Abstract
The two genes encoding DNA gyrase in Mycobacterium tuberculosis are present next to each other in the genome, with gyrB upstream of gyrA. We show that the primary transcript is dicistronic. However, in addition to the principal promoter, there are multiple weaker promoters that appear to fine-tune transcription. With these and other mycobacterial promoters, we propose consensus promoter sequences for two distinct sigma factors. In addition to this, the gyr genes in M. tuberculosis, as in other species, are subject to autoregulation, albeit with slower kinetics, probably reflecting the slower metabolism of the organism.
Most of our understanding of prokaryotic transcription initiation is based on extensive analysis of promoter architecture in Escherichia coli. Since the regions in σ70 involved in contacting the promoter show extensive conservation across the prokaryotic world (13), a similar picture for transcription initiation is expected in all bacteria. However, this does not appear always to be the case. For instance, results of earlier random promoter screens indicate that only a small fraction of mycobacterial promoters are recognized by the E. coli machinery (6, 26). Furthermore, a random promoter screen in Mycobacterium paratuberculosis detected only promoters that were highly GC-rich in both their −10 and −35 regions (2). Thus, the features that define species-specific promoters are not clear.
Here we present the analysis of the transcription of the DNA gyrase genes in Mycobacterium tuberculosis. As the sole supercoiling activity in the cell, DNA gyrase faces the daunting task of opposing the relaxing activities of both topoisomerases I and IV (29). As a result, DNA gyrase is essential in all eubacterial cells that have been tested so far, and the final topology of DNA is maintained by the equilibrium achieved by these divergent forces. Since DNA gyrase needs to oppose the relaxation induced by other topoisomerases, it regulates its own synthesis by a unique mechanism. In general, transcription of most genes is induced by increased negative supercoiling. In contrast, negative supercoiling represses transcription of the gyrase genes in E. coli (15). This phenomenon, referred to as relaxation-stimulated transcription, is believed to be the cell's strategy to homeostatically maintain the topology of DNA (15). Thus, increased gyrase levels lead to an increase in supercoiling, which, in turn, repress the expression of gyrase and allow other topoisomerases to bring the topology of the DNA back to its optimum state. Relaxation-stimulated transcription appears to be conserved in all organisms tested so far (14, 23, 25, 28); however, the underlying mechanism appears to vary (27).
Therefore, there are multiple reasons to analyze the transcription of the gyr genes in M. tuberculosis, especially since the genome lacks both topoisomerases III and IV (5). In addition, since the expression of many virulence genes is dependent on the topology of DNA in many pathogenic bacteria (8), understanding the regulation of DNA gyrase in M. tuberculosis might help decipher the various players involved in the infection process. Our analysis revealed that while the majority of the gyr message is dicistronic, additional promoters are present that appear to be regulatory in function. From these as well as other promoters identified previously in mycobacteria, we have developed two potential consensus sequences specific to mycobacterial promoters. In addition to this, we found that although the gyr genes were subject to relaxation-stimulated transcription, the kinetics of the process was significantly slower than in other species such as E. coli and Mycobacterium smegmatis, probably reflecting the overall slow metabolism of the organism.