid, or plasmid | Description | Source or reference |
| pRU941 | 2.3-kb PCR product (primers P340 and P341; mctP) from pRU3147 cloned into pCR2.1-TOPO | This study |
| pRU942 | 1.5-kb PCR product (primers P342 and P344; mctR) from pRU3147 cloned into pCR2.1-TOPO | This study |
| pRU947 | 2.3-kb HindIII fragment from pRU941 in pRK415-1 with mctP under the control of the lac promoter | This study |
| pRU948 | 2.3-kb HindIII fragment from pRU941 in pRK415-1 with mctP not under the control of the lac promoter | This study |
| pRU950 | 1.8-kb EcoRI fragment from pRU935 in pRK415-1 with mctS under the control of the lac promoter | This study |
| pRU951 | 1.5-kb EcoRI fragment from pRU942 in pRK415-1 with mctR under the control of the lac promoter | This study |
| pRU952 | 2.7-kb EcoRI fragment from pRU936 in pRK415-1 with mctSR under the control of the lac promoter | This study |
| pRU953 | 1.5-kb EcoRI fragment from pRU942 in pRK415-1 with mctR not under the control of the lac promoter | This study |
| pRU974 | 2.7-kb EcoRI fragment from pRU936 in pRK415-1 with mctSR not under the control of the lac promoter | This study |
| pRU971 | 1.8-kb PCR product (primers P353, P355, P357 and P358; aapJ promoter-gltP fusion) cloned into pCR2.1-TOPO | This study |
| pRU976 | 1.8-kb XbaI/HindIII fragment from pRU971 (aapJ promoter-gltP fusion) in pJP2 | This study |
| pRU980 | 1.8-kb PCR product (primers P354, P356, P357 and P359; aapJ promoter-gltS fusion) cloned into pCR2.1-TOPO | This study |
| pRU986 | 1.8-kb XbaI/HindIII fragment from pRU976 (aapJ promoter-gltS fusion) in pJP2 | This study |
Oligonucleotide primers used in this study were as follows: P69, GAGAGAGAACTAGTGGAGGAAGAAAAAATGAGTAAAGGAGAAGAAC; P237, TCACATGCGTTACATCAGCTTCCG; P238, CGCCCTGATCTCTCTCCAGCTTATG; P266, TGTAGAGCTCATCCATGCCATG; P285 TGCAAAATAATCTTCCTCGTGCGG; P316, GGGGACCACTTTGTACAAGAAAGCTGGGTCTTGGCGCGTTTCCACAGCA; P317, GGGGACAAGTTTGTACAAAAAAGCAGGCTCGGCAGGAGCAACAAGGAGA; P340, AAGCTTCGCCCTGATCTCTCTCCAGCTTATG; P341, TTCGAACGGCAGGAGCAACAAGGAGA; P342, AAGAGACGGCTCGCGGACATTGTAA; P343, TGGGCGGAGAAGCAGGATCA; P344, TGGAGGCACTCTGCTATCATTTTGC; P345, TCTCCAGTCCCGTTTGGGCA; P353, CGGAAAAGGTTGGGAAAATGAAAAATATAAAATTCAG; P354, CGGAAAAGGTTGGGAAAATGTTTCATCTCGATACTTTAG; P355, CTGAATTTTATATTTTTCATTTTCCCAACCTTTTCCG; P356, CTAAAGTATCGAGATGAAACATTTTCCCAACCTTTTCCG; P357, CGCGCGTCCCGTAAGTGTAT; P358, TCCCCGGCAATCTTCAATTATT; and P359, AATTATGAATCAATACGCAGGCTTG.
Genetic modification of bacterial strains.
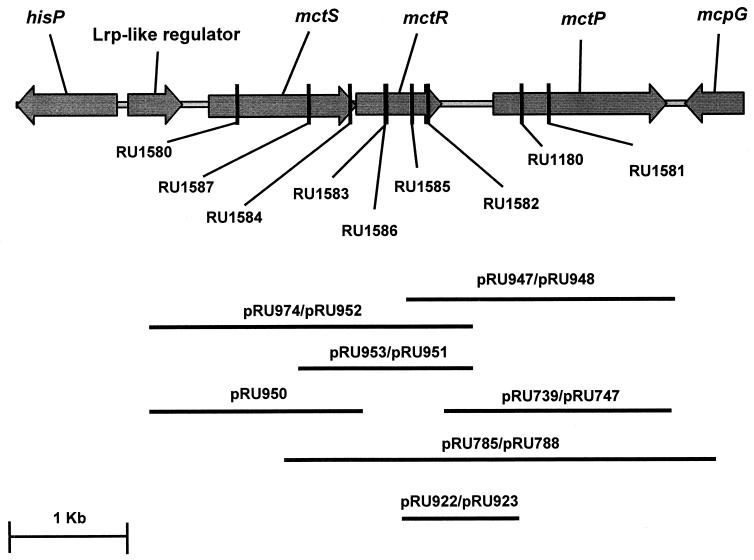
All DNA and genetic analyses and Tn5 mutagenesis were carried out as described previously (57). Transduction was carried out using the lytic phage RL38 as previously described (7). The sequenced region of the R. leguminosarum genome that includes the mctPSR is shown in Fig. 1, with a representation of fragments located on the plasmids used in this study and the sites of transposon insertion in mutant strains. Plasmids were conjugated into R. leguminosarum as described previously (36).
FIG. 1.
Map of the mct region. The region of the R. leguminosarum genome containing the mct permease is represented here (the sequence has been submitted to the EMBL database under accession number AJ421944). Vertical lines represent the locations of the insertions used to construct mutants used in this study. The lines beneath the map indicate the region located on the plasmids used in this study.
To obtain mutants in mctSR and mctP, plasmid pRU890 (Table 1) was mutated in vitro by GeneJumper transposon (Kmr) insertion as described by the manufacturer (Invitrogen Life Technologies). Chromosomal insertion mutants were constructed by conjugating the mutated plasmids into R. leguminosarum 3841 and selecting for Gms Kmr homogenates on TY containing 10% sucrose. All mutants were confirmed by Southern blotting.
Construction of aapJ promoter-gltS/gltP fusion plasmids.
In order to ensure that E. coli gltS and gltP were expressed in R. leguminosarum, the gltS and gltP genes were fused to the R. leguminosarum aapJ promoter region by two PCRs using overlapping primers (18). The intergenic region between aapJ and the upstream metC gene (accession number X82596) was first amplified using primer P357 (which has identity to the metC region) and either primer P355 or P356, which have identity to the region immediately upstream of aapJ and a 5′ extension with identity to the N-terminal coding region of E. coli gltP or gltS, respectively. Similarly, gltP and gltS were amplified using primers specific to sequence downstream of gltP or gltS (P358 and P359, respectively) and primers specific to the N terminus coding region of gltP or gltS and the region immediately upstream of aapJ (P353 and P354, respectively). The PCR products from these reactions were purified by gel extraction (QIAquick gel extraction kit; QIAGEN Ltd.), mixed (i.e., P357-P355 product with P353-P358 product or P357-P356 product with P354-P359 product), and used as template for a second round of PCR. The second PCRs used primers specific for the aapJ-metC intergenic region and a sequence downstream of gltP or gltS (i.e., P357 with P358 for aapJ promoter-gltP fusion and P357 with P359 for aapJ promoter-gltS fusion). The resulting PCR products were cloned by TOPO reaction into pCR2.1 TOPO (Invitrogen Life Technologies), and fusion junctions were confirmed by sequencing, before subcloning into the broad-host-range vector pJP2, using the XbaI and HindIII sites located on the multicloning site of these two vectors. Thus, the resulting plasmids, pRU976 and pRU986, contain aapJ promoter-gltP and aapJ promoter-gltS fusions, respectively.
