Abstract
The flagellar transcriptional regulator FleQ appears to be the highest-level regulator in the hierarchical regulatory cascade of flagellar biogenesis in Pseudomonas aeruginosa. Except for the posttranslational downregulation of FleQ activity by FleN, an antiactivator, not much is known about the regulation of the fleQ gene or its gene product. Some FleQ homologs in other bacterial species either are positively regulated by another regulator (e.g., CtrA, the master regulator regulating FlbD in Caulobacter crescentus) or are expressed from a σ70-dependent promoter (e.g., FlgR of Helicobacter pylori). In this study we demonstrated that Vfr, an Escherichia coli CRP homolog known to function as an activator for various genes, including lasR, regA, and toxA, in P. aeruginosa, is capable of repressing fleQ transcription by binding to its consensus sequence in the fleQ promoter. In a DNase I footprint assay, purified Vfr protected the sequence 5′-AATTGACTAATCGTTCACATTTG-3′. When this putative Vfr binding site in the fleQ promoter was mutated, Vfr was unable to bind the fleQ promoter fragment and did not repress fleQ transcription effectively. Primer extension analysis of the fleQ transcript revealed two transcriptional start sites, t1 and t2, that map within the Vfr binding site. A putative −10 region (TAAAAT) for the t2 transcript, with a five-of-six match with the E. coli σ70 binding consensus, overlaps with one end of the Vfr binding site. A 4-bp mutation and an 8-bp mutation in this −10 region markedly reduced the activity of the fleQ promoter. The same mutations led to the disappearance of the 203-nucleotide fleQ transcript in an in vitro transcription assay. Vfr probably represses fleQ transcription by binding to the Vfr binding site in the fleQ promoter and preventing the sigma factor from binding to the −10 region to initiate transcription.
Flagellar biogenesis in Pseudomonas aeruginosa is an intricate process in which various transcriptional activators (FleQ, FleR) and alternative sigma factors (RpoN/σ54, RpoF/σ28) participate in the transcriptional regulation of the flagellar genes and operons (3, 23, 31, 33). Of the two transcriptional activators identified in our laboratory, FleQ appears to be the highest-level positive regulator of flagellar biogenesis in P. aeruginosa.
FleQ is homologous to the NtrC group of response regulators, which are part of a two-component signal transduction system, that works in concert with the alternative sigma factor RpoN (3). Typically, a two-component system consists of a phosphorelay between a sensor kinase and a response regulator pair (5). The sensor kinase is activated by autophosphorylation when an environmental stimulus is sensed, and the response regulator is subsequently phosphorylated at the receiver domain. The response regulators are usually DNA binding proteins, and phosphorylation activates them to promote activation or repression of the downstream genes (5). However, FleQ and several of its homologs that regulate flagellar biogenesis in other bacterial species, including FlrA of Vibrio cholerae (16), FlaK of Vibrio parahaemolyticus (15), and FlgR of Helicobacter pylori (30), do not contain the hallmark residues in their receiver domains for phosphorylation and do not have a cognate sensor kinase encoded as a part of the same operon. Therefore, phosphorelay signaling does not appear to be the modulating mechanism that regulates the activities of these response regulators. Attempts to detect phosphorylation of FleQ in vitro were not successful in our laboratory, thus raising further doubts about the existence of a putative sensor kinase for FleQ (unpublished data). One of the mechanisms employed to accomplish posttranslational modulation of FleQ activity in P. aeruginosa involves direct protein-protein interactions with FleN, an antiactivator that represses FleQ-dependent transcriptional activation (8). Whether fleQ is subject to transcriptional activation or repression in addition to posttranslational modulation has been a question. Preliminary data have demonstrated that FleQ synthesis occurs from the early log phase to the stationary phase of growth in shaken cultures of P. aeruginosa PAK (unpublished data), which suggests that constitutive expression occurs under these conditions. Whether the constitutive expression observed was driven by a constitutively expressed activator or merely by the housekeeping RNA polymerase holoenzyme is not clear.
The flagellar regulon is subject to hierarchical regulation in most of the flagellated bacteria. Among the flagellar regulators of monoflagellate bacteria, flgR, the fleQ homolog of H. pylori, has been proposed to be transcribed from a σ70-like promoter (30). No information has been published concerning the transcriptional regulation of the fleQ homolgs flrA and flaK in V. cholerae and V. parahaemolyticus, respectively. However, flbD, the fleQ homolog of Caulobacter crescentus, along with the gene encoding its cognate sensor kinase, flbE, is positively regulated by CtrA, the master regulator which controls the initiation of DNA replication, DNA methylation, cell division, and flagellar biogenesis (10, 35). In the peritrichous organisms Escherichia coli and Salmonella enterica serovar Typhimurium, transcriptional activators encoded by flhC and flhD are at the top of the hierarchy and are essential for the expression of flagellar genes (6). The flhDC operon is controlled by a σ70 promoter and a number of global regulatory factors (17). There is evidence which suggests that the cyclic AMP receptor protein (CRP) activates and the histone-like nucleoid-structuring protein (H-NS) represses transcription of flhDC in E. coli (28). In S. enterica serovar Typhimurium it has been demonstrated that the flhDC operon carries six possible transcriptional start sites, one of which is CRP dependent and one of which is CRP repressible (36). Thus, flagellar biogenesis may be under both positive regulation and negative regulation in some bacteria.
Based on the known mechanisms of transcriptional regulation of FleQ homologs described above, fleQ could be envisaged to be regulated by one or more similar mechanisms. In this report, we provide evidence that fleQ and thus flagellar biogenesis in P. aeruginosa are σ70 dependent and are negatively regulated by Vfr. Vfr is a homolog of E. coli CRP that functions as an activator for many genes, including lasR, that have been implicated in virulence and quorum sensing (2, 32, 34). The physiological implications are discussed below.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this paper are listed in Table 1. All strains were routinely maintained by using Luria-Bertani (LB) (25) medium at 37°C. Antibiotics were used as necessary at the following concentrations: 100 μg of ampicillin per ml and 20 μg of tetracycline per ml for E. coli and 300 μg of carbenicillin per ml and 100 μg of tetracycline per ml for P. aeruginosa.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant information | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | hsdR recA lacZYA φ80 lacZΔM15 | GIBCO-BRL |
| XL1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lac1qZΔM15 Tn10 (Tetr)] | Stratagene |
| S17-1 | RP4-2 Tc::Mu Km::Tn7 Tpr Smr Pro Res− Mod+ | 27 |
| P. aeruginosa strains | ||
| PAK | Wild-type clinical isolate | D. Bradley |
| PAK fleQp-lacZ | PAK with fleQp-lacZ in the attB site | This study |
| PAK fleQpmutV-lacZ | PAK with fleQpmutV-lacZ in the attB site | This study |
| PAK lacZ | PAK with lacZ in the attB site | This study |
| PAO1 | Wild type | 14 |
| PAO fleQp-lacZ | PAO with fleQp-lacZ in the attB site | This study |
| PAO fleQpmutV-lacZ | PAO with fleQpmutV-lacZ in the attB site | This study |
| PAO lacZ | PAO with lacZ in the attB site | This study |
| PA103 | Prototroph, exotoxin A hyperproducer, elastase deficient | 18 |
| PAO9001 | Δvfr::Gentr derivative of strain PAO1 | 2 |
| PAO9001 fleQp-lacZ | PAO9001 with fleQp-lacZ in the attB site | This study |
| Plasmids | ||
| pMMB66HE/EH | Broad-host-range tacP expression vector, lac1q Apr Cbr | 11 |
| pWNP28 (ptac917Δp) | pMMB66HE carrying the 0.9-kb NarI fragment containing vfr region from positions −77 to +999, Apr Cbr | 2 |
| pDN19lacΩ | Promoterless lacZ oriV oriT Tetr StrrΩ fragment | 7 |
| placΩQ | pDN19lacΩ containing the fleQ promoter region as a 720-bp EcoRI-BamHI insert | 3 |
| placΩQmutS8 | placΩQ with 8 bp in the −10 region of the fleQ promoter mutated | This study |
| placΩQmutS4 | placΩQ with 4 bp in the −10 region of the fleQ promoter mutated | This study |
| pBluescript II/KS(+) | General-purpose cloning vector, Apr | Stratagene |
| pBS-fleQp | pBluescript II/KS(+) containing the fleQ promoter region as a 720-bp EcoRI-BamHI nsert | This study |
| pBS-fleQpmutV | pBS-fleQp with an ACA→CGC mutation in the Vfr binding site | This study |
| mini-CTXlacZ | Integration-proficient vector for single-copy chromosomal lacZ fusion | 4 |
| CTX fleQp-lacZ | Insert from pBS-fleQp cloned into mini-CTXlacZ | This study |
| CTX fleQpmutV-lacZ | Insert from pBS-fleQpmutV cloned into mini-CTXlacZ | This study |
| pFLP2 | Plasmid encoding the FLP recombinase, Cbr | 4 |
| Primersa | ||
| fleQ2F | 5′ ATCGGTCGACTTTTTTATTGCCTCT 3′ | |
| fleQ2R | 5′ CGATCTCGAGATCGAATGGGTCTCGCTC 3′ | |
| fleQ1F | 5′ CAGCAAAAAAAACACGCC 3′ | |
| fleQ1R | 5′ GCCTATTTCTAATAAGCAGCC 3′ | |
| Qpmut1 | 5′ CCGCCATAAATTGACTAATCGTTCCGCTTTGACTTAACTAGTGGCC 3′ | |
| Qpmutcom2 | 5′ GGCCACTAGTTAAGTCAAAGCGGAACGATTAGTCAATTTTATGGCGTG 3′ | |
| Qp70mut1 | 5′ GAAACAGCAAAAAAAACACGCCGCGCGCGCGACTAATCGTTCACATTTGACTTA 3′ | |
| Qp70mutcom2 | 5′ TAAGTCAAATGTGAACGATTAGTCGCGCGCGCGGCGTGTTTTTTTTGCTGTTTC 3′ | |
| Qp70mut3 | 5′ CAGCAAAAAAAACACGCCATAGCGCTGACTAATCGTTCACATTTGAC 3′ | |
| Qp70mutcom4 | 5′ GTCGTTTTTTTTGTGCGGTATCGCGACTGATTAGCAAGTGTAAACTG 3′ | |
| RT2 | 5′ CAGCTGCCTTGCATCGAATGGGTCTCGC 3′ |
Bases mutated in the primers are underlined.
Homology searches.
The Vfr binding sequence (AAATGTGATCTAGATCACATTT) from the lasR promoter was used to search the complete P. aeruginosa genome (www.pseudomonas.com) with the program FindPattern in the Genetics Computer Group package (Accelrys, San Diego, Calif.). Nine mismatches were allowed.
Site-directed mutagenesis.
Primers Qpmut1 and Qpmutcomp2 (Table 1) (Genomechanix Inc., Alachua, Fla.) were used with a Quick-change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) to mutate the Vfr binding site in the fleQ promoter according to the supplier's protocol. Primers Qp70mut1 and Qp70mutcomp2 and primers Qp70mut3 and Qp70mutcomp4 (Table 1) (Genomechanix Inc.) were similarly used to introduce 8- and 4-bp mutations in the putative −10 region of the fleQ promoter. Briefly, 20 ng of column-purified (Plasmid mini kit; Qiagen, Valencia, Calif.) plasmid template (pBS-fleQp) was used in a 50-μl amplification reaction mixture consisting of 1 μl of a solution containing each deoxynucleoside triphosphate at a concentration of 10 mM, 1 μl of Pfu polymerase, 1.25 μl of a 20 μM solution of each primer, and 5 μl of 10× reaction buffer. The mixture was subjected to a cycling profile consisting of initial denaturation for 30 s at 95°C, followed by 13 cycles (Qpmut1 and Qpmitcomp2) or 18 cycles (Qp70mut1 and Qp70mutcomp2 or Qp70mut3 and Qp70mutcomp4) of denaturation at 95°C for 30 s, annealing at 60°C for 1 min, and extension at 68°C for 10 min. The reaction mixture contents were then treated with DpnI to digest the original plasmid template. One microliter of the postdigestion amplification reaction mixture was used to transform E. coli XL1 Blue cells, and transformants were selected on LB agar plates containing ampicillin. Several clones were sequenced, and a clone with the desired site-specific mutation was subsequently used for further subcloning. The plasmid with the mutated Vfr binding site (ACA → CGC) was designated pBS-fleQpmutV. The plasmids with the 8-bp (ATAAAATT → GCGCGCGC) and 4-bp (AAAT → GCGC) mutations in the −10 region were designated pBS-fleQpmutS8 and pBS-fleQpmutS4, respectively. The inserts from pBS-fleQpmutS8 and pBS-fleQpmutS4 were subcloned into pDN19lacΩ, generating placΩQmutS8 and placΩQmutS4, respectively.
Transformation and electroporation.
Unless indicated otherwise, E. coli DH5α was the strain used for transformation (25). Electroporation of P. aeruginosa was carried out as described elsewhere (7).
PCR amplification of fragments for EMSAs and DNase I footprint assays.
For electrophoretic mobility shift assays (EMSAs), 88-bp DNA fragments containing the fleQ promoter and the mutant fleQ promoter (ACA → CGC) were generated by PCR amplification by using oligonucleotide primers fleQ1F and fleQ1R (Table 1) (Integrated DNA Technologies, Inc., Coralville, Iowa). placΩQ and pBS-fleQpmutV (Table 1) served as the templates. The high-fidelity polymerase Pfx (Stratagene) was used in the amplification reactions, which were performed according to the manufacturer's specifications. Radiolabeling of the DNA fragments was accomplished by inclusion of [α-32P]dCTP (800 Ci/mmol; Perkin-Elmer Life Sciences Inc., Boston, Mass.) in the PCR mixtures. Following PCR amplification, the wild-type and mutant fleQ promoter fragments were purified by using the Promega Wizard DNA Cleanup system (Promega Corp., Madison, Wis.).
For the DNase I footprint assays, oligonuclotide primers fleQ2F and fleQ2R (Table 1) (Integrated DNA Technologies) were used to PCR amplify a 158-bp fragment containing the fleQ promoter region by using placΩQ as a template. The first 10 bp of each primer contained either a SalI (fleQ2F) or XhoI (fleQ2R) restriction site. PCR amplification and fragment purification were carried out as described above. The amplified fragment was cut with either SalI or XhoI to generate a 5′ overhang that was end labeled with a single 5′ [α-32P]TTP (800 Ci/mmol; Perkin-Elmer Life Sciences Inc.) by using T7 polynucleotide kinase (Amersham Biosciences, Piscataway, N.J.) according to the manufacturer's directions.
Vfr purification.
Vfr purification from P. aeruginosa PA103(pWNP28) was based on cAMP affinity chromatography and additional procedures described by Albus et al. (2), with modifications. Briefly, cultures were grown to an optical density at 600 nm (OD600) of ∼0.5, and then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce overexpression of Vfr. After an additional 6 h of incubation, the cells were harvested by centrifugation at 5,000 × g for 15 min. The cell pellet was resuspended in a solution containing 100 mM Tris-HCl (pH 8.0), 50 mM KCl, 2 mM EDTA, 5 mM β-mercaptoethanol, and 1 mM sodium azide. A crude lysate was prepared by incubation of the solution with 200 μg of lysozyme per ml for 20 min at room temperature. The lysate was adjusted to 125 mM Tris-HCl (pH 8.0), 25 mM MgSO4, and 1% Brij 35, incubated at 4°C for 10 min, and then passed through a 40K SLM-Aminco French pressure cell (Thermo Electron Corp., Waltham, Mass.) at a pressure of approximately 15,000 lb/in2. Cellular debris was removed by centrifugation at 16,000 × g for 30 min at 4°C. An ammonium sulfate fractionation step was performed next; most of the Vfr protein precipitated between 50 and 65% ammonium sulfate saturation. This fraction was centrifuged at 27,000 × g for 30 min at 4°C. The pellet was resuspended in a solution containing 50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 5% glycerol, and 5 mM β-mercaptoethanol and dialyzed against the same buffer to remove the ammonium sulfate. The resulting solution was then applied to a cAMP-agarose column (Sigma Chemical Co., St. Louis, Mo.) as described by Ghosaini et al. (12). Eluted fractions containing Vfr were pooled and dialyzed into a solution containing 10 mM sodium phosphate (pH 7.2), 2 mM EDTA, 5 mM β-mercaptoethanol, and 5% glycerol. The resulting solution was applied to an SP-Sephadex cation-exchange column. Vfr-containing fractions were pooled and dialyzed into a solution containing 50 mM Tris-HCl (pH 7.2), 500 mM KCl, 0.2 mM EDTA, 0.2 mM dithiothreitol (DTT), and 50% glycerol for storage.
EMSAs.
Binding reactions were performed as described by Devault et al. (9), with minor modifications. Briefly, the 20-μl reaction mixtures contained the radiolabeled DNA fragments, with the radioactivity standardized to approximately 1,000 cpm (15 to 40 pM), and various concentrations of purified Vfr in a solution containing 10 mM Tris-HCl (pH 7.4), 100 mM KCl, 1 mM EDTA, 5% glycerol, 1 mM DTT, 50 μg of bovine serum albumin per ml, 20 μM cAMP, and 10 μg of poly(dI-dC)-poly(dI-dC) (Amersham Biosciences Inc.) per ml. The reaction mixtures were incubated for 20 min at room temperature. Samples were electrophoresed on a preequilibrated 4% acrylamide-45 mM Tris-HCl-45 mM borate gel at 10 V/cm and 4°C. The upper buffer chamber contained 45 mM Tris-HCl, 45 mM borate (pH 8.0), and 20 μM cAMP. The gels were dried and exposed to Kodak X-Omat AR5 film overnight at −80°C with an intensifying screen.
DNase I footprint assay.
The DNase I footprint assay was based on the procedure of Ross et al. (24). Binding reactions were performed by incubating purified Vfr with the radiolabeled DNA fragments in a solution containing 10 mM Tris-HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 5% glycerol, 1 mM DTT, 50 μg of bovine serum albumin per ml, and 20 μM cAMP for 20 min at room temperature. DNase I (0.75 to 1.5 μg/ml) was then added to the samples. The DNase I digestion reaction mixtures were incubated for 30 s at room temperature, and then the reactions were terminated by adding 12.5 mM (final concentration) EDTA, 200 mM (final concentration) sodium acetate, and 30% (vol/vol) (final concentration) phenol. The digestion products were ethanol precipitated and then resuspended in 0.5× TBE buffer (45 mM Tris-HCl, 45 mM borate) containing 8 M urea, 0.5% bromophenol blue, and 0.5% xylene cyanol. The samples were analyzed on a 7 M urea-8% polyacrylamide gel to determine the regions protected by Vfr. Maxam-Gilbert sequencing reactions (19) were also performed with the radiolabeled DNA fragments to obtain sequence ladders for comparison with the footprint reaction mixtures.
Construction of promoter-lacZ fusions in the mini-CTX lacZ plasmid and chromosomal insertion of these fusions into the attB site.
The wild-type and mutated fleQ promoter-containing inserts in pBS-fleQp and pBS-fleQpmutV were cloned as EcoRI-BamHI fragments in mini-CTX lacZ (4), generating CTX-fleQplacZ and CTX-fleQpmutVlacZ. E. coli S17-1 was transformed with these plasmids and the vector control mini-CTXlacZ. Matings were set up with the transformed E. coli S17-1 strain as the donor and P. aeruginosa PAK or PAO1 as the recipient. The exconjugants were selected on tetracycline-containing plates, and one colony was cultured further to electroporate the pFLP2 plasmid into it. Transformants were selected on carbenicillin-containing plates. Two colonies from each strain background were propagated in LB broth, and serial dilutions of the cultures were plated on LB medium and LB medium containing sucrose (5%). A few colonies from the sucrose-containing plate were examined for tetracycline sensitivity and carbenicillin sensitivity to ensure excision of the plasmid backbone and curing of the pFLP2 plasmid, respectively. The chromosomal integrants with the fleQp-lacZ fusion in PAK and PAO1 were designated PAK fleQp-lacZ and PAO fleQp-lacZ, respectively. The fleQpmutV-lacZ derivatives were designated PAK fleQpmutV-lacZ and PAO fleQpmutV-lacZ. The controls with a promoterless lacZ integrated were designated PAK lacZ and PAO lacZ.
Motility assay.
Plain 0.3% LB agar plates were used to test the motility of P. aeruginosa PAO9001, a vfr deletion mutant. To examine the effect of Vfr overexpression on the motility of strain PAK harboring either pMMB66EH or pWNP28, 0.3% LB agar plates containing carbenicillin without or with various concentrations of IPTG were used. The plates were stabbed with the strains and incubated at 37°C.
β-Galactosidase assays.
P. aeruginosa cultures were grown in LB broth with the appropriate antibiotic (if needed) to an OD600 of ∼0.8. The β-galactosidase assay was performed as described previously (20). To study the effect of Vfr overexpression on the fleQ promoter activity, cultures were grown in LB broth with carbenicillin to an OD600 of ∼0.2, induced with 1 mM IPTG, and grown for an additional 6 h at 37°C with shaking.
SDS-polyacrylamide gel electrophoresis and Western analysis.
Bacteria were pelleted from 1 ml of each culture grown for 6 h (noninduced and IPTG induced). The cells were denatured by boiling in 2% sodium dodecyl sulfate (SDS)-1% β-mercaptoethanol-50 mM Tris-Cl (pH 7.5). Aliquots (5 μl) of the samples were resolved on a 12.5% polyacrylamide gel, and the proteins were transferred to a polyvinylidene difluoride membrane by Western blotting. Following blocking in 3% skim milk for 2 h, the blot was treated with either anti-FleQ (1:2,000) or anti-Vfr (1:5,000) antibody as the first antibody and anti-rabbit immunoglobulin G-alkaline phosphatase conjugate as the second antibody. The blot was developed by using nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate) as the color substrate (8).
RNA extraction.
A 100-ml LB broth culture of P. aeruginosa PAK was grown at 37°C to an OD600 of 0.6 with shaking, and the cells were harvested by centrifugation at 3,000 rpm for 20 min at 4°C (JA-10 rotor; Beckman). Total RNA was isolated from the cells by using the TRIZOL reagent (Invitrogen Life Technologies, Carlsbad, Calif.) and the supplier's protocol for isolating total bacterial RNA.
Primer extension analysis.
Oligonucleotide RT2 was 5′ end labeled in the presence of [γ-32P]ATP (3,000 Ci/mmol; Perkin-Elmer Life Sciences) and T4 polynucleotide kinase (Invitrogen Life Technologies). The labeled oligonucleotide (1 × 106 cpm) was used in a primer extension reaction mixture (total volume, 20 μl) consisting of 30 μg of denatured (incubation at 65°C for 20 min) total RNA, 5 μl of 5× first-strand buffer, 1 μl of a solution containing each deoxynucleoside triphosphate at a concentration of 10 mM, 1 μl of a 100 mM DTT solution, 1 μl of RNaseOUT, and 200 U of Superscript II RT. The reaction mixture was incubated at 42°C for 1 h, treated with 20 U of RNase H, phenol-chloroform extracted, and ethanol precipitated. The precipitate was resuspended in 6 μl of sequencing loading buffer, denatured at 95°C for 2 min, and subjected to 8% urea-polyacrylamide gel electrophoresis along with a sequencing ladder. The sequencing ladder was generated by Sanger's dideoxy sequencing method by using a Sequenase version 2.0 kit (U.S. Biochemical Corp., Cleveland, Ohio) with pBS-fleQpmutV as the template and RT2 as the primer. The gel was dried and subsequently autoradiographed.
In vitro transcription assay with E. coli RNA polymerase σ70 holoenzyme.
In vitro single-round runoff transcription analysis was performed by using E. coli RNA polymerase holoenzyme saturated with σ70 (Epicentre Technologies, Madison, Wis.) and a DNA fragment containing the fleQ promoter. ScaI-BamHI fragments (289 bp) excised from placΩQ and placΩQmutS4 served as templates for the transcription assays. Briefly, 0.1 pmol of the DNA template was incubated with 0.2 U of the RNA polymerase holoenzyme, 10 μCi of [α-32P]CTP (3,000 Ci/mmol; Perkin-Elmer Life Sciences), 0.4 mM ATP, 0.4 mM GTP, 0.4 mM UTP, 11 μM CTP, 1× transcription buffer (40 mM Tris-HCl [pH 7.9], 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl), and 1 μl of RNaseOUT at 37°C for 1 h in a 10-μl (total volume) mixture. Subsequently, the samples were treated with 1 U of RNase-free DNase for 15 min at 37°C, phenol-chloroform extracted, and ethanol precipitated. Each pellet was dissolved in RNA gel loading dye, and samples were electrophoresed in a 6% denaturing polyacrylamide gel containing 7 M urea and analyzed by autoradiography. A 50-bp DNA ladder served as the molecular weight marker.
RESULTS
Vfr binds to the fleQ promoter.
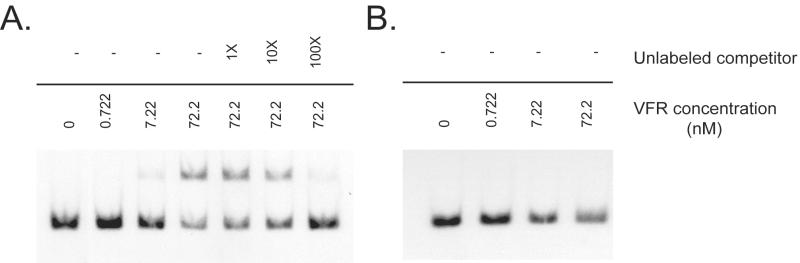
Vfr has been shown to bind to specific sequences in the lasR, regA, and toxA promoters (K. J. Kanack, E. P. Ferrell, L. J. Runyen-Janecky, and S. E. H. West, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. D140, p. 258, 2000; L. J. Runyen-Janecky, A. M. Albus, B. H. Iglewski, and S. E. H. West, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. B-35, p. 160, 1996). To identify additional Vfr binding sites, homology searches of the P. aeruginosa genome were conducted by using the Vfr binding site present in the lasR promoter. These searches revealed the putative Vfr binding site sequence AATTGACTAATCGTTCACATTT centered 116.5 bp upstream of the fleQ translational start (Fig. 1). This sequence is 62.5% similar to the lasR Vfr binding site sequence. In order to authenticate the putative Vfr binding site identified in the fleQ promoter, EMSAs were performed by using an 88-bp fragment containing this sequence and purified Vfr. The mobility of the 88-bp fleQ promoter fragment was retarded. The retardation was partial in the presence of 72.2 nM Vfr (Fig 2A) and complete in the presence of 722 nM Vfr (data not shown). This demonstrated that the promoter region did indeed contain a Vfr binding site. Inclusion of a 100-fold excess of the unlabeled fleQ promoter fragment in the binding reaction mixture inhibited the retardation of the labeled fragment (Fig. 2A). This result indicated that the observed binding of Vfr to the fleQ promoter fragment was specific.
FIG. 1.
Schematic representation of the fleQ promoter region: 238-bp region including the upstream region of fleQ and the first six codons (boldface type) of its coding region. The deduced amino acid sequence is shown under the codons. The Vfr binding site is enclosed in a box. The putative −10 and −35 sequences for the σ70 binding site are underlined. Transcriptional start sites t1 and t2, as determined by primer extension analysis, are shown. The bases mutagenized in the Vfr binding site and the −10 region are italicized and in boldface type, and the designations of the plasmids in which they were generated are indicated. The location of the RT2 primer used for primer extension is indicated by an arrow. The ScaI site is underlined.
FIG. 2.
EMSA of Vfr on the fleQ promoter. (A) An 88-bp radiolabeled fleQ promoter fragment was incubated either with different concentrations of Vfr or with a fixed amount (72.2 nM) of Vfr and different amounts of the unlabeled fleQ promoter fragment as described in Materials and Methods. (B) The mutated fleQ promoter fragment in which CGC was substituted for ACA in the putative Vfr binding site was incubated with different concentrations of Vfr. The amounts of the unlabeled competitor fleQ DNA are indicated above the bars. The concentrations of purified Vfr are indicated below the bars.
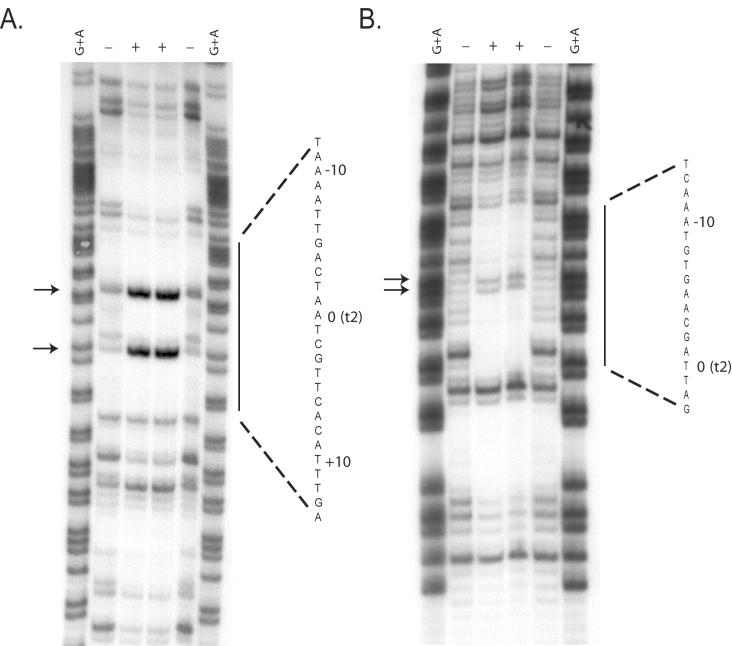
To confirm the identity of the Vfr DNA binding site in the fleQ promoter, DNase I footprinting was performed for a 158-bp fleQ promoter fragment that included the 88-bp region used in the EMSAs (Fig. 1 and 3). In the presence of Vfr, the 5′-AAATTGACTAATCGTTCACATTTG-3′ site was protected from DNase I cleavage. This region contains hypersensitive sites located at positions 11 and 19 on the coding strand and at positions 9 and 10 on the antisense strand of the protected sequence. This observation is similar to previous observations of DNase I hypersensitive sites in the promoters of other Vfr-regulated genes (Kanack et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol.).
FIG. 3.
DNase I footprint of Vfr bound to the fleQ promoter. The fleQ promoter radiolabeled on the coding (A) or antisense (B) strand was incubated with or without purified Vfr and was treated with DNase I as described in Materials and Methods. Lanes G+A, sequencing ladder of the fleQ promoter; lanes +, Vfr present; lanes −, Vfr absent. The region protected from DNase I digestion by Vfr is indicated by a bar and the sequence of the protected region. The DNase I hypersensitive sites are indicated by arrows. The numbers indicate the positions of the nucleotides with respect to the t2 transcriptional start site.
Of the eight known and putative Vfr binding sites identified in the P. aeruginosa genome by subsequence search analysis, the CACA motif was highly conserved. To further confirm that the AAATTGACTAATCGTTCACATTTG site is the Vfr DNA binding site in the fleQ promoter, we substituted CGC for ACA by performing site-directed mutagenesis of pBS-fleQp. EMSAs were performed for a 88-bp fleQ promoter fragment containing this mutation. At a Vfr concentration of 72.2 nM (Fig. 2B), Vfr failed to bind to the fleQ promoter, probably because of its reduced affinity for the mutated site.
fleQ promoter activity is not downregulated in a PAO1 vfr mutant.
Vfr activates transcription of toxA, regA, and lasR (2, 34). To test whether the same was true for fleQ, the promoter activity of fleQ was assessed in PAO1 and the vfr deletion mutant PAO9001. The CTX-fleQplacZ plasmid containing the fleQ promoter cloned upstream of a promoterless lacZ gene and the mini-CTXlacZ plasmid were integrated into the attB site of the chromosome of each strain, and their β-galactosidase activities were quantified (Table 2). The fleQ promoter activities were comparable in the wild-type and vfr mutant strains. The absence of Vfr in PAO9001 had no effect on the activity of the promoter, indicating that the fleQ promoter was not dependent on Vfr for transcriptional activation. To further confirm that the activation of fleQ transcription was not Vfr dependent, PAO1 and the vfr mutant PAO9001 were tested in a motility plate assay (data not shown). There was no motility defect in vfr mutant PAO9001.
TABLE 2.
Activity of the fleQ promoter in PAO1 and its isogenic vfr mutant, PAO9001
| Promoter-lacZ fusion (chromosomal insertion) | β-Galactosidase activity (Miller units)
|
|
|---|---|---|
| PAO1 | PAO9001 (vfr mutant) | |
| fleQp-lacZ (wild type) | 1,981 ± 59a | 1,992 ± 36 |
| Promotrless lacZ | 69 ± 16 | 99 ± 20 |
The values are means ± standard deviations.
vfr overexpression from a plasmid renders P. aeruginosa nonmotile.
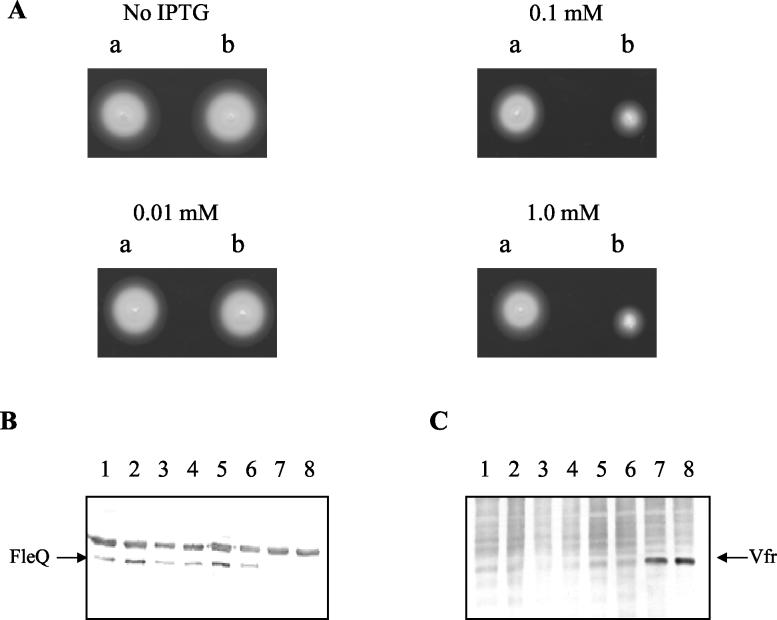
As Vfr binds to the fleQ promoter in vitro and does not activate fleQ transcription, we explored the possibility that Vfr may function as a repressor. To test whether overexpression of Vfr from an IPTG-inducible promoter would repress flagellar motility through fleQ, the vfr overexpression plasmid pWNP28 and the vector control pMMB66EH were electroporated into PAK and PAO1. The carbenicillin-resistant transformants were tested on motility agar plates containing carbenicillin and various concentrations of IPTG (0, 0.01, 0.1, and 1 mM). At IPTG concentrations of 0.1 and 1 mM, the PAK and PAO1 strains containing the inducible Vfr expression plasmid pWNP28 initially appeared to be nonmotile (6 h) (data not shown), but they displayed small motility zones upon further incubation (22 h), which were markedly reduced compared to that of the vector control. The motility profile of PAK under these conditions is shown in Fig. 4A.
FIG. 4.
Vfr overexpression downregulates FleQ, thereby reducing motility. (A) Motility assay conducted on 0.3% agar plates containing carbenicillin and various IPTG concentrations, as indicated. Spots a and b, PAK harboring the vector control pMMB66EH and the vfr overexpression plasmid pWNP28, respectively. (B and C) Western analysis of the strains used in the motility assay with different IPTG concentrations and with anti-FleQ (B) and anti-Vfr (C) antibodies. Lanes1 to 4 contained lysates from cultures of PAK harboring pMMB66EH grown with 0, 0.01, 0.1, and 1.0 mM IPTG, respectively. Lanes 5 to 8 contained lysates from cultures of PAK harboring pWNP28 grown with 0, 0.01, 0.1, and 1.0 mM IPTG, respectively.
FleQ production is downregulated following Vfr overproduction.
To examine whether the repressive effect of Vfr on motility was due to repression of fleQ, a semiquantitative Western analysis of FleQ production was performed. Lysates of equivalent numbers of bacterial cells from IPTG-induced cultures of PAK containing either pWNP28, the vfr overexpression plasmid, or pMMB66HE, the vector control, were electrophoresed on an SDS-polyacrylamide gel and subjected to Western analysis with anti-FleQ (Fig. 4B) and anti-Vfr (Fig. 4C) polyclonal antibodies. The specificities of the anti-FleQ and anti-Vfr antibodies were examined prior to being used (data not shown). FleQ was barely detectable in PAK(pWNP28) (0.1 mM and 1.0 mM IPTG) compared to the control strain containing the vector. This indicated that downregulation of fleQ was one possible reason for the reduced motility observed with PAK(pWNP28). The amount of Vfr produced increased as the IPTG concentration increased (Fig. 4C). For further experiments under Vfr overexpression conditions we used 1 mM as the inducing concentration of IPTG.
To ascertain that the observed lower level of expression of FleQ in the vfr overexpression strain PAK(pWNP28) was due to transcriptional repression and not due to instability of FleQ under vfr-overexpressing conditions, the activities of the fleQ promoter-lacZ fusions integrated into the attB sites in strains PAK fleQp-lacZ and PAO fleQp-lacZ were assessed by performing a β-galactosidase reporter assay with vfr overexpression from pWNP28. PAK lacZ and PAO lacZ with promoterless lacZ inserted into the attB site served as controls. Strains overexpressing vfr repressed transcription from the wild-type fleQ promoter four- and twofold in PAK(pWNP28) and PAO1(pWNP28), respectively (Table 3). The basal level of activity of the wild-type promoter in PAO1(pMMB66EH) was higher than that in PAK(pMMB66EH).
TABLE 3.
Effect of vfr overexpression on the activity of the wild-type fleQ promoter and its mutagenized derivative
| Promoter-lacZ fusion (chromosomal insertion) | β-Galactosidase activities (Miller units) in the following strains:
|
|||
|---|---|---|---|---|
| PAK(pMMB66EH) (vector) | PAK(pWNP28) (vfr) | PAO(pMMB66EH) (vector) | PAO(pWNP28) (vfr) | |
| fleQp-lacZ | 1,007 ± 15a | 253 ± 7 | 2,649 ± 37 | 1,267 ± 6 |
| fleQpmutV-lacZ | 1,339 ± 4 | 680 ± 49 | 3,341 ± 76 | 3,005 ± 59 |
| Promoterless lacZ | 33 ± 3 | 54 ± 1 | 92 ± 60 | 150 ± 32 |
The values are means ± standard deviations.
If downregulation of the fleQ promoter was due to repression by Vfr, an ACA-to-CGC mutation in the Vfr binding site of the fleQ promoter with reduced affinity for Vfr in an EMSA would be expected to be unresponsive or less responsive to Vfr-dependent downregulation. Therefore, we assessed the activity of an fleQpmutV-lacZ fusion that had been integrated into the attB sites of PAK and PAO1 containing the vfr overexpression plasmid pWNP28 (Table 3). The activity of fleQpmutV was higher than the activity of the wild-type fleQ promoter in both PAK and PAO1 carrying either the vfr overexpression plasmid pWNP28 or the vector pMMB66EH. The activity of the mutated promoter was not repressed by overproduction of Vfr in PAO1, indicating that Vfr was not able to bind to the fleQ promoter in vivo in strain PAO1. However, in PAK, the fleQpmutV promoter activity still displayed a twofold reduction under vfr overexpression conditions, indicating that the 3-bp ACA-to-CGC mutation in the Vfr binding site with reduced affinity for purified Vfr in vitro (Fig. 2B) was still responsive to partial repression by Vfr in vivo.
Vfr binding site overlaps the putative −10 region and the transcriptional start sites of fleQ.
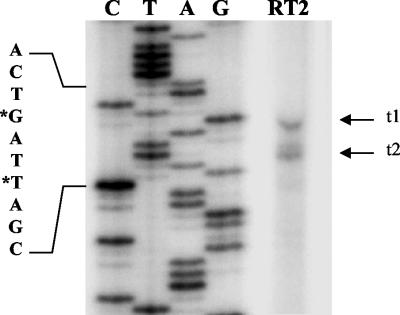
We used primer extension analysis with PAK total RNA to determine the transcriptional start sites of fleQ. The transcriptional start sites mapped 121 bp (t1) and 118 bp (t2) upstream of the ATG initiation codon (Fig. 1 and 5), and both of these sites mapped within the Vfr binding site. Analysis of the nucleotide sequence upstream of the transcriptional start sites revealed a −10 region centered 9.5 bp upstream of the t2 transcript and a −35 region centered 34 bp upstream of the t2 transcript with five of six and three of six matches with the E. coli σ70 consensus sequence, respectively (Fig. 1). Four of the six bases comprising the −10 region were also part of the Vfr binding consensus region. These findings were consistent with a model in which the t2 transcript of fleQ is σ70 regulated and occupancy of the Vfr binding site by Vfr could sterically hinder binding of the sigma factor (σ70) in the −10 region and deny access to the RNA polymerase to initiate transcription at either t1 or t2, thereby repressing fleQ transcription. However, it was uncertain whether fleQ was σ70 regulated.
FIG. 5.
Primer extension analysis of fleQ. Thirty micrograms of total RNA isolated from PAK was subjected to primer extension analysis with the RT2 primer and loaded in lane RT2. A sequencing reaction mixture (lanes C, T, A, and G) containing the same primer with pBS-fleQpmutV as the template was loaded and run on a denaturing urea-polyacrylamide gel simultaneously. The sequence of the region spanning the transcriptional start site is shown on the left. The residues marked with an asterisk are the probable start sites determined in this experiment, designated t1 and t2.
Mutations in the putative −10 region of the fleQ promoter result in significant reductions in its promoter activity.
Two mutated derivatives of the fleQ promoter carrying an 8-bp mutation (ATAAAATT → GCGCGCGC) and a 4-bp mutation (AAAT → GCGC) in the −10 region were cloned into pDN19lacΩ, generating placΩQmutS8 and placΩQmutS4, respectively. placΩQmutS8 and placΩQmutS4 were electroporated into PAK, and their β-galactosidase activities were measured and compared. Both the mutated derivatives displayed a sixfold reduction in promoter activity compared to the wild-type promoter activity (Table 4). Moreover, the residual β-galactosidase activities of the 8- and 4-bp mutated promoters were comparable to the residual activity of the promoterless vector control, indicating that there was total loss of transcriptional activity even with the less extensive 4-bp mutation.
TABLE 4.
Effects of mutations in the putative −10 region of the fleQ promoter on promoter activity
| Plasmid | β-Galactosidase activity (Miller units) |
|---|---|
| placΩQ | 253 ± 21a |
| placΩQmutS8 | 41 ± 4 |
| placΩQmutS4 | 39 ± 21 |
| placΩ | 38 ± 1 |
The values are means ± standard deviations.
E. coli RNA polymerase σ70 holoenzyme initiates transcription from the fleQ promoter in an in vitro transcription assay.
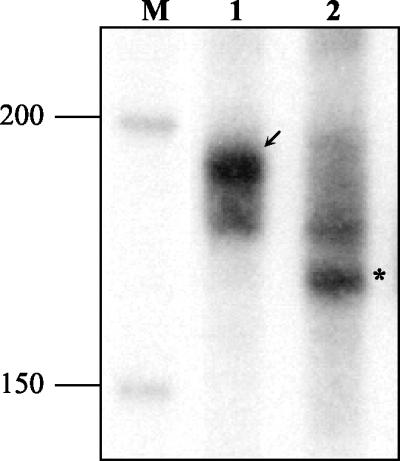
In order to determine whether transcription from the fleQ promoter could be initiated solely by the σ70-associated RNA polymerase, the fleQ promoter region was subjected to an in vitro transcription assay. Promoter sequences cloned in placΩQ and placΩQmutS4 were excised as ScaI-BamHI fragments and used in a cell-free in vitro transcription assay with the E. coli RNA polymerase holoenzyme saturated with σ70 (Fig. 6). The presence of a major transcript migrating at the expected position of the fleQ transcript (203 nucleotides) (Fig. 6, lane 1) and the absence of this transcript in the reaction performed with the −10 region mutated template (Fig. 6, lane 2) confirmed that σ70 was capable of transcribing fleQ. A smaller transcript (lane 2) specific to the mutated template could have resulted from an alternative transcription initiation site.
FIG. 6.
In vitro transcription analysis of fleQ. DNA templates containing the wild-type fleQ promoter (lane 1) and the 4-bp mutation in its −10 region (lane 2) were analyzed in a transcription reaction by using the E. coli RNA polymerase σ70 holoenzyme. 32P-labeled DNA size markers were electrophoresed in lane M. The sizes of the individual DNA fragments (in bases) are indicated on the left. The arrow indicates the position of the σ70-dependent fleQ transcript in lane 1. The asterisk indicates the position of the smaller transcript visible exclusively in lane 2.
DISCUSSION
fleQ appears to be the highest-level activator in the hierarchical regulatory cascade involved in the flagellar biogenesis pathway of P. aeruginosa. It is essential for flagellar motility as an fleQ mutant does not assemble a flagellum nor does it synthesize detectable amounts of flagellin (3). Moreover, at least eight of the various flagellar genes and operons studied to date appear to be directly and positively regulated by FleQ (7). Any mechanism that downregulates FleQ activity would have a negative impact on flagellar biogenesis and thereby motility and chemotaxis in this pathogen. We have identified FleN, an antiactivator for FleQ which posttranslationally downregulates FleQ activity by direct binding (8). Whether in addition to this level of regulation there is any other mechanism that upregulates or downregulates the transcription of fleQ remains to be determined.
Based on sequence homology, we found a putative Vfr binding site in the fleQ promoter, and we set out to ascertain its functional significance. The data presented here indicate that the Vfr binding site is an authentic site, as purified Vfr was able to bind to it, as demonstrated in the EMSAs (Fig. 2), and to protect it from DNase I (Fig. 3). The sequence of the protected region on both strands was consistent with the predicted consensus binding site.
Vfr is known to function as a transcriptional activator for the quorum-sensing regulatory protein lasR (2). To determine the consequences of Vfr binding to the fleQ promoter, the promoter activity of FleQ was tested in PAO1 and a vfr mutant derivative of this strain, PAO9001 (Table 2). As there was no appreciable decline in the promoter activity in the vfr mutant, activation of fleQ transcription was not dependent on Vfr. We next explored whether repression by Vfr was a possibility. Since fleQ promoter activity in the vfr mutant was not greater than fleQ promoter activity in the wild-type strain, there was no evidence that suggested a repressor function for Vfr. We hypothesized that the amount of Vfr synthesized under our culture conditions was probably not sufficient for repression. Overexpression of vfr from pWNP28 under IPTG induction conditions in PAK (Fig. 4) and PAO1 (data not shown) repressed motility and downregulated the transcription of fleQ (Table 3), indicating the ability of Vfr to repress fleQ as speculated. A comparison of the promoter activities in PAK and PAO1 (Table 3) revealed that although the fleQ promoter regions analyzed in strains PAK and PAO1 were identical, the basal activity of the promoter appeared to be greater in PAO1. Repression of the fleQ wild-type promoter by Vfr appears to be more effective in PAK (fourfold downregulation) than in PAO1 (twofold downregulation). As the sources of Vfr and the Vfr binding site sequences in the two strains tested are the same, involvement of an accessory factor (corepressor) that makes Vfr a better repressor in PAK than in PAO1 remains a possibility. The effector molecule that regulates Vfr activity in P. aeruginosa has not been identified. cAMP is one possibility and is required for Vfr activity in E. coli (32, 34). Not much is known about the levels of cAMP in P. aeruginosa (21, 26) and specifically about quantitative differences (if any) in the cAMP levels in PAK and PAO1 which would enable us to explain our observations.
To understand how Vfr represses fleQ transcription, we mapped the fleQ transcriptional start sites. Two transcriptional start sites, t1 and t2, mapped within the Vfr binding site in PAK (Fig. 5). Inspection of the sequences upstream of the transcriptional start sites revealed a σ70 binding consensus (Fig 1) for the t2 transcript, indicating that it may be σ70 dependent. In order to determine whether transcription of fleQ was σ70 dependent in vivo, a 4-bp mutation and an 8-bp mutation were generated in the putative −10 region of the fleQ promoter, and the activities of the mutated promoters were assessed with a β-galactosidase assay by using a plasmid-based lacZ reporter (Table 4). Both of the mutated derivatives showed low promoter activity comparable to that of the promoterless lacZ construct. Thus, the AAAT-to-GCGC substitution probably eliminated fleQ promoter activity due to the inability of the σ70 RNA polymerase holoenzyme to bind to the −10 region of the promoter and to initiate transcription effectively in P. aeruginosa. In an in vitro transcription assay (Fig. 6), the ability of the σ70-saturated RNA polymerase holoenzyme of E. coli to initiate transcription from the fleQ promoter suggested the presence of an authentic σ70-recognizing sequence in the fleQ promoter. The same holoenzyme was unable to generate the 203-nucleotide fleQ-specific transcript from a DNA template with a 4-bp mutation in the −10 region of the fleQ promoter, which is predicted to be important in σ70 binding. Another, smaller transcript seen in the assays probably arose from an alternative start site in the mutated template but did not appear to contribute to any residual β-galactosidase activity in a reporter plasmid carrying the same mutation (placΩQmutS4) (Table 4). The results of the primer extension analysis, β-galactosidase assay, and in vitro transcription assay are consistent with involvement of σ70 in the transcription of fleQ. Possible involvement of σ54 in the transcriptional regulation of fleQ in another pseudomonad (29) has been suggested. However, in P. aeruginosa, the fleQ promoter retains activity in an rpoN (σ54) mutant (3). These differences suggest that the promoters of fleQ and its homologs are probably complex structures with multiple regulatory mechanisms functioning under different environmental conditions; repression by Vfr is probably one of these mechanisms. The involvement of σ70 in transcriptional control of fleQ suggests that fleQ could either be constitutively expressed in a manner similar to the housekeeping genes using the σ70 holoenzyme or be activated by an activator that works in concert with σ70 (13).
Introduction of mutations into the Vfr binding site to eliminate Vfr binding and thereby repress fleQ would add credence to the proposed role of Vfr as a repressor of fleQ. As the Vfr binding site overlaps the proposed −10 region and transcriptional start sites, the sites that could be mutated that would eliminate Vfr binding but not transcription of fleQ were limited. As the CACA motif appeared to be conserved in eight putative Vfr binding sites, CGC was substituted for ACA and tested in an EMSA (Fig. 2B) for Vfr binding in vitro. The mutated Vfr binding site did not exhibit binding in an EMSA with purified Vfr. The same mutation was integrated into the chromosomes of PAK and PAO1 as an fleQpmutV-lacZ fusion, and the activity of the fleQ promoter was measured under vfr-overexpressing conditions with a β-galactosidase assay (Table 3). In PAO1 repression by Vfr was apparently eliminated as there was no significant reduction in the promoter activity when Vfr was overproduced. In PAK, the same mutation in the Vfr binding site still resulted in twofold downregulation of the mutated promoter (compared to 4-fold downregulation with the wild-type promoter), indicating that this mutation only partially eliminated repression in this strain. A detailed analysis of various mutations is needed to determine the best possible combination that completely eliminates repression of the fleQ promoter by Vfr in PAK without compromising the initiation of fleQ transcription.
Whether any environmental or cultural conditions enhance vfr expression from its own promoter enough to result in substantial repression of fleQ, like that seen in the overexpression experiments, is difficult to predict. Nevertheless, as there are at least eight flagellar genes or operons that are FleQ dependent (7, 8), even a small quantitative reduction in the free FleQ pool within the cell could potentially retard the process of flagellar biogenesis and have significant physiological implications.
Repression of flagellar biogenesis by Vfr is new to Pseudomonas research because to the best of our knowledge our report is the first evidence that a nonflagellar regulator (Vfr) affects the topmost regulator (FleQ) in the flagellar biogenesis hierarchy by downregulating its transcription. Moreover, previously, Vfr has been shown to be a transcriptional activator of lasR and toxA, which are involved in quoroum sensing (2) and exotoxin A production, respectively (34). Evidence obtained in this study suggests that like CRP, its E. coli homolog (13), Vfr is also capable of functioning as a repressor, depending on the position of its binding site in the promoter region with respect to the transcriptional start site. In general, in σ70-dependent promoters activator binding sites do not appear in positions downstream from the −30 region, whereas repressor binding sites are very common in this region (13). For example, in the malT promoter CRP binds around the −70 region and functions as an activator (22), whereas in the cya promoter the CRP binding site encompasses the +1 site and functions as a repressor instead (1). Similarly, in the lasR promoter of P. aeruginosa, where the Vfr binding site is located around position −47, Vfr is reported to function as an activator (2), whereas in the fleQ promoter, where Vfr binds to DNA from position −10 to position +14 with respect to the t2 transcript (Fig. 1), it functions as a repressor (this study).
Induction of many virulence genes occurs in P. aeruginosa at a time after quorum sensing has been initiated. We hypothesize that in order for Vfr to promote quorum sensing, it would be necessary to inhibit flagellar motility through repression of fleQ in order for bacteria to reach the appropriate cell density. Appropriate environmental cues could induce Vfr production and simultaneously downregulate flagellar biogenesis to prevent the bacteria from swimming away from a site of infection. If quorum-sensing mechanisms are important for virulence in specific infections, then a similar mechanism should be initially operative to prevent motility-dependent dissemination of P. aeruginosa from infection sites until the full repertoire of virulence genes is induced.
Acknowledgments
This work was supported by National Institutes of Health research grants AI45014 (to R.R.) and AI31477 to (S.E.H.W).
REFERENCES
- 1.Aiba, H. 1985. Transcription of the Escherichia coli adenylate cyclase gene is negatively regulated by cAMP-cAMP receptor protein. J. Biol. Chem. Mar. 10:3063-3070. [PubMed] [Google Scholar]
- 2.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-950. [DOI] [PubMed] [Google Scholar]
- 5.Bourret, R. B., K. A. Borkovich, and M. I. Simon. 1991. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60:401-411. [DOI] [PubMed] [Google Scholar]
- 6.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasgupta, N., and R. Ramphal. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 183:6636-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devault, J. D., W. Hendrickson, J. Kato, and A. M. Chakrabarty. 1991. Environmentally regulated algD promoter is responsive to the cAMP receptor protein in Escherichia coli. Mol. Microbiol. 5:2503-2509. [DOI] [PubMed] [Google Scholar]
- 10.Domian, I. J., A. Reisenauer, and L. Shapiro. 1999. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. USA 96:6648-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 12.Ghosaini, L. R., M. Brown, and J. M. Sturtevant. 1988. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry 27:5257-5261. [DOI] [PubMed] [Google Scholar]
- 13.Gralla, J. D., and J. Collado-Vides. 1996. Organization and function of transcription regulatory elements, p. 1232-1245. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 14.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, Y. K., and L. L. McCarter.2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 17.Kutsukake, K. 1997. Autogenous and global control of the flagellar master operon, flhDC, in Salmonella typhimurium. Mol. Gen. Genet. 24:440-448. [DOI] [PubMed] [Google Scholar]
- 18.Liu, P. V. 1973. Exotoxins of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J. Infect. Dis. 128:506-513. [DOI] [PubMed] [Google Scholar]
- 19.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-650. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Phillips, A. T., and L. M. Mulfinger. 1981. Cyclic adenosine 3′,5′-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J. Bacteriol. 145:1286-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raibaud, O., D. Vidal-Ingigliardi, and A. Kolb. 1991. Genetic studies on the promoter of malT, the gene that encodes the activator of the Escherichia coli maltose regulon. Res. Microbiol. 142:937-942. [DOI] [PubMed] [Google Scholar]
- 23.Ritchings, B. W., E. C. Almira, S. Lory, and R. Ramphal. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross, W., J. F. Thompson, J. T. Newlands, and R. L. Gourse. 1990. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 9:3733-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Siegel, L. S., P. B. Hylemon, and P. V. Phibbs, Jr. 1977. Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,5′-monophosphate phosphodiesterase in Pseudomonas and Bacteroides. J. Bacteriol. 129:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon, R., U. Preifer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 28.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soutourina, O. A., E. A. Semenova, V. V. Parfenova, A. Danchin, and P. Bertin. 2001. Control of bacterial motility by environmental factors in polarly flagellated and peritrichous bacteria isolated from Lake Baikal. Appl. Environ. Microbiol. 67:3852-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is co-coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starnbach, M. N., and S. Lory. 1992. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6:459-469. [DOI] [PubMed] [Google Scholar]
- 32.Suh, S.-J., L. J. Runyen-Janecky, T. C. Maleniak, P. Hager, C. H. MacGregor, N. A. Zielinski-Mozny, P. V. Phibbs, Jr., and S. E. H. West. 2002. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148:1561-1569. [DOI] [PubMed] [Google Scholar]
- 33.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West, S. E. H., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wingrove, J. A., and J. W. Gober. 1996. Identification of an asymmetrically localized sensor histidine kinase responsible for temporally and spatially regulated transcription. Science 274:597-601. [DOI] [PubMed] [Google Scholar]
- 36.Yanagihara, S., S. Iyoda, K. Ohnishi, T. Iino, and K. Kutsukake. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet. Syst. 74:105-111. [DOI] [PubMed] [Google Scholar]