Abstract
In Pseudomonas aeruginosa, flagellar genes are regulated in a cascade headed by FleQ, an NtrC/NifA-type activator. FleQ and RpoN positively regulate expression of flhA, fliE, fliL, and fleSR genes, among others. Direct interaction of FleQ with flhA, fliE, fliL, and fleSR promoters was demonstrated by gel shift assay, along with experiments to conclusively determine the specificity of its binding. DNase I footprinting was performed to determine the FleQ binding sites on flhA, fliE, fliL, and fleSR promoters. No sequence conservation among these binding sites was observed. Primer extension analysis revealed the transcription start sites (TSSs) to be localized above the FleQ binding sites in flhA, fliE, and fliL promoters. Analysis of the above data revealed FleQ binding to be in the leader sequence of these promoters, whereas FleQ binding was 67 bp upstream of the TSS in the fleSR promoter. Mutagenesis of the FleQ binding site in the flhA promoter confirmed its functionality in vivo. Deletion of the flhA promoter upstream of the RNA polymerase binding site did not result in a significant loss of promoter activity. These results point to two modes of regulation by an NtrC-type regulator in the flagellar hierarchy in P. aeruginosa, the first being the typical model of activation from a distance via looping in the fleSR promoter and the second involving flhA, fliE, and fliL promoters, where FleQ binds in the downstream vicinity of the promoter and activates transcription without looping.
In gram-negative bacteria, the alternate sigma factor σ54, working in concert with transcriptional activators that belong to the NtrC superfamily, activates a variety of genes that are regulated in response to external stimuli. For example, in various bacteria, σ54 is required for expression of the enzymatic pathways responsible for nitrogen utilization, dicarboxylate transport, xylene degradation, and hydrogen utilization (7, 16, 19, 21, 35). σ54-regulated genes may be involved in RNA modification (13), chemotaxis, development, energy transduction, fructose assimilation (21), response to heat and phage shock (33), and expression of alternate sigma factors such as σH (rpoH) (23) and σS (rpoS) (29). In Pseudomonas aeruginosa, σ54 is also involved in the regulation of expression of virulence factors including pilin (14), flagellin (31), and alginate (34). There is no obvious theme in the repertoire of functions carried out by σ54-dependent transcripts.
Flagellar biogenesis in P. aeruginosa involves more than 40 genes intertwined in a complex regulatory cascade. Its flagellar hierarchy appears to be different from the FlhDC-dependent Salmonella enterica serovar Typhimurium hierarchy (20) and resembles more closely that in Vibrio cholerae, which involves both σ28- and σ54-dependent genes (24). FleQ, a NifA/NtrC-type σ54-dependent activator, is at the highest level of the flagellar hierarchy in P. aeruginosa. Homologues of FleQ have been identified in Caulobacter crescentus (25), V. cholerae (17), and Helicobacter pylori (28) and play important roles in flagellar protein synthesis and secretion. Structurally FleQ lacks the highly conserved phospho-acceptor Asp54 and instead has a serine residue. Phosphorylation could also occur at the serine residue, but lack of evidence to that effect and absence of any cognate sensor kinase probably indicate that FleQ does not require phosphorylation for its activation (9). This is also observed in NifA of Klebsiella pneumoniae, which lacks an N-terminal phospho-acceptor domain, and FlrA of V. cholerae, which lacks the same aspartate residue.
Previous studies showed that FleQ positively regulates many flagellar genes (1, 2, 10, 31). These include flhA and fliLMNOPQ involved in flagellar export; flhF involved in the localization of the flagellar apparatus; fleSR, a two-component sensor and regulator involved in flagellin synthesis; fliEFG, encoding the flagellar basal body MS ring and motor switch complex; fliDS, encoding the flagellar cap and export proteins; and flgA, involved in P-ring formation of the flagellar basal body (N. Dasgupta, unpublished data). The mechanism by which FleQ activates these flagellar genes has not been elucidated. Also, it is unknown whether FleQ acts in the typical manner as other NtrC-like regulators by binding to consensus upstream activating enhancer elements. In this study we have randomly selected four of the above promoters and identified the binding sites of FleQ in order to derive a consensus, if evident. Four different FleQ binding sites were identified which revealed a lack of any consensus sequence among these sites. Furthermore, three of the sites were located downstream in close proximity to the σ54 RNA polymerase (RNAP) binding sites, and the other was located in the typical upstream position for NtrC-like regulators. This suggests two varied mechanisms of σ54-dependent activation in the flagellar regulation in P. aeruginosa. One fits the paradigm of transcription initiation by upstream binding of activators and their interaction with bound RNAP via looping, and in the second novel mechanism the activator binds adjacent to the polymerase site and probably touches the RNAP without looping.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were maintained at −70°C in Luria-Bertani (LB) medium containing 25% glycerol. All strains were propagated in LB at 37°C at 250 rpm or on LB agar plates unless stated otherwise. P. aeruginosa strains harboring plasmids were grown with either 300 μg of carbenicillin/ml, 100 μg of tetracycline/ml, or 300 μg of streptomycin/ml, while the plasmid-containing Escherichia coli cells were grown with either 200 μg of ampicillin/ml or 25 μg of tetracycline/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | hsdR recA lacZYA f80 lacZDM15 | GIBCO-BRL |
| XL1 Blue | recA1 endA1 gyrA96thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZDM15 Tn10 (Tetr)] | Stratagene |
| P. aeruginosa strains | ||
| PAK | Wild-type clinical isolate | D. Bradley |
| PAK-Q | PAK fleQ::Gmr | 1 |
| PAK-NIG | PAK rpoN::Gmr | 15 |
| Plasmids | ||
| pDN19lacΩ | Promoterless lacZ oriV oriT Tetr Strr Ω fragment | 30 |
| placΩA | pDN19lacΩ containing a 588-bp Eco-Bam fragment of the flhA promoter region | 9 |
| placΩA(Δ1-362bp) | pDN19lacΩ containing a 231-bp Eco-Bam fragment of the flhA promoter region | This study |
| placΩE | pDN19lacΩ containing a 332-bp Eco-Bam fragment of the fliE promoter region | 3 |
| placΩS | pDN19lacΩ containing a 355-bp Eco-Bam fragment of the fleSR promoter region | 1 |
| placΩL2 | pDN19lacΩ containing a 412-bp Eco-Bam fragment of the fliLMNOPQ promoter region | This study |
| pUC-SR | pUC19 containing insert of placΩS | This study |
| pUC-E | pUC19 containing insert of placΩE | This study |
| pBSK-A | pBSK containing insert of placΩA | This study |
| pBSK-L2 | pBSK containing insert of placΩL2 | This study |
| pBSK-AmutA | pBSK-A but with mutA in the FleQ binding site | This study |
| placΩ-AmutA | placΩA but with mutA in the FleQ binding site | This study |
| pIH1119 | oriV in pGEM3Zf(+) | NEBa |
| pIH-fleQ | FleQ clone into Eco-Bam sites of pIH1119 to express MBP-FleQ | 10 |
NEB, New England Biolabs.
Construction of recombinant plasmids.
In order to obtain high plasmid yields, inserts from placΩA (9), placΩE (3), and placΩS (1), containing the flhA, fliE, and fleSR promoters, were cloned into EcoRI-BamHI (E-B) sites of pBSK or pUC19 to yield pBSK-A, pUC-E, and pUC-SR, respectively. A two-step PCR using P. aeruginosa PAK genomic DNA and primers 5PfliLβgal and JJ37 (Table 2) was performed to amplify a 412-bp fragment containing the fliLMNOPQ promoter region. The reaction cocktail consisted of primers at a concentration of 0.2 mM (Genomechanix, Alachua, Fla.), 5 mM MgCl2, 4% dimethyl sulfoxide, 100 ng of template, 1× buffer, and 3 U of Taq DNA polymerase (Gibco-BRL)/100 μl. The template genomic DNA used was purified using cetyltrimethylammonium bromide (4). The template was initially denatured at 95°C for 5 min followed by 35 cycles at 95°C for 1 min and 70°C for 2 min. A final extension for 10 min was done at 72°C. PCR products were analyzed on a 0.7% agarose gel, eluted, digested with E-B, and cloned in pBSK or pDN19lacΩ vectors.
TABLE 2.
Primers used in this study
| Primer name | Primer sequencea |
|---|---|
| JJ2 | 5′ gatccATGGCGGGATCGGCAG 3′, part of BamHI site |
| JJ5 | 5′ gatccCGCATTTCCAGCATC 3′, part of BamHI site |
| JJ8 | 5′ gatccCGTTGAGGGCTGGTTGC 3′, part of BamHI site |
| JJ26 | 5′ GGTTGCTGCGCACATTGCTGATCAGTTGC 3′ |
| JJ27 | 5′ CCAGCATCAGACGATTGAACTCGACACCC 3′ |
| JJ48 | 5′ CTCTTGCCGCCGCTCGGCGCCCCCGCG 3′ |
| JJ37 | 5′ cccaaaggatccCAGGATCAGCTTCAGCTTG 3′, BamHI site incorporated |
| JJ38 | 5′ gatccCAGGATCAGCTTCAGCTTGCTCTTG 3′, part of BamHI site |
| JJ39 | 5′ CATTCCGGCGTtctcAGTTTGTTCGGC 3′ |
| JJ40 | 5′ GCCGAACAAACTgagaACGCCGGAATG 3′ |
| 5PfliLβgal | 5′ cccaaagaattcCTCGGGCGATGAGGAAC 3′, EcoRI site incorporated |
| JJ61 | 5′ cccaaagaattcAACCCTTTTCAATCAATGAATTG 3′ |
| RER62 | 5′ cccaaaggatccATGGCGGGATCGGCAGGG 3′ |
Lowercase letters denote nucleotides added or modified to facilitate restriction digestion or mutagenesis.
Transformations and electroporations.
E. coli transformations were performed using the RbCl2 method (27a). Column-purified plasmids were used for electroporation in P. aeruginosa as described previously (10).
β-Galactosidase assay.
The β-galactosidase activity was measured by a modified method of Miller (22) as described previously (1). The strains were grown to late log phase (A600 of 0.7 to 1.0) in LB medium containing streptomycin.
Expression and purification of MBP-FleQ fusion protein.
FleQ was overexpressed as a fusion protein with maltose binding protein (MBP) in E. coli as described elsewhere (9), using a previously constructed gene fusion of fleQ with the malE gene of E. coli in pIH1119 to yield pIH-fleQ. To overexpress MBP-FleQ, E. coli DH5α containing pIH-fleQ was grown at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside induction, and MBP-FleQ was purified using affinity chromatography as described in the product manual (New England Biolabs, Beverly, Mass.). Eluted fractions were electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel to check for the presence and purity of MBP-FleQ. The eluted protein was ≥95% pure; it was dialyzed at 4°C against 10 mM Tris (pH 7.9), 5% glycerol, and 1 mM dithiothreitol and quantitated, and aliquots were stored at 4°C.
Gel-shift assay.
Binding of FleQ to flhA, fliE, fleSR, fliL, and flhAmutA promoters was studied using E-B inserts from plasmids pBSK-A, pUC-E, pUC-SR, pBSKL2, and pBSK-AmutA, respectively. These promoter fragments were radiolabeled by fill-in using Klenow polymerase (Promega Inc., Madison, Wis.) and [α-32P]dATP. The promoter probes were purified by passing through a G-50 Sephadex microspin column (Amersham Pharmacia Biotech Inc.). Various amounts of MBP-FleQ were incubated with the promoter probes (≈1,500 cpm) in a binding reaction mixture containing 1 μl of deoxyinosine-deoxycytosine (2 μg/μl), 0.5 μl of bovine serum albumin (10 mg/ml), and 0.8 μl of magnesium acetate (0.1 M) to a final volume of 20 μl with 10 mM Tris (pH 7.9). The reaction mixtures were incubated on ice for 30 min and separated on a 4% low-ionic-strength polyacrylamide gel with 8 mM MgCl2 in Tris-acetate-EDTA buffer at 4°C.
Sequencing.
Sanger's dideoxy sequencing was performed using [α-32P]dATP with unlabeled primer on a denatured plasmid template as described in the manufacturer's protocol (Sequenase DNA sequencing kit, version 2.0; USB Corporation, Cleveland, Ohio).
DNase I footprinting.
DNA probes labeled at the BamHI end (EB∗) were made by linearizing the plasmids (pBSK-A, pUC-E, pUC-SR, and pBSK-L2) with BamHI and dephosphorylating the ends by using calf intestinal alkaline phosphatase (Promega). The enzyme was heat inactivated, and the digested products were extracted by phenol chloroform and ethanol precipitated. BamHI-digested dephosphorylated DNA was end labeled with [γ-32P]ATP using T4 polynucleotide kinase at 37°C for 30 min. The reaction was stopped by adding 8 mM EDTA, followed by phenol extractions and ethanol precipitation. DNA fragments radiolabeled at the BamHI end only were released by digestion with EcoRI and purification of the DNA probe by electrophoresis and gel elution (Strataprep kit; Stratagene, La Jolla, Calif.). The binding reactions were performed as described above using 20,000 to 60,000 cpm of EB∗ probe in a 40-μl reaction volume. After 30 min on ice, 1.5 μl of 1:10 diluted RQI DNase (Promega) was added for 1 min at 30°C. The reaction was stopped by adding 20 mM EGTA (Sigma), extracted by phenol-chloroform, and ethanol precipitated. Samples were suspended in stop solution (Sequenase, version 2.0). A sequencing reaction with a primer from the BamHI site was run alongside the footprinted samples. All samples were heated to 95°C for 3 min and subjected to electrophoresis in a denaturing 8% acrylamide-8 M urea sequencing gel.
Primer extension analysis.
Primer extension analysis was performed as described previously (11). Briefly, RNA was prepared from PAK and PAK-Q mutant strains by using Trizol reagent according to the manufacturer's protocol. The primer (7.5 μM) was labeled at the 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase at 37°C for 30 min. The kinase was heat inactivated by adding 40 mM EDTA (pH 7.5), and the labeled primer was purified through a G-25 spin column (Amersham Pharmacia Biotech, Inc). The labeled primer was annealed to 50 μg of RNA in first-strand buffer and RNaseOUT RNase inhibitor at 65°C for 1 h. After slow cooling to room temperature, deoxynucleoside triphosphates (0.5 mM), dithiothreitol (10 mM), RNaseOUT, and SuperscriptII (RNase H− RT) were added for reverse transcription at 42°C for 1 h. RNase H treatment at 37°C for 20 min after cDNA synthesis was followed by phenol-chloroform extractions and ethanol precipitations. DNA was resolved on an 8% polyacrylamide-8 M urea gel.
Site-directed mutagenesis.
Primers JJ39 and JJ40, each containing four base mutations (see Fig. 4B), were used in the QuikChange site-directed mutagenesis kit (Stratagene) to mutate the FleQ binding site in the flhA promoter (the mutation referred to as mutA). The primers used in this study are listed in Table 2. Briefly, 20 ng of column-purified (QIAGEN, Valencia, Calif.) pBSK-A was used in a 50-μl amplification reaction mix containing 0.1 mM concentrations of deoxynucleoside triphosphates, 2.5 U of Pfu Turbo DNA polymerase, 125 ng of each primer, and 1× buffer. The cycling parameters used were as follows: initial denaturation at 95°C for 30 s followed by 18 cycles of denaturation (95°C for 30 s), annealing (55°C for 1 min), and extension (68°C for 10 min). This was followed by a final extension at 72°C for 10 min. Aliquots of the amplified products were examined by gel electrophoresis. The contents were then treated with DpnI to digest the parent plasmid template. One microliter of the above digested mix was used to transform E. coli XL1 blue cells, and transformants were selected on LB-ampicillin plates. The mutation was confirmed by sequencing using the forward and reverse universal primers.
FIG. 4.
(A) Promoter organization of flhA, fliE, and fliL flagellar promoters of P. aeruginosa. (B) Sequence comparison of the FleQ binding sites obtained by DNase I footprinting analysis of fleSR, flhA, fliE, and fliL promoters. A half site of the fliE inverted repeat is underlined. There is no recognizable consensus. The mutated flhA binding site is shown in bold, and the residues of the wt flhA promoter that have been mutated are underlined.
Deletion analysis.
To conclusively rule out the presence of any additional FleQ binding sites upstream of the RNAP binding site, the flhA promoter was deleted upstream of the RNAP binding site. Primers JJ61 and RER62 were used to PCR amplify a 231-bp fragment from pBSK-A. After digestion with E-B, this fragment was cloned in pDN19lacΩ to generate placΩA(Δ1-362). The β-galactosidase activity of placΩA(Δ1-362) was compared to that of PAK wild type (wt) (placΩA) and the PAK lacΩ control.
RESULTS
FleQ and RpoN positively regulate expression of flagellar genes.
Previous studies from this laboratory showed that FleQ as well as RpoN positively regulate many flagellar genes (1, 10). Since FleQ is an NtrC-like transcriptional activator, it probably works in concert with RpoN in playing a role in activating flagellar genes. As representatives of FleQ- and RpoN-regulated genes, we randomly selected four promoters, flhA, fliE, fleSR, and fliL, for this study. The promoter regions of these genes were cloned upstream of a promoterless lacZ, and their activities were measured in PAK wt, rpoN, and fleQ mutants (1, 10) (Table 3). The results of the β-galactosidase assays confirmed that all these promoters were FleQ and RpoN regulated and that flhA was the strongest promoter. There was a dramatic reduction in the activities of all the promoters in the rpoN (2.4- to 32-fold) and fleQ (2.5- to 22-fold) mutants.
TABLE 3.
RpoN and FleQ regulate promoter activities of four flagellar genes of P. aeruginosa
| Plasmid | β-Galactosidase activity (mean ± SD in Miller units)
|
||
|---|---|---|---|
| PAK | PAK (rpoN) | PAK (fleQ) | |
| pDN19lacΩ | 24 ± 3 | 30 ± 5 | 26 ± 5 |
| placΩA | 1,314 ± 89 | 40 ± 1 | 58 ± 3 |
| placΩE | 128 ± 18 | 32 ± 5 | 47 ± 2 |
| placΩSR | 736 ± 150 | 64 ± 2 | 96 ± 9 |
| placΩL2 | 346 ± 32 | 139 ± 12 | 138 ± 10 |
FleQ directly and specifically regulates transcription at flagellar promoters.
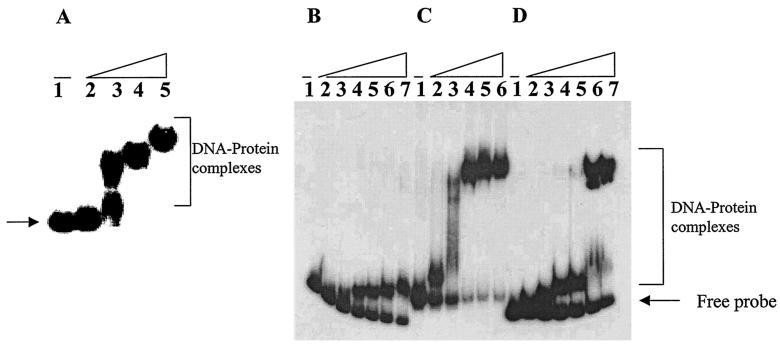
The carboxy terminus of FleQ contains a sequence similar to the helix-turn-helix motif present in many DNA binding proteins (1). To investigate its potential DNA binding ability, MBP-FleQ was expressed in E. coli and its binding to the flagellar promoters was examined by gel shift assay. Radiolabeled E-B fragments containing the promoter regions of flhA, fliE, fliL, and fleSR were incubated with various amounts of MBP-FleQ in the binding reaction buffer and assayed for formation of protein-DNA complexes (Fig. 1A, B, C, and D, respectively). Slower-migrating probe-protein complexes were observed upon incubation with increasing amounts of MBP-FleQ, which subsequently increased in molecular weight upon increases in protein amount (Fig. 1). These results indicate that MBP-FleQ recognizes and binds to sequences within these promoter-probe fragments. The complexes of retarded mobility indicate either multiple binding sites of FleQ or its oligomerization at higher protein concentrations leading to high-molecular-weight complexes.
FIG. 1.
(A) Gel shift assay to show binding of MBP-FleQ to 588-bp flhA promoter. Lane 1, free DNA probe; lanes 2 to 5, DNA incubated with 0.5, 2, 5, and 15 μM MBP-FleQ. (B to D) Gel shift assays with 332-bp fliE promoter (B), 412-bp fliL promoter (C), or 355-bp fleSR promoter (D). Lane 1, free DNA probe; lanes 2 to 6, 0.5, 1, 2.5, 3.75, and 5 μM MBP-FleQ; lane 7 (panels B and D only), 7 μM MBP-FleQ.
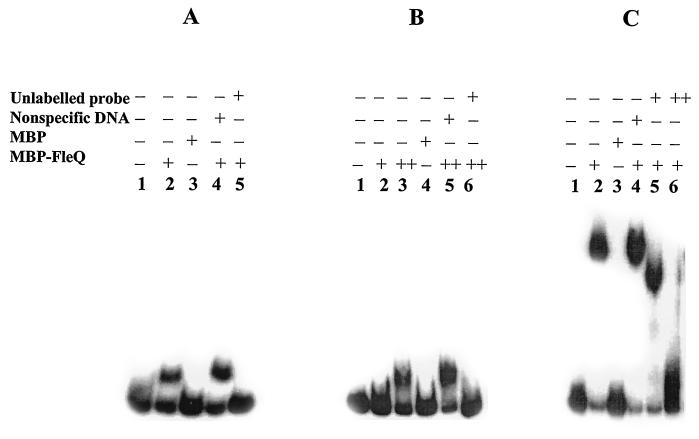
The specificity of binding by purified MBP-FleQ to promoter regions of the fleSR, fliE, and fliL promoters was examined by addition of MBP (the fusion partner in MBP-FleQ), unlabeled probe DNA (specific competitor), and herring sperm DNA (HS-DNA; a nonspecific competitor) to the binding reaction mixtures. The specificity of binding of MBP-FleQ to the flhA promoter has been shown as part of an earlier study (9). Addition of MBP did not shift the fleSR, fliE, and fliL promoter fragments (Fig. 2A, B, and C, respectively). Including HS-DNA in the binding reaction had no discernible effect on binding of FleQ to these promoters (Fig. 2). In contrast, addition of an excess of unlabeled probe DNA completely disassociated the probe DNA-protein complex formation and abolished the shift. All of the above findings support specific binding of FleQ to the promoter regions of fliE, fliL, and fleSR as well as flhA (9).
FIG. 2.
Gel shift assays showing that FleQ binding to fleSR (A), fliE (B), and fliL (C) promoters is specific. For panels A and C: lane 1, free DNA probe; lanes 2 to 5, DNA probe incubated with 2.5 μM MBP-FleQ (lane 2), 4 μM MBP (lane 3), 2.5 μM MBP-FleQ and 1 μg of HS-DNA (lane 4), or 2.5 μM MBP-FleQ with 500 ng of unlabeled probe (lane 5); lane 6 in panel C, 2.5 μM MBP-FleQ with 750 ng of unlabeled probe. For panel B (fliE promoter): lane 1, free DNA probe; lanes 2 to 6, DNA probe incubated with 2.5 μM MBP-FleQ (lane 2), 10 μM MBP-FleQ (lane 3), 10 μM MBP (lane 4), 10 μM MBP-FleQ and 2 μg of HS-DNA (lane 5), or 10 μM MBP-FleQ and 750 ng of unlabeled probe (lane 6).
Identification of FleQ binding sites by DNase I footprinting.
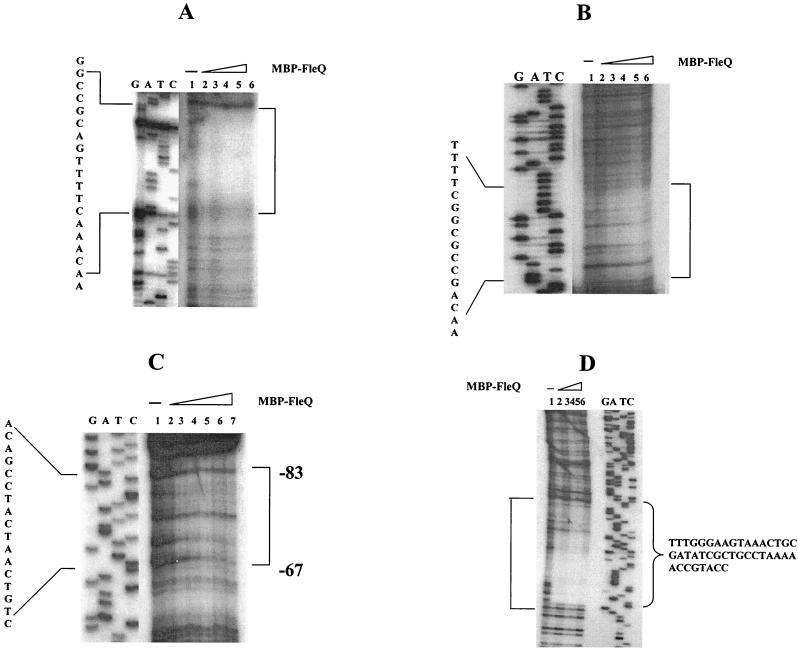
To precisely determine the FleQ binding sites in the above promoters, DNase I footprinting assays were carried out using the same E-B promoter fragments as used in gel shift assays. The E-B promoter fragments end labeled at the BamHI end on the lower nontemplate or antisense strand were incubated with MBP-FleQ (hereafter referred to as FleQ) and then partially digested with DNase I. The protected regions were compared against a no-protein control, on a denaturing polyacrylamide gel, along with a DNA sequencing ladder generated using primers beginning at the labeled promoter ends.
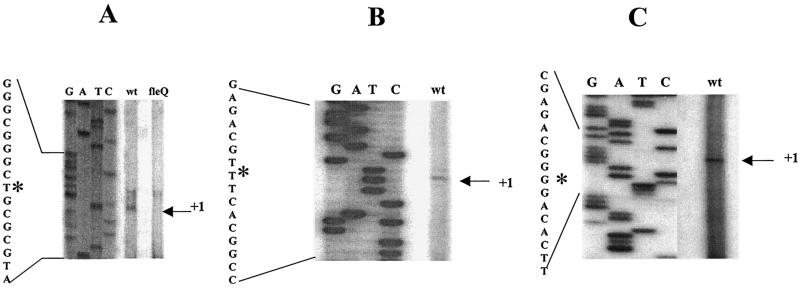
The 588-bp E-B* flhA promoter fragment gave a 19-bp protection, reading 5′-AACAAACTTTTGACGCCGG-3′, upon binding with FleQ (Fig. 3A and 4A). This region mapped 45 bp upstream of the translational start site. A similar binding site was obtained using the flhA promoter fragment labeled at the EcoRI end. Subsequently, for the other promoters only E-B* fragments showing protection on the lower strand were used. The 332-bp E-B* fliE promoter fragment showed a 16-bp protection from DNase I digestion (Fig. 3B and 4A). The FleQ binding sequence in the fliE promoter, 5′-AACAGCCGCGGCTTTT-3′, was found only 12 bp upstream of the ATG of fliE. The FleQ binding sequence was an inverted repeat sequence and had only one mismatch in the repeat sequence. This suggests that FleQ binds at the fliE promoter as a dimer. The 355-bp E-B* fleSR promoter generated a 17-bp-protected sequence upon DNase I footprinting. This 17-bp sequence extended from −67 to −83 bp upstream of the transcription start site (TSS) (Fig. 3C). The FleQ binding sequence herein was 5′-CTGTCAATCATCCGACA-3′, which matched the NifA binding sequence (27). The 412-bp E-B* fliL promoter fragment exhibited a 42-bp-long protection upon incubation with FleQ followed by DNase I digestion (Fig. 3D and 4A). This footprint overlapped and extended 1 base into the start codon of fliL. No evident consensus emerged upon comparing the above four FleQ binding sites (Fig. 4B).
FIG. 3.
DNase I footprinting analysis. Results of footprinting reactions with the lower strand labeled with [γ-32P]ATP are shown. Sequencing reactions (GATC) with the primers beginning at the labeled end are shown next to the footprinting reactions. Open brackets represent regions protected from DNase I digestion. The protected sequence is shown next to the sequencing lanes. Binding sites of FleQ on the following promoters were determined. (A) flhA promoter, using primer JJ2 for the sequencing reaction. Lane 1, free DNA probe; lanes 2 to 6, 2, 5, 7.5, 10, and 15 μM MBP-FleQ. (B) fliE promoter using primer JJ5 for the sequencing reaction. Lane 1, free DNA probe; lanes 2 to 6, 5, 10, 15, 20, and 25 μM MBP-FleQ. (C) fleSR promoter using JJ8 for the sequencing reaction. Position with respect to +1 is shown. Lane 1, free DNA probe; lanes 2 to 7, 1, 2.5, 3.75, 5, 7, and 15 μM MBP-FleQ. (D) fliL promoter using JJ38 for the sequencing reaction. Lane 1, free DNA probe; lanes 2 to 6, 2.5, 3.75, 5, 7.5, and 15 μM MBP-FleQ.
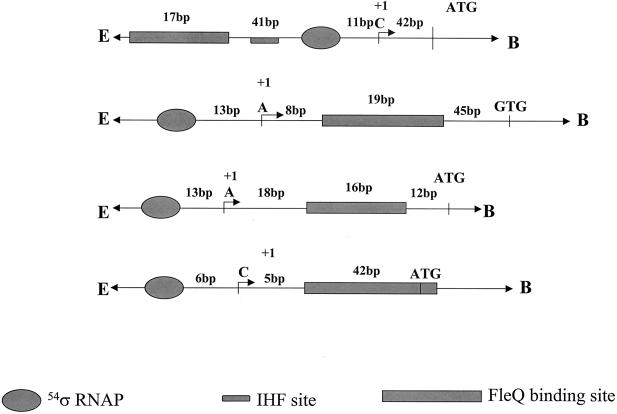
FleQ binds downstream and in close proximity to the σ54 binding site in flhA, fliE, and fliL promoters.
Since there was no commonality observed in the binding site sequences, we closely examined the positioning of these binding sites in flhA, fliE, and fliL promoters. Primer extension analyses of the flhA, fliE, and fliL promoters were performed to determine the TSSs in these promoters. The TSS for the fleSR promoter has been reported earlier and is a cytosine, 12 bp downstream of the −24/−12 RpoN binding site (1). There is also an integration host factor binding site present (−31 to −63) between the RpoN and the FleQ binding sites (−67 to −83) in the fleSR promoter (1) (see Fig. 6). In this case, the binding of FleQ represents a typical model for regulation of σ54 promoters, wherein an activator binds far upstream of the promoter and contacts σ54 by looping. Primer extension analysis of the flhA promoter was done using total RNA from the wt PAK and the fleQ mutant strains. Multiple primer pairs (data not shown) were designed to detect transcripts starting in the vicinity of the putative σ54 binding sites. With primer JJ26, two transcripts were detected in the wt, of which one was missing in the fleQ mutant. Since flhA is FleQ regulated, this transcript initiating at thymine (adenosine on the coding strand) appeared to be FleQ dependent (Fig. 5A). This TSS was 13 bp downstream of the putative σ54 binding site which had highest homology to the σ54 binding site consensus (5). The FleQ binding site in the flhA promoter is 8 bp downstream of the TSS and thus downstream of the RpoN binding site (Fig. 4A and 6).
FIG. 6.
Schematic representation of the fleSR, flhA, fliE, and fliL (top to bottom) promoter regions showing the relative positions of the FleQ binding sites with respect to the TSSs and the RpoN binding and translational start sites. The integration host factor (IHF) binding site in the fleSR promoter is shown.
FIG. 5.
Primer extension analysis of P. aeruginosa RNAs. (A) flhA promoter using primer JJ26; (B) fliE promoter using primer JJ27; (C) fliL promoter using primer JJ48. The TSSs are marked as +1 and are shown with an asterisk on the sequence readout from the sequencing lanes using the same primers. RNA was prepared from PAK wt or a fleQ mutant strain.
The primer extension analysis of fliE using primer JJ27 detected a single transcript ending at a thymine 13 bp downstream of the putative σ54 binding site with highest homology to the consensus σ54 binding site (Fig. 5B). The FleQ binding site maps 18 bp downstream of the TSS in fliE (Fig. 4A and 6), demonstrating that in this promoter, too, the FleQ binding site is downstream of the RpoN binding site.
In the fliL promoter we could detect a transcript 6 bp downstream of the best-matched putative σ54 binding site by using primer JJ48 (Fig. 5C). The FleQ binding site fell 5 bp downstream of this TSS and thus here, too, the FleQ binding was downstream of the RpoN binding site (Fig. 4A and 6).
Thus, in three of the four promoters, the FleQ binding site is located within the leader sequence, downstream of the TSS and above the translational start site (Fig. 6). Based on these findings two models of regulation in the σ54-regulated flagellar promoters in P. aeruginosa can be envisaged. The first model, fleSR, fits the typical model of regulation of activation from a distance, and the second model involves flhA, fliE, and fliL genes, where activation occurs by mechanisms different from the paradigm.
Mutagenesis of the FleQ binding site confirms functionality in vivo.
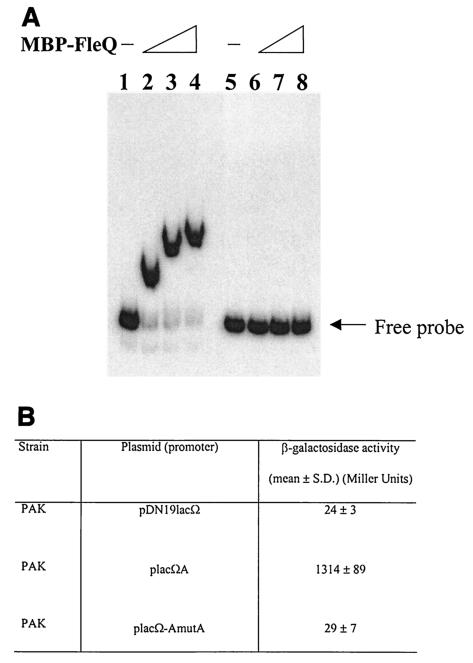
In view of the above findings, we needed to confirm the binding sites observed in the second class of promoters comprising flhA, fliE, and fliL were functional in vivo. To demonstrate this, we selected the flhA promoter for further studies since it was the strongest promoter in this group (based on the β-galactosidase assay). The FleQ binding site in the flhA promoter was mutated at four bases in roughly the core area (Fig. 4B). The sequence GTTT in the FleQ binding site of the flhA promoter was mutated to AGAG, and corresponding complementary mutations were made on the other strand (mutA). The changes in the FleQ binding site in the flhA promoter completely abolished binding of FleQ in the gel shift assay (Fig. 7A). Further confirmation was sought by comparing the β-galactosidase activities of the wild-type flhA promoter with that of the mutated flhA promoter. As shown in Fig. 7B, there was a 45-fold reduction in promoter activity. Taken together, these results consolidate our findings that FleQ binds downstream in the flhA promoter and in close proximity to the RpoN binding site and that this site is functionally active in vivo as a downstream activating element. The same probably holds for fliE, fliL, and the other FleQ-regulated promoters whose regulation is unlike that in the fleSR promoter.
FIG. 7.
(A) Gel shift analysis using the mutagenized flhA promoter (mutated at its FleQ binding site). Lanes 1 to 4 have wt flhA promoter and lanes 5 to 8 have the mutated flhA promoter. Lanes 1 and 5, free DNA probe; lanes 2 and 6, 2 μM MBP-FleQ; lanes 3 and 7, 5 μM MBP-FleQ; lanes 4 and 8, 7.5 μM MBP-FleQ. (B) β-Galactosidase assay results comparing the promoter activities of the mutagenized flhA promoter with the wt promoter.
Deletion analysis reveals lack of any additional FleQ binding site.
Some transcriptional regulators are known to bind to multiple sites for activation of target genes. Upon searching the Pseudomonas genome, no homologous sequences or sequences similar to the FleQ binding sites in the flhA, fliL, and fliE promoters were found. To conclusively rule out the possibility of additional FleQ binding sites, the region upstream of the RNAP binding site was deleted in the flhA promoter. The construct placΩA(Δ1-362) had regions −34 to +323 bp with respect to the TSS. Measurements of promoter activity using β-galactosidase assays did not result in a large difference in its activity when compared to that of the wild-type promoter in placΩA; the activity levels (mean ± standard deviation) were 23.93 ± 8.98, 1,079.57 ± 213.44, and 727.82 ± 73.02 in pDN19lacΩ, placΩA, and placΩA(Δ1-362), respectively.
DISCUSSION
FleQ is at the top of the flagellar regulatory cascade in P. aeruginosa.
Identification of its binding sites and derivation of a consensus binding site were the major objectives of this study. Four promoters (flhA, fliE, fliL, and fleSR) that were RpoN and FleQ regulated were selected for this study. Binding of FleQ directly to these promoters was demonstrated in a gel shift assay. FleQ appears to be a weak DNA binding protein, since very large amounts of FleQ were required for its binding to the promoter fragments in vitro. This could be due to the lack of a glycine residue that plays an important role in the C-terminal helix-turn-helix motif DNA binding domain (LR465 instead of LG). Another plausible explanation could be that the conditions for binding were not perfectly optimal. However, the binding was convincingly specific, since MBP could not cause a shift, nor could HS-DNA abolish the shift. It was only with an excess of specific promoter fragment that the probe-protein complex was totally disassociated.
DNase I footprinting showed that FleQ had different binding sites in the promoters that were studied. Of note there exists a certain sequence homology between the FleQ binding site in fliL and flhA sequences when aligning flhA with the reverse-complement sequence of fliL. This sequence is also coincident with the region that was mutated in mutA. In fliE, there was an almost-perfect inverted repeat, but there were no direct or inverted repeats found in FleQ binding sites of other promoters. The FleQ binding site in fleSR matched that of the NifA binding site. We searched the sequences of the other flagellar operons in the P. aeruginosa genome database (http://www.pseudomonas.com) for the presence of any other NifA-like binding sites, but we could not find any matches. Homology searches were also done with the FleQ binding sequences of flhA, fliE, and fliL promoters. No other homologous sequences could be identified among the flagellar genes, reaffirming our conclusion about the lack of any consensus sequence among FleQ binding sites. It is therefore possible that FleQ does not recognize a limited number of specific bases but rather a sequence with a specific secondary structure. Indeed, certain protein-DNA interactions have been reported to be the result of conformational differences in DNA structure instead of a specific nucleotide sequence (32).
There were several putative σ54 binding sites in the flhA, fliE, and fliL promoter regions, based on homology with the consensus σ54 sequence (5). Therefore, without a primer extension analysis it was impossible to dissect the regulation of these genes. The TSSs were found upstream of the FleQ binding sites in the flhA, fliE, and fliL promoters (Fig. 5 and 6). This is a unique organization not seen for NtrC-like regulators, where FleQ binding sites were part of the leader sequence and bound in close proximity to the σ54 binding site. Since the σ54-bound RNAP and FleQ would be placed adjacent to one another, the possibility of any looping mechanism would be remote. FleQ could interact with σ54 RNAP directly, contacting the polymerase either with the FleQ molecule bound to DNA or by contacting through FleQ oligomers. In the gel shift assays, we did see oligomerization of FleQ; therefore, it is possible that protein may be bound by protein-protein interactions in addition to direct binding to DNA (36). These protein-protein-interacting FleQ molecules could trigger the open complex formation by increasing the local concentration of the activator molecules near the promoter (18).
In algC and algD promoters in P. aeruginosa, three binding sites each are found for the AlgR1 regulator (12). These binding sites range from far upstream of the promoters to downstream and also in the structural genes. However, the sites closest to and downstream of the promoter are the weakest binding sites, and the far upstream site offers the strongest activation (12). This is in contrast to our findings, where we observe single binding sites which are not weak activator sites, based on our β-galactosidase results. The presence of an enhancer element located downstream of the major glutamate dehydrogenase gene (rocG) of Bacillus subtilis has been reported (6), but this element is located beyond the end of the rocG coding region. We are unaware of any reports describing the location of functional enhancer elements adjacent to the RNAP binding site.
The enhancer-like activity of NtrC binding sites confers a high level of activation of the glnAp2 promoter when it binds tightly to an intact high-affinity site(s) present on either side of the DNA helix, as long as the binding sites are separated from the RNAP binding site by at least 30 bp (26). In the case of flhA, fliE, and fliL promoters, we see binding sites downstream of the promoter, and in two of these they are at a distance of less than 30 bp from their promoters. This implies that there is a different model for their regulation. The distance between the TSS and the FleQ binding site in fleSR, 67 bp upstream, is more typical of NtrC-like activators.
Most binding sites present within transcriptional units belong to repressors. Rarely are activators known to bind in these locations and, when they do, additional binding sites far upstream or downstream of the coding region accompany them. We did not find any other FleQ binding sites similar to those reported on searching the Pseudomonas genome sequence. We could attribute functional activity of these promoters to the binding sites by the β-galactosidase assay results, since mutations of a few bases in the binding site of flhA could drastically drop the activity of the promoter and binding of FleQ was lost, as seen in the gel shift assay. Deleting the promoter region upstream of the RNAP binding site in the flhA promoter resulted in no significant loss of promoter activity, indicating that there are no FleQ binding sites upstream of the RNAP binding site. A recent report showed that enhancer-promoter interaction and the initiation complex must be formed de novo during each round of transcription and that no protein remains bound to the promoter after RNAP escapes into elongation (8). In the light of the above findings, the probability that FleQ would remain bound in the vicinity of the promoter, blocking transcription, would seem remote.
In summary, this paper describes two deviations from that seen in activation by NtrC-like regulators, i.e., the lack of consensus sequences for activation and the atypical location of probably the majority of enhancer binding sites. Whether this is peculiar to FleQ of P. aeruginosa is unclear, since such information does not exist for most of its homologues. The proposed model would suggest the existence of other pathways by which smaller organisms with compact genomes could use σ54 and circumvent the need for activation across large distances.
Acknowledgments
We thank S. K. Arora for helpful discussions and critical reading of the manuscript.
This work was supported by NIH grant AI 45014 to R.R.
REFERENCES
- 1.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1996. Cloning and characterization of Pseudomonas aeruginosa fliF, necessary for flagellar assembly and bacterial adherence to mucin. Infect. Immun. 64:2130-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 5.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 22:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belitsky, B. R., and A. L. Sohenshein. 1999. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:10290-10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, L. K., and R. J. Maier. 1995. IHF- and RpoN-dependent regulation of hydrogenase expression in Bradyrhizobium japonicum. Mol. Microbiol. 16:405-413. [DOI] [PubMed] [Google Scholar]
- 8.Bondarenko, V., Y. Liu, A. Ninfa, and V. M. Studitsky. 2002. Action of prokaryotic enhancer over a distance does not require continued presence of promoter-bound σ54 subunit. Nucleic Acids Res. 30:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasgupta, N., and R. Ramphal. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 183:6636-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisk, A., J. Jyot, S. K. Arora, and R. Ramphal. 2002. Identification and functional characterization of flgM, a gene encoding the anti-sigma 28 factor in Pseudomonas aeruginosa. J. Bacteriol. 184:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara, S., N. A. Zielinski, and A. M. Chakrabarty. 1993. Enhancer-like activity of A1gR1-binding site in alginate gene activation: positional, orientational, and sequence specificity. J. Bacteriol. 175:5452-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genschik, P., K. Drabikowski, and W. Filipowicz. 1998. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J. Biol. Chem. 273:25516-25526. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg, J. B., and T. Dahnke. 1992. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol. Microbiol. 6:59-66. [DOI] [PubMed] [Google Scholar]
- 15.Ishimoto, K. S., and S. Lory. 1989. Formation on pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc. Natl. Acad. Sci. USA 86:1954-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler, B., S. Marqués, T. Köhler, J. L. Ramos, K. N. Timmis, and V. de Lorenzo. 1994. Cross talk between catabolic pathways in Pseudomonas putida: XylS-dependent and -independent activation of the TOL meta operon requires the same cis-acting sequences within the Pm promoter. J. Bacteriol. 176:5578-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 18.Kustu, S., A. K. North, and D. S. Weiss. 1991. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem. Sci. 16:397-402. [DOI] [PubMed] [Google Scholar]
- 19.Macaluso, A., E. A. Best, and R. A. Bender. 1990. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J. Bacteriol. 172:7249-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 21.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor σ54 (σN). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Pallen, M. 1999. RpoN-dependent transcription of rpoH? Mol. Microbiol 31:393.. [DOI] [PubMed] [Google Scholar]
- 24.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 25.RamaKrishnan, G., and A. Newton. 1990. FlbD of Caulobacter crescentus is a homologue of the NtrC (NRI) protein and activates sigma 54-dependent flagellar gene promoters. Proc. Natl. Acad. Sci. USA 87:2369-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reitzer, L. J., B. Movsas, and B. Magasanik. 1989. Activation of glnA transcription by nitrogen regulator I (NRI)-phosphate in Escherichia coli: evidence for a long-range physical interaction between NRI-phosphate and RNA polymerase. J. Bacteriol. 171:5512-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchings, B. W., E. C. Almira, S. Lory, and R. Ramphal. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is co-coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studholme, D. J., and M. Buck. 2000. Novel roles of σN in small genomes. Microbiology 146:4-5. [DOI] [PubMed] [Google Scholar]
- 30.Totten, P. A., and S. Lory. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 12:7188-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Travers, A. A. 1989. DNA conformation and protein binding. Annu. Rev. Biochem. 58:427-452. [DOI] [PubMed] [Google Scholar]
- 33.Weiner, L., J. L. Brissette, and P. Model. 1991. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on σ54 and modulated by positive and negative feedback mechanisms. Genes Dev. 5:1912-1923. [DOI] [PubMed] [Google Scholar]
- 34.Woods, D. E., D. C. Straus, W. G. Johanson, Jr., V. K. Berry, and J. A. Bass. 1980. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect. Immun. 29:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, Z.-L., T. C. Charles, H. Wang, and E. W. Nester. 1992. The ntrA gene of Agrobacterium tumefaciens: identification, cloning, and phenotype of a site-directed mutant. J. Bacteriol. 174:2720-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyman, C., I. Rombel, A. K. North, C. Bustamante, and S. Kustu. 1997. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science 275:1658-1661. [DOI] [PubMed] [Google Scholar]