Construction and detection of the mctP promoter reporter plasmid.
The promoter reporter plasmids used were derived from the pOT family of vectors described previously (2). Another plasmid in this family, pOT2gfp+, has been constructed as part of a separate project, and full details will be described elsewhere. Essentially, the gfp+ gene was amplified from pMN402 using primers P69 and P266 and cloned into pCR2.1 TOPO. A SpeI/XhoI fragment from this was used to replace the same fragment in pOT2, creating pOT2gfp+. gfp+ has been compared directly against lacZ as a reporter gene and shown to give almost identical results (47). Plasmids pOT2 and pOT2gfp+ were modified for Gateway technology by cloning the Gateway vector conversion cassette (reading frame A; Invitrogen Life Technologies) into the two SmaI sites in the multicloning site to create pOT3gfpuv and pOT3gfp+. The orientation of the Gateway cassette was confirmed by DNA sequencing.
The mctR-mctP intergenic region, which contains a putative mctP promoter, was amplified by PCR using primers P316 and P317. To create a Gateway entry clone (pRU917), the resulting PCR product was cloned into pDONR201 by using the Invitrogen Life Technologies BP cloning mixture (Int and IHF). The mctP promoter reporter plasmids (pRU922 and pRU923) were formed from pRU917 and either pOT3gfpuv or pOT3gfp+ by using the Invitrogen Life Technologies LR cloning mixture (Int, IHF, and Xis).
Gfp+ expression in bacterial cultures was quantified in microtiter plates with a GENios plate reader (Tecan), using a 485-nm-wavelength excitation filter and a 510-nm-wavelength emission filter. Cell optical density was measured at 595 nm. The data are expressed as relative fluorescence (fluorescence emission at 510 nm/optical density at 595 nm).
Transport assays.
R. leguminosarum uptake assays were performed by the rapid filtration method as previously described (35). Uptake assays were performed with a final concentration of 500 μM (0.5 μCi) l-[U-14C]alanine, 500 μM (0.5 μCi) [1-14C] acetate, 5 μM (0.125 μCi) l-[U-14C]lactate, or 5 μM (0.125 μCi) [1-14C] pyruvate. Competing solutes were added to a final concentration of 5 mM for inhibition of alanine or 0.5 mM for inhibition of lactate or pyruvate. The kinetic constants of solute uptake by R. leguminosarum strains were determined using various 14C-labeled-solute concentrations in standard uptake assays.
To determine the affect of sodium on solute uptake, transport assays were carried out in sodium-free RMS (10 mM potassium phosphate buffer [pH 7], 1 mM MgSO4). Sodium chloride or lithium chloride was added to a final concentration of 5 mM.
Plant nodulation phenotype.
Seeds of Pisum sativum cv. avola were surface sterilized and then added to cotton wool-plugged 250-ml conical flasks containing 230 ml of sterile vermiculite, which was wetted with nitrogen-free rooting solution (34). The seeds were inoculated with a bacterial culture, and the flask was wrapped with foil to exclude light from the roots. Plants were incubated at 25°C in a grow room with illumination provided by a Philips Sont-Agro grow light. When the shoot reached the cotton wool plug it was pulled through. Four weeks after inoculation, plants were harvested and acetylene reduction was carried out on whole plants as described previously (51). Sample nodules were removed, surface sterilized in calcium hypochlorite (0.7%), and crushed, and bacteria were streaked on TY. Isolated bacteria were then replica plated and screened for appropriate antibiotic and nutritional markers.
Nucleotide sequence accession number.
The sequence of the region of the R. leguminosarum genome containing the mct operon was submitted to the EMBL database under accession no. AJ421944.
RESULTS
Identification of a novel alanine transporter in R. leguminosarum.
To identify the pathways of alanine catabolism, an R. leguminosarum Tn5 library (approximately 15,000 mutants) was screened for growth on 20 mM alanine as a sole carbon and nitrogen source. The characterization of one alanine growth mutant with an insertion in dadR, which encodes a regulator of alanine catabolic genes (dadX and dadA), has been reported previously (1). Another mutant of interest was RU1180, as it formed only small colonies on alanine minimal medium, indicating a decreased growth rate. This mutant also displayed slow growth on pyruvate-ammonia minimal medium, but grew as wild type on glucose-ammonia and succinate-ammonia. To confirm that the alanine growth defect of this mutant is tightly linked to the transposon insertion, the transposon was transduced into wild-type 3841 and eight kanamycin-resistant transductants were tested for growth on alanine or glucose as the sole carbon source. These transductants displayed the same slow growth on alanine observed in the original mutant.
Mutant RU1180 was characterized by cloning the genome fragment containing the transposon and sequencing the region flanking the transposon insertion. BLAST (3) searches identified similarity to sodium/solute symporters, including the sodium/proline symporter (ecPutP) and sodium/pantothenate symporter (ecPanF) from E. coli (22, 31, 43). Mutant RU1180 was complemented for growth on alanine by cosmid pRU3147, and direct sequencing of this cosmid confirmed it contained the region of DNA corresponding to the transposon insertion site of RU1180. Further sequence of this region was obtained (Fig. 1) and submitted to the EMBL database under accession number AJ421944. The sequence confirmed that the gene inactivated in RU1180 encodes a putative permease with significant identity to members of the sodium/solute symporter family (T.C. number 2.A.21 [44]).
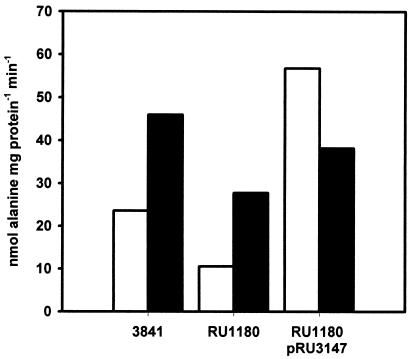
To confirm the role of this permease in alanine uptake, the transport properties of the wild type, RU1180 and RU1180 pRU3147 were compared (Fig. 2). Alanine uptake rates were lower in RU1180 than in wild-type 3841 and enhanced by the cosmid pRU3147. Therefore, this permease is an alanine transporter required for the utilization of alanine as a sole carbon and nitrogen source. This permease was named MctP (for monocarboxylate transport permease protein) as it also transports other monocarboxylates, such as lactate and pyruvate (see below).
FIG. 2.
Uptake of alanine by strains of R. leguminosarum. Uptake of 500 μM (0.125 μCi) l-[U-14C] alanine was assayed by the rapid filtration method for different R. leguminosarum strains grown in minimal medium (AMS) with added glucose (10 mM) plus ammonium chloride (10 mM) (white bars) or alanine (20 mM) (black bars). Data shown are the means of three independent experiments.
Topology prediction software (TopPred 2, HMMTOP, and TMHMM [29, 52, 54]) indicates MctP is a membrane protein with 13 transmembrane helices, with the N terminus located in the periplasm. This prediction supports the relationship of MctP and the sodium/solute symporters as it is in accordance with the experimentally determined topology of the sodium/proline transporter of E. coli (25).
Complementation of mctP mutants.
To confirm that the phenotype observed in the mctP mutant (RU1180) can be complemented in trans by mctP, a number of plasmids were constructed and conjugated into RU1180, and growth on minimal media containing alanine or pyruvate and ammonia was determined. A PCR product containing the complete mctP gene and part of the upstream noncoding region was cloned into a broad-host-range vector in both orientations (pRU739 and pRU747). Plasmid pRU739, which contained the PCR product in the same orientation as the lac promoter (plac), did restore growth on minimal media containing alanine or pyruvate-ammonia (Table 2) to the rates observed in the wild-type strain. Therefore, the decreased apparent growth rates observed in RU1180 are caused by mutation of mctP. However, pRU747, which contains the same insert in the opposite orientation, failed to complement the mctP mutation (Table 2). Thus, the region of DNA cloned in pRU747 cannot include an active promoter. Therefore, it is probable that the portion of the mctR-mctP intergenic region that is not located on the cloned PCR product is required for promoter activity. A convenient SalI restriction site is present in the mctS gene. This was utilized to clone a SalI fragment containing mctR, mctP, and the intergenic region between these genes. Growth on alanine or pyruvate and ammonia was enhanced in RU1180 by the presence of plasmids containing this SalI fragment regardless of orientation (pRU785 and pRU788; Table 2). Therefore, this fragment contains an active mctP promoter. As the mctS and mctR genes overlap (Fig. 1) and are probably cotranscribed, it is unlikely that there is a promoter immediately upstream of mctR on the cloned SalI fragment. The presence of an mctP promoter in the mctR-mctP intergenic region was confirmed by the complementation of the alanine and pyruvate-ammonia growth phenotype of RU1180 by a PCR product containing this intergenic region, mctP, and only a small portion of mctR, cloned in both orientations in pRK415 (pRU947 and pRU948) (Table 2).
TABLE 2.
Growth of mutant strains containing various plasmids on minimal medium (AMA) containing either alanine or pyruvate and ammonia as a sole C and N source
| Plasmid | Growth of strainb
|
|||
|---|---|---|---|---|
| RU1180 (mctP::Tn5) | RU1581 (mctP::GJ) | RU1580 (mctS::GJ) | RU1582 (mctR::GJ) | |
| pRK415 | ± | ± | + | + |
| pRU785 (mctRP) | +++ | ND | ND | ND |
| pRU788 (mctRP)a | +++ | ND | ND | ND |
| pRU739 (mctP)a | +++ | ND | ND | ND |
| pRU747 (mctP) | + | ND | ND | ND |
| pRU947 (mctP) | +++ | +++ | ND | ND |
| pRU948 (mctP)a | ++ | +++ | ND | ND |
| pRU950 (mctS)a | ND | ND | ± | ND |
| pRU951 (mctR)a | ND | ND | ND | +++ |
| pRU953 (mctR) | ND | ND | ND | + |
| pRU952 (mctSR)a | ND | ND | +++ | +++ |
| pRU974 (mctSR) | ND | ND | +++ | +++ |
The mct gene is under the control of the lac promoter, which is constitutive in R. leguminosarum. Thus, no IPTG was required to induce the expression of the cloned genes.
There was no discernible difference between growth of strains on minimal medium with alanine (20 mM) or pyruvate (20 mM) and ammonium chloride (10 mM). Symbols: ±, negligible growth; +, positive for growth; +++, strongly positive for growth. ND, not determined.
Distribution of MctP orthologues.
BLAST (3) searches of the sequence databases (including the unfinished microbial genome sequences) indicates that putative MctP orthologues are present in a broad range of microorganisms, including gram-negative β-proteobacteria (Burkholderia cepacia), gram-positive eubacteria (Bacillus subtilis, Bacillus cereus, and Streptomyces coelicolor A3) and archaea (Ferroplasma acidarmanus, Sulfolobus solfataricus, Sulfolobus tokodaii, Thermoplasma volcanium, and Thermoplasma acidophilum). However, the completed genome sequences of the closely related α-proteobacteria (i.e., S. meliloti, M. loti, A. tumefaciens, and Brucella melitensis) did not reveal any genes with significant similarity to R. leguminosarum mctP.
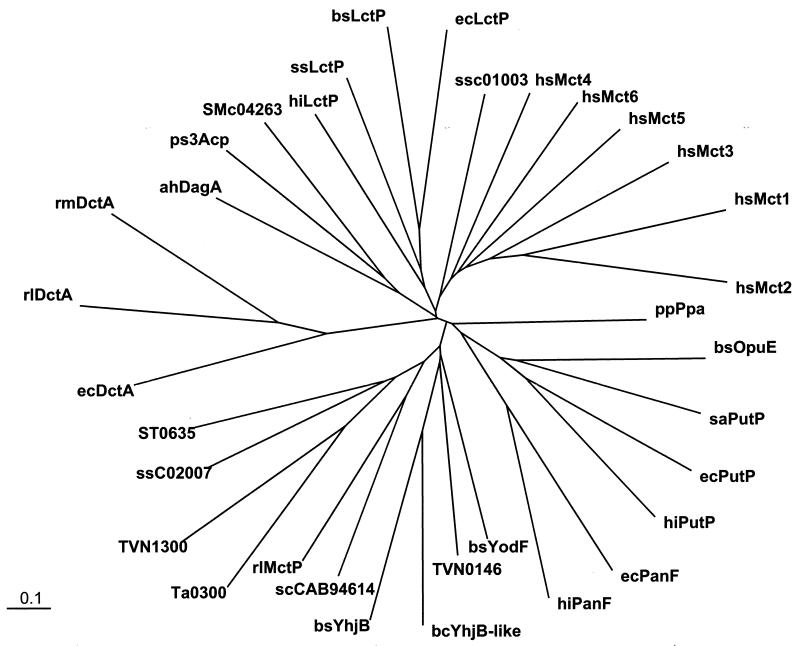
Amino acid sequence alignments indicate that the orthologues of MctP form a distinct group from the sodium/proline, sodium/pantothenate, and sodium/phenylacetate symporters (Fig. 3). This new subfamily of permeases is also distinct from other transporters of related MctP solutes, such as the monocarboxylate porter family (e.g., hsMCT1 to -6), the alanine/glycine/cation symporter family (e.g., ahDagA), the lactate permease family (e.g., ecLctP), and the dicarboxylate/amino acid/cation symporter family (e.g., rlDctA) (Fig. 3).
FIG. 3.
Phylogenetic tree indicating the relationship between rlMctP and related permeases. A phylogenetic tree was constructed from the amino acid sequences of rlMctP and a number of related transporters using Vector NTI Suite (version 6), which uses the ClustalW algorithm, and Treeview (version 1.6.1). Protein designations and accession numbers are as follows: scCAB94614, S. coelicolor A3 putative permease (CAB94614); Ta0300, T. acidophilum conserved hypothetical protein (CAC11445); ST0635, S. tokodaii conserved hypothetical protein (BAB65633); ssC02007, S. solfataricus conserved hypothetical protein c02007 (CAA69485); bsYodF, B. subtilis YodF (C69903); TVN0146, T. volcanium proline permease (BAB59289); TVN1300, T. volcanium Na+/pantothenate symporter; bsYhjB, B. subtilis metabolite permease (B69833); bcYhjB-like, B. cereus YhjB-like protein (AF387344); ecPanF, E. coli Na+/pantothenate symporter (P16256); hiPanF, Haemophilus influenzae Rd Na+/pantothenate symporter (P44963); ecPutP, E. coli Na+/proline symporter (P07117); hiPutP, H. influenzae Rd Na+/proline symporter (P45174); bsOpuE, B. subtilis osmoregulated Na+/proline symporter (O06493); ppPpa, P. aeruginosa phenylacetic acid transporter (CAA94864); hsMCT1 to -6, human monocarboxylic acid transporters 1 to 6 (P53985, AF049608, O15427, O15374, O15375, and O15403); ssc01003, S. solfataricus orf c01003 (CAA69453); rlDctA, R. leguminosarum C4-dicarboxylate transporter (Q01857); rmDctA, S. meliloti C4-dicarboxylate transporter (P20672); ecDctA, E. coli C4-dicarboxylate transporter (P37312); ps3Acp, thermophilic bacterium PS3 alanine carrier protein (D12512); ahDagA, Alteromonas haloplanktis d-alanine/glycine permease (M59081); ssLctP, S. solfataricus l-lactate permease (CAA69452); bsLctP, B. subtilis l-lactate permease (BAA21776); ecLctP, E. coli l-lactate permease (AAC76627); hiLctP, H. influenzae Rd l-lactate permease (AAC22871).
MctSR is a two-component system with putative orthologues in α-proteobacteria.
Immediately upstream of mctP are two genes (mctS and mctR [for monocarboxylate transporter sensor and regulator, respectively) with homology to the sensor and response-regulator components of bacterial two-component systems (17). MctS has similarity to histidine protein kinases, including the conserved histidine (His266), asparagine (Asn326), and aspartate (Asp353) residues, which are characteristic of this family of sensor proteins (49). MctS also has one putative transmembrane helix (amino acid 160 to 182), with the N terminus probably located in the periplasm and, constitutes a hypothetical sensory domain. MctR has similarity to bacterial regulators and contains a helix-turn-helix signature of the LuxR family (Prosite accession number PS00622) from amino acid 164 to 191. These helix-turn-helix signatures are involved in DNA binding (12, 15). MctR also possesses the conserved amino acids (Asp13, Asp59, and Lys109) associated with regulators of the two-component systems, with Asp59 being the probable site of phosphorylation (49).
BLAST searches of the completed microbial genome sequences indicated that A. tumefaciens and B. melitensis contain putative MctSR orthologues (accession numbers AAL52763, AAL52764, AAL45956, and AAL45955). These putative orthologues are more than 60% identical to R. leguminosarum MctS and MctR. This is the same level of identity as the A. tumefaciens orthologue has to the B. melitensis orthologue. However, in A. tumefaciens and B. melitensis, no mctP orthologue could be identified. Rather, in both these species, the two-component response regulators are adjacent to transporters of the tripartite ATP-independent periplasmic transporter family (T.C. number 2.A.56) (28, 44). The relevance of the MctSR orthologues in these organisms and any possible role for the tripartite ATP-independent periplasmic transporters in the uptake of monocarboxylates requires further investigation.
MctS differs from other sensors of two-component systems, including the putative A. tumefaciens and B. melitensis orthologues, as it has only one putative transmembrane domain. Other sensors have an additional membrane-spanning domain at the extreme N terminus (48). A closer comparison of the R. leguminosarum mctS sequence with that of the putative A. tumefaciens and B. melitensis orthologues reveals significant amino acid similarity upstream of the putative ATG start codon of mctS, but no alternative start codon is present between this ATG and the first upstream stop codon. This possible truncation of mctS in R. leguminosarum could be the result of a single base (adenine) insertion, 136 nucleotides upstream of the ATG start codon. Repeated sequencing of different clones indicates this sequence is authentic.
Inhibition of alanine uptake by competing solutes.
As R. leguminosarum3841 has two high-affinity transporters of alanine (i.e., Aap and Bra) (19, 20, 56), characterization of alanine uptake by the MctP was performed using a previously constructed aap bra double mutant strain (RU1357). To determine the solute specificity of MctP, a number of solutes were tested for inhibition of alanine uptake (Fig. 4). The known solutes of sodium/solute symporters (i.e., proline and pantothenate) did not inhibit alanine uptake. Therefore, the solute specificity of MctP is distinct from these previously characterized symporters. Uptake of [14C]alanine was only partially inhibited (between 48 and 62% inhibition) by a 10-fold excess of alanine (l- and d-isomers), cysteine, or histidine, while other amino acids tested did not significantly inhibit alanine uptake. In contrast, monocarboxylates (acetate, pyruvate, lactate, propionate, butyrate, and α-hydroxybutyrate) were strong inhibitors of alanine uptake (between 77 and 97% inhibition), while dicarboxylates (malonate and succinate) and some C4-monocarboxylates (β-hydroxybutyrate and γ-hydroxybutyrate) did not inhibit alanine uptake. Therefore, inhibition studies indicate that MctP is a monocarboxylic acid transporter.
FIG. 4.
Inhibition of alanine uptake by other solutes. Uptake of 500 μM (0.125 μCi) l-[U-14C]alanine by RU1357, grown on minimal medium (AMS) with alanine (20 mM), was assayed by the rapid filtration method. Competing solutes were added to a final concentration of 5 mM. Data shown are the means of at least three independent experiments. Abbreviations: AIB, 2-aminoisobutyric acid; HB, hydroxybutyrate.
Uptake of lactate, pyruvate, and acetate by MctP.
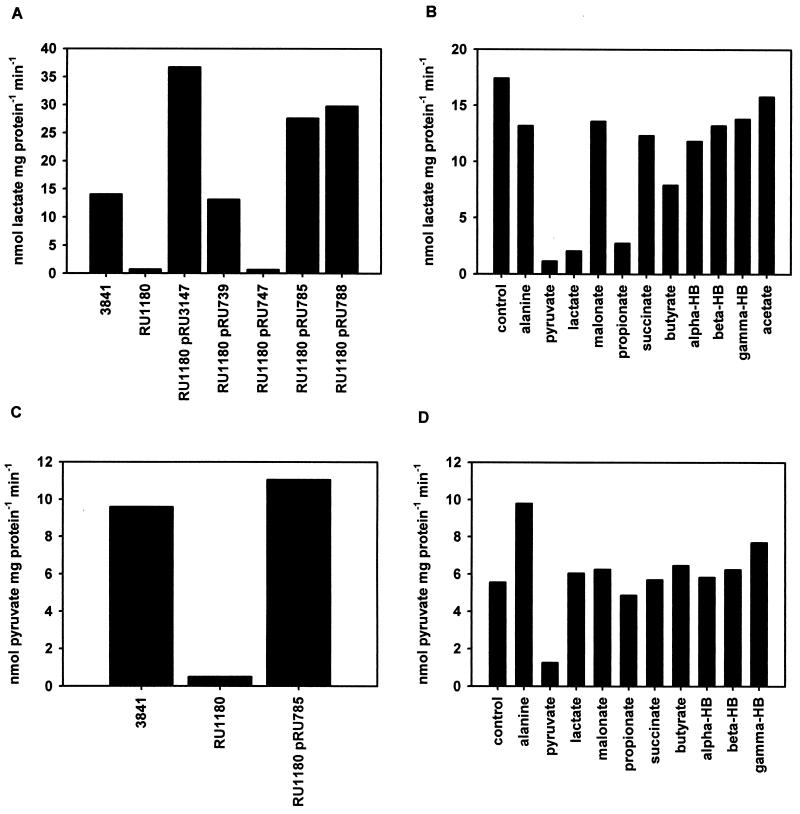
Although competitive inhibition of solute uptake can indicate which solutes may be transported by a permease, not all competitive inhibitors are transported. Therefore, to confirm that MctP is indeed a transporter of monocarboxylates, uptake of lactate, pyruvate, and acetate was measured in wild-type and mctP mutant strains. Mutation of mctP (RU1180) reduced the rate of uptake of lactate and pyruvate by 95% relative to wild-type 3841 (Fig. 5A and C). Furthermore, plasmids that complemented the growth phenotype of RU1180, also restored lactate and pyruvate uptake rates in this mutant to at least the rates observed in wild type 3841 (Fig. 5A and C). Therefore, MctP transports lactate and pyruvate, and is the dominant transporter of these solutes in free-living R. leguminosarum.
FIG. 5.
Uptake of lactate and pyruvate by strains of R. leguminosarum. Uptake of 5 μM (0.125 μCi) l-[U-14C]lactate (A) and [1-14C]pyruvate (C) was assayed by the rapid filtration method for different R. leguminosarum strains grown in minimal medium (AMS) with added glucose (10 mM) plus ammonium chloride (10 mM). The apparent specificity of lactate (B) and pyruvate (D) uptake by strain 3841 was determined by the addition of competing solutes to a final concentration of 0.5 mM. Data shown are the means of at least three independent experiments. HB, hydroxybutyrate.
In contrast, mutation of mctP did not affect the rate of acetate uptake by free-living R. leguminosarum. Both 3841 (wild type) and RU1180 (mctP) transport acetate with Km values of 20.1 ± 10.4 and 16.5 ±3.6 μM and Vmax values of 41.7 ± 4.3 and 37.8 ± 1.6 nmol of acetate mg of protein−1 min−1, respectively (unless otherwise noted, results are presented as means ± standard errors of the means). Therefore, MctP is not the dominant transporter of acetate in free-living R. leguminosarum. However, it is possible that MctP can transport acetate, but the unidentified acetate permease prevents this being detected in whole-cell assays.
The apparent affinity of MctP for lactate and pyruvate is higher than that for alanine, as alanine did not inhibit uptake of either of these solutes (Fig. 5B and D). Indeed, other than pyruvate, no solute tested inhibited the uptake of [14C]pyruvate (Fig. 5D). Therefore, of the solutes tested, MctP has the greatest apparent affinity for pyruvate. Further competition assays indicated that uptake of lactate is inhibited by pyruvate and propionate (95 and 78% inhibition, respectively) but only partially by butyrate (38% inhibition). Other C4-monocarboxylates (including α-hydroxybutyrate), C2-monocarboxylates (acetate), and dicarboxylates (C3 and C4) did not inhibit lactate uptake (Fig. 5B). Therefore, MctP has a preference for C3-monocarboxylates, with the following apparent relative affinity order (greatest to least): pyruvate, lactate, propionate, butyrate, α-hydroxybutyrate, alanine (acetate).
Kinetics of solute uptake by MctP.
As R. leguminosarum 3841 has two high-affinity transporters of alanine (Aap and Bra), the kinetics of alanine uptake by MctP were determined using the aap bra mutant RU1357. Preliminary experiments using cells grown in minimal media containing glucose and ammonia gave low alanine uptake rates at the concentrations tested. Therefore, RU1357 was grown on alanine minimal medium, as this increased the rate of transport (data not shown). Although the rate of alanine uptake was also increased in strain RU1180 by growth on alanine (Fig. 2), this can be attributed to up-regulation of aap or bra expression on the basis of specificity and Km values (data not shown). The kinetic constants obtained indicate that MctP has a low affinity for alanine (Km = 0.56 ± 0.16 mM), but the Vmax for alanine uptake is high (122 ± 15 nmol of alanine mg of protein−1 min−1).
At high lactate concentrations, an unidentified lactate permease, which was induced by growth on alanine, became apparent (data not shown). To overcome this, apparent kinetic constants were determined by subtracting uptake rates obtained from RU1180 from those for wild-type 3841, with each strain grown on glucose and ammonia minimal media. MctP has an apparent Km for lactate uptake of 4.4 ± 0.7 μM (Vmax = 22.4 ± 1.01 nmol of lactate mg of protein−1 min−1), while the lactate permease retained in the mctP mutant, RU1180, has a Km of 38.1 ± 7.3 μM lactate (Vmax = 8.5 ± 0.7 nmol of lactate mg of protein−1 min−1).
As the characterization of pyruvate uptake had indicated that the uptake of pyruvate is entirely dependent on MctP when R. leguminosarum 3841 is grown on glucose and ammonia, this strain and growth conditions were used for kinetic analysis of pyruvate uptake. The Km for pyruvate uptake by MctP is 3.8 ± 1.1 μM, and the Vmax is 9.8 ± 1.0 nmol of pyruvate mg of protein−1 min−1.
The Km values obtained for lactate and pyruvate are more than 100-fold lower than that for alanine. Therefore, MctP is a high-affinity transporter of lactate and pyruvate but a low-affinity transporter of alanine. Although the Vmax values obtained for lactate and pyruvate are significantly lower than that for alanine, this reflects the different growth conditions used (i.e., glucose and ammonia rather than alanine).
Sodium concentration does not affect solute uptake by MctP.
As MctP has significant similarity to sodium/solute symporters, the affect of sodium on transport by MctP was investigated. Lactate and succinate were selected as representative solutes transported by MctP and DctA (a proton-coupled dicarboxylate transporter [6, 23, 50]), respectively, and the uptake of these solutes by strain 3841 was determined in sodium-free transport buffer (RMS) with and without the addition of sodium or lithium. The addition of sodium or lithium had no affect on the uptake of either lactate or succinate (Fig. 6).
FIG. 6.
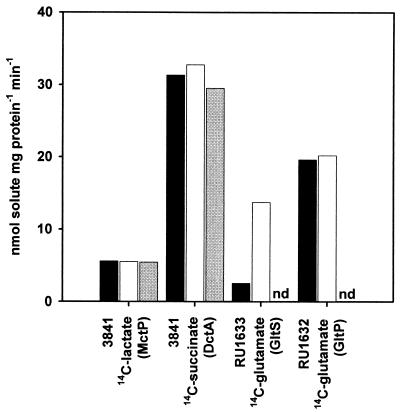
Affect of sodium concentration on solute uptake. Uptake of 5 μM (0.125 μCi) l-[U-14C]lactate or 25 μM (0.125 μCi) [2,3-14C]succinate or 25 μM (0.125 μCi) l-[U-14C]glutamic acid was assayed by the rapid filtration method for R. leguminosarum strains 3841, RU1632 (RU1357 pRU976),or RU1633 (RU1357 pRU986) grown in minimal medium (AMS) with added succinate (10 mM) plus ammonium chloride (10 mM). Uptake assays were performed in sodium-free RMS with no additions (black columns), with added NaCl (5 mM) (white columns), or with added LiCl (5 mM) (grey columns). Data shown are the means of at least three independent experiments. nd, not determined.
However, there is, to our knowledge, no characterized sodium-dependent transporter in Rhizobium spp. which can be used as a control. Therefore, two glutamate transporters of E. coli, GltS (a sodium-coupled permease [9]) and GltP (a proton-coupled permease [9, 55]), were used as controls to confirm that R. leguminosarum is able to use sodium ions as a source of energy for transport. Also, it is notoriously difficult to remove sodium from assay buffers. Therefore, GltS was also a control of the experimental conditions. GltS and GltP were expressed in R. leguminosarum RU1357 (aap bra) under the control of the aapJ promoter, and glutamate uptake was determined with or without the addition of sodium. Uptake of glutamate by GltS was increased fivefold by the addition of sodium, whereas, the rate of glutamate uptake by GltP was not altered by sodium concentration (Fig. 6). Hence, the maximum rate of glutamate uptake by GltS is dependent on the addition of sodium, so the level of any contaminating sodium in the sodium-free assay buffer is not sufficient to mask the sodium dependence of this permease.
Therefore, although known sodium-coupled transporters can function in R. leguminosarum, uptake by MctP is not apparently influenced by sodium concentration. However, a caveat is that if MctP were to use sodium with much higher affinity than GltS, our experiments may not have detected sodium dependence. The transport of solutes by MctP does require metabolic energy, as lactate uptake was inhibited 97% by 5 μM carbonylcyanide m-chlorophenylhydrazone (CCCP). While this is consistent with proton dependence, sodium-dependent transporters (for example, GltS) are also inhibited by CCCP (9). At present we conclude there is no evidence for sodium as opposed to proton coupling, but absolute proof is difficult to acquire in studies with whole cells.
Symbiotic phenotype of mctP mutants.
Strain RU1180 formed normal red nodules on peas, and the plants were healthy and green. Plants inoculated with the wild type versus strain RU1180 strain reduced acetylene at rates of 2 ± 0.6 μmol h−1 plant−1 and 1.5 ± 0.1 μmol h−1 plant−1 (means ± standard errors of the means), respectively, all of which indicates a Fix+ phenotype for the mutant.
The MctSR two-component system regulates mctP expression.
The proximity of the mctSR genes to mctP suggests that the two-component response system may regulate expression of mctP. To investigate this, a 4.4-kb DNA fragment containing mctSRP was mutated in vitro by GeneJumper insertion. Eight plasmids with transposon insertions in mctS (three plasmids), mctR (four plasmids), or mctP (one plasmid) were identified by restriction mapping, and the location of the transposon insertion was confirmed by DNA sequencing. The mutated plasmids were then used to obtain chromosomal insertion mutants by homologous recombination. The position of the GeneJumper transposon insertion in each of the resulting strains is shown on Fig. 1. The mctS::GeneJumper, mctR::GeneJumper, and mctP::GeneJumper mutants each displayed the same low growth rate on alanine or pyruvate and ammonia minimal media as observed for the original mctP transposon mutant, RU1180. Therefore, in addition to mctP, mctS and mctR are required for optimal growth on alanine or pyruvate and ammonia.
As with RU1180, the mctP::GeneJumper mutant (RU1581) can be complemented for growth by plasmids containing mctP and the mctR-mctP intergenic region (Table 2). The mctS::GeneJumper and mctR::GeneJumper mutants (RU1580 and RU1582, respectively) can be complemented by plasmids which contain mctSR (pRU952 and pRU974) (Table 2). However, RU1580 cannot be complemented by mctS alone, and complementation of mctR mutants requires the gene to be expressed with plac (Table 2). This can be explained by mctS and mctR being transcriptionally linked, with insertion mutations of mctS having a polar affect on mctR expression and no promoter being present immediately upstream of mctR.
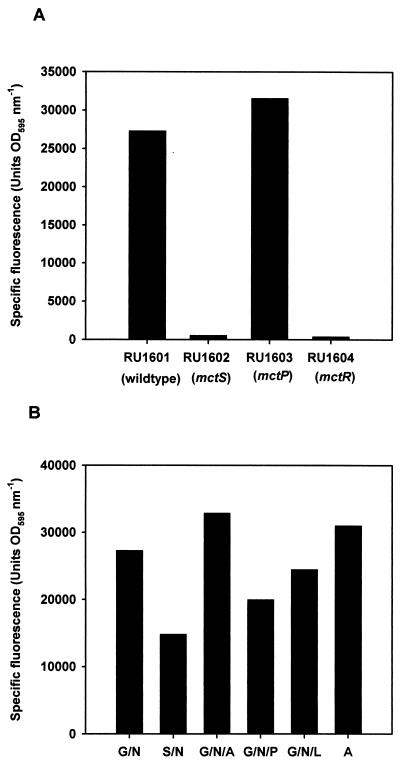
The mctR-mctP intergenic region, which contains the putative mctP promoter (pmctP), was amplified by PCR and cloned by Gateway technology into the transcription reporter vector pOT3gfp+ (pRU923). Promoter activity (pmctP) could not be detected in mctS or mctR mutant strains but was detected in wild-type and mctP mutant strains (Fig. 7A). Therefore, the MctSR two-component sensor-regulator is an essential activator of mctP expression and mctP is not involved in self-regulation.
FIG. 7.
Regulation of the mctP promoter. Specific fluorescence (fluorescence [excitation at 485 nm; emission at 510 nm] optical density at 595 nm−1) was used as a measure of expression of the mctP promoter-green fluorescent protein fusion in different strains grown in AMS minimal medium with 10 mM glucose and 10 mM ammonium chloride (A) or in RU1601 (3841 pRU923) grown on different carbon and nitrogen sources (B). Data shown are the means of at least three independent experiments. Abbreviations: G, 10 mM glucose; N, 10 mM ammonium chloride; S, 10 mM succinate; A, 20 mM alanine; P, 20 mM pyruvate; L, 20 mM lactate.
To determine the affect of solutes on mctP expression, wild-type 3841 containing the pmctP reporter plasmid was grown under different growth conditions. Approximately twofold-lower pmctP activity was recorded when succinate, rather than glucose, was present as a carbon source. However, there was no increase in mctP expression in response to alanine, pyruvate, or lactate in the growth media (Fig. 7B). Therefore, although MctSR is an activator of mctP, no environmental signal for this regulator has yet been identified. Indeed, mctP is expressed at high levels in both nutrient-rich medium (TY) and minimal medium (AMA) containing only glucose and ammonia as carbon and nitrogen sources, indicating that mctP expression is constitutive. The mctP reporter vector, pRU922, was conjugated into two other wild-type strains of R. leguminosarum (A34 and VF39), which are distinct from 3841. As in 3841, the fluorescence derived from the mctP-gfp fusion was high on TY and minimal media with glucose and ammonia, indicating the constitutive expression of mctP is not strain dependent.
DISCUSSION
The data presented here indicate that the MctP of R. leguminosarum is a novel monocarboxylate transporter. This permease has a low affinity for alanine (Km = 560 μM) but a higher affinity for lactate and pyruvate (Km = 4.4 μM and 3.8 μM, respectively). Uptake inhibition studies indicate that acetate, propionate, butyrate, and α-hydroxybutyrate are also solutes transported by MctP, with the apparent affinity indicating a preference for C3-monocarboxylates. Dicarboxylates (malonate and succinate) did not inhibit uptake by MctP. Therefore, the specificity of MctP is distinct from those of previously characterized bacterial carboxylate transporters. Typically, bacterial carboxylate transporters transport a narrow range of related compounds. For example, the characterized members of lactate permease family (i.e., two E. coli transporters, GlcA [YghK] and LctP [LldP]) are specific for 2-hydroxymonocarboxylates (l-lactate, d-lactate and glycolate) (32). However, the data suggest MctP transports a broad range of monocarboxylates, including 2-hydroxymonocarboxylates (lactate and α-hydroxybutyrate), 2-aminomonocarboxylates (alanine), and 2-ketomonocarboxylates (pyruvate), in addition to monocarboxylates with no side group (propionate and butyrate).
Amino acid sequence comparisons also indicate MctP is distinct from other transporters of alanine or monocarboxylates, such as members of the d-alanine/glycine/cation symporter family (T.C. number 2.A.25) (42, 44), the LctP family (T.C. number 2.A.14) (32, 44), and the proton-linked monocarboxylate porter family (T.C. number 2.A.1.13) (14, 44) (Fig. 3). However, MctP does have significant similarity to the sodium/solute symporter family (42). In accordance with this, in silico predictions indicate MctP is a protein with 13 transmembrane helices, the same number demonstrated experimentally for E. coli PutP (sodium/proline symporter), the best-characterized member of this family (25). Other bacterial members of this family include the sodium/pantothenate symporter from E. coli, the sodium/phenylacetate symporter from Pseudomonas putida, and the sodium/glucose symporter from Vibrio parahaemolyticus (22, 45, 46). Therefore, the solutes transported by permeases of this family are diverse.
Although similar to members of the sodium/solute symporter family, the putative MctP orthologues form a distinct group from the previously characterized sodium/solute symporters (Fig. 3). Also, we were unable to demonstrate any dependence on sodium for transport of solutes by MctP. However, as it is difficult to ensure that transport buffers are truly sodium free, it is possible that small levels of sodium present in the assay buffers were sufficient to allow transport to proceed if MctP has a high affinity for sodium. Uptake was inhibited by CCCP (5 to 10 μM), indicating that transport by MctP requires metabolic energy and may be proton linked, as it is with the monocarboxylate porter family (14). Although the previously characterized members of the sodium/solute symporter family use sodium under physiological conditions, it has been demonstrated that the human Na+/glucose transporter can utilize H+, although the affinity for glucose was 10-fold lower than when Na+ was used (40). Therefore, it is not surprising that MctP may act as a proton symporter, although this requires confirmation.
Putative MctP orthologues have been identified in a wide taxonomic range of prokaryotes (Fig. 3), and two species (B. subtilis and T. volcanium) have two putative MctP-like paralogues. It is noteworthy that a gene encoding a putative MctP orthologue in B. cereus (YhjB-like protein) is adjacent to the alanine spore germination operon, gerL (4). It is possible that this permease is required for the uptake of alanine during outgrowth from endospores and therefore has a similar specificity to MctP.
Two-component systems are a well-established mechanism for signal transduction in bacteria (17). The sensor component senses an environmental signal and, through a phosphorylation reaction, transmits this signal to the response regulator. Many of the response regulators of this family act by binding to a promoter and activate or repress gene expression (17). Specific two-component sensor regulators are known to activate expression of dicarboxylate and tricarboxylate transporter genes (23). For example, the DctBD two-component system activates expression of dctA in R. leguminosarum (41), and CitST activates citM expression in B. subtilis (60). The data presented indicate the MctSR two-component system of R. leguminosarum is required for activation of the monocarboxylate transporter gene mctP.
Typically, solutes transported by a permease act as inducers for the two-component regulators of the permease. However, we were unable to identify any solute that induced expression of mctP; indeed, it is constitutive. The putative orthologues of MctS identified in A. tumefaciens and B. melitensis have two transmembrane domains toward the N terminus, whereas R. leguminosarum MctS has only one. This appears to have resulted from a single nucleotide insertion upstream of the apparent start codon, leading to a truncation of the protein and loss of the N terminus transmembrane domain. The consistency of repeated sequencing reactions indicates the authenticity of this apparent nucleotide insertion. It is possible that the loss of the N-terminal transmembrane region has resulted in the sensor being inactivated in such a way as to constitutively activate the regulator, which in turn activates expression of MctP. The expression of the pmctP-gfp fusion (pRU922) was the same in R. leguminosarum strains A34 and VF39. This suggests that R. leguminosarum has adapted the regulation of mctP to allow constitutive expression and that the putative truncation observed in 3841 is not a specific mutation of this strain.
MctP is not required for R. leguminosarum to form symbiotic nodules and fix atmospheric nitrogen, which is consistent with the lack of putative orthologues in the genomes of sequenced rhizobia (i.e., S. meliloti and M. loti) (13, 26). However, MctP is required for optimal growth of free-living R. leguminosarum on alanine or pyruvate and ammonia. Furthermore, the characterization of MctP has resulted in the identification of a new subfamily of C3-monocarboxylate transporters in bacteria.
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council of the United Kingdom.
REFERENCES
- 1.Allaway, D., E. Lodwig, L. A. Crompton, M. Wood, T. R. Parsons, T. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36:508-515. [DOI] [PubMed] [Google Scholar]
- 2.Allaway, D., N. A. Schofield, M. E. Leonard, L. Gilardoni, T. M. Finan, and P. S. Poole. 2001. Use of differential fluorescence induction and optical trapping to isolate environmentally induced genes. Environ. Microbiol. 3:397-406. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlass, P. J., C. W. Houston, M. O. Clements, and A. Moir. 2002. Germination of Bacillus cereus spores in response to L-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology 148:2089-2095. [DOI] [PubMed] [Google Scholar]
- 5.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 6.Bhandari, B., and D. J. D. Nicholas. 1985. Proton motive force in washed cells of Rhizobium japonicum and bacteroids from Glycine max. J. Bacteriol. 164:1383-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan-Wollaston, V. 1979. Generalized transduction in Rhizobium leguminosarum. J. Gen. Microbiol. 112:135-142. [Google Scholar]
- 8.Day, D. A., P. S. Poole, S. D. Tyerman, and L. Rosendahl. 2001. Ammonia and amino acid transport across symbiotic membranes in nitrogen-fixing legume nodules. Cell. Mol. Life Sci. 58:61-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deguchi, Y., I. Yamato, and Y. Anraku. 1989. Molecular cloning of gltS and gltP, which encode glutamate carriers of Escherichia coli B. J. Bacteriol. 171:1314-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downie, J. A., Q. S. Ma, C. D. Knight, G. Hombrecher, and A. W. B. Johnston. 1983. Cloning of the symbiotic region of Rhizobium leguminosarum: the nodulation genes are between the nitrogenase genes and a nifA-like gene. EMBO J. 2:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freidman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of the broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lalaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 14.Halestrap, A. P., and N. T. Price. 1999. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem. J. 343:281-299. [PMC free article] [PubMed] [Google Scholar]
- 15.Henikoff, S., J. C. Wallace, and J. P. Brown. 1990. Finding protein similarities with nucleotide-sequence databases. Methods Enzymol. 183:111-132. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch, P. R., and J. E. Beringer. 1984. A physical map of pPH1JI and pJB4JI. Plasmid 12:139-141. [DOI] [PubMed] [Google Scholar]
- 17.Hoch, J. A., and T. J. Silhavy (ed.). 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 18.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene-splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 19.Hosie, A. H. F., D. Allaway, H. A. Dunsby, C. S. Galloway, and P. S. Poole. 2002. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 184:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosie, A. H. F., D. Allaway, M. A. Jones, D. L. Walshaw, A. W. B. Johnston, and P. S. Poole. 2001. Solute-binding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 40:1449-1459. [DOI] [PubMed] [Google Scholar]
- 21.Hosie, A. H. F., and P. S. Poole. 2001. Bacterial ABC transporters of amino acids. Res. Microbiol. 152:259-270. [DOI] [PubMed] [Google Scholar]
- 22.Jackowski, S., and J. H. Alix. 1990. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. J. Bacteriol. 172:3842-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 24.Johnston, A. W. B., and J. E. Beringer. 1975. Identification of the Rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 87:343-350. [DOI] [PubMed] [Google Scholar]
- 25.Jung, H., R. Rubenhagen, S. Tebbe, K. Leifker, N. Tholema, M. Quick, and R. Schmid. 1998. Topology of the Na+/proline transporter of Escherichia coli. J. Biol. Chem. 273:26400-26407. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 27.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 28.Kelly, D. J., and G. H. Thomas. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25:405-424. [DOI] [PubMed] [Google Scholar]
- 29.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., R. Parsons, D. A. Day, and F. J. Bergersen. 2002. Reassessment of major products of N2 fixation by bacteroids from soybean root nodules. Microbiology 148:1959-1966. [DOI] [PubMed] [Google Scholar]
- 31.Nakao, T., I. Yamato, and Y. Anraku. 1987. Nucleotide-sequence of putP, the proline carrier gene of Escherichia coli K12. Mol. Gen. Genet. 208:70-75. [DOI] [PubMed] [Google Scholar]
- 32.Nunez, M. F., M. T. Pellicer, J. Badia, J. Aguilar, and L. Baldoma. 2001. The gene yghK linked to the glc operon of Escherichia coli encodes a permease for glycolate that is structurally and functionally similar to L-lactate permease. Microbiology 147:1069-1077. [DOI] [PubMed] [Google Scholar]
- 33.Poole, P. S., and D. A. Allaway. 2000. Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 43:117-163. [DOI] [PubMed] [Google Scholar]
- 34.Poole, P. S., A. Blyth, C. J. Reid, and K. Walters. 1994. myo-inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv viciae. Microbiology 140:2787-2795. [Google Scholar]
- 35.Poole, P. S., M. Franklin, A. R. Glenn, and M. J. Dilworth. 1985. The transport of L-glutamate by Rhizobium leguminosarum involves a common amino acid carrier. J. Gen. Microbiol. 131:1441-1448. [Google Scholar]
- 36.Poole, P. S., N. A. Schofield, C. J. Reid, E. M. Drew, and D. L. Walshaw. 1994. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology 140:2797-2809. [DOI] [PubMed] [Google Scholar]
- 37.Prell, J., B. Boesten, P. S. Poole, and U. B. Priefer. 2002. The Rhizobium leguminosarum bv. viciae VF39 gamma-aminobutyrate (GABA) aminotransferase gene (gabT) is induced by GABA and highly expressed in bacteroids. Microbiology 148:615-623. [DOI] [PubMed] [Google Scholar]
- 38.Priefer, U. B. 1989. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar Viciae VF39. J. Bacteriol. 171:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 40.Quick, M., D. D. F. Loo, and E. M. Wright. 2001. Neutralization of a conserved amino acid residue in the human Na+/glucose transporter (hSGLT1) generates a glucose-gated H+ channel. J. Biol. Chem. 276:1728-1734. [DOI] [PubMed] [Google Scholar]
- 41.Reid, C. J., and P. S. Poole. 1998. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J. Bacteriol. 180:2660-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reizer, J., A. Reizer, and M. H. Saier. 1994. A functional superfamily of sodium/solute symporters. Biochim. Biophys. Acta-Rev. Biomembr. 1197:133-166. [DOI] [PubMed] [Google Scholar]
- 43.Reizer, J., A. Reizer, and M. H. Saier. 1990. The Na+ pantothenate symporter (panF) of Escherichia coli is homologous to the Na+ proline symporter (putP) of Escherichia coli and the Na+ glucose symporters of mammals. Res. Microbiol. 141:1069-1072. [DOI] [PubMed] [Google Scholar]
- 44.Saier, M. H. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarker, R. I., W. Ogawa, T. Shimamoto, and T. Tsuchiya. 1997. Primary structure and properties of the Na+/glucose symporter (SglS) of Vibrio parahaemolyticus. J. Bacteriol. 179:1805-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schleissner, C., E. R. Olivera, M. Fernandezvalverde, and J. M. Luengo. 1994. Aerobic catabolism of phenylacetic acid in Pseudomonas putida U: biochemical characterization of a specific phenylacetic acid transport system and formal demonstration that phenylacetyl coenzyme A is a catabolic intermediate. J. Bacteriol. 176:7667-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scholz, Q., A. Thiel, W. Hillen, and M. Niederweis. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur. J. Biochem. 267:1565-1570. [DOI] [PubMed] [Google Scholar]
- 48.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 49.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationship and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 50.Tremblay, P. A., and R. W. Miller. 1984. Cytoplasmic membrane of Rhizobium meliloti bacteroids. II. Functional-differentiation and generation of membrane-potentials. Can. J. Biochem. Cell Biol. 62:592-600. [Google Scholar]
- 51.Trinick, M. J., M. J. Dilworth, and M. Grounds. 1976. Factors affecting the reduction of acetylene by root nodules of Lupinus species. New Phytol. 77:359-370. [Google Scholar]
- 52.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 53.Vanrhijn, P., and J. Vanderleyden. 1995. The rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 55.Wallace, B., Y.-J. Yang, J. Hong, and D. Lum. 1990. Cloning and sequencing of a gene encoding a glutamate and aspartate carrier of Escherichia coli K-12. J. Bacteriol. 172:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walshaw, D. L., and P. S. Poole. 1996. The general L-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that influences efflux of solutes. Mol. Microbiol. 21:1239-1252. [DOI] [PubMed] [Google Scholar]
- 57.Walshaw, D. L., A. Wilkinson, M. Mundy, M. Smith, and P. S. Poole. 1997. Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology. 143:2209-2221. [DOI] [PubMed] [Google Scholar]
- 58.Waters, J. K., B. L. Hughes, L. C. Purcell, K. O. Gerhardt, T. P. Mawhinney, and D. W. Emerich. 2042. 1998. Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proc. Natl. Acad. Sci. USA 95:12038-12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, L. Woo, Y. C. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. N. Wu, P. Romero, D. Gordon, S. P. Zhang, H. Y. Yoo, Y. M. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto, H., M. Murata, and J. Sekiguchi. 2000. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 37:898-912. [DOI] [PubMed] [Google Scholar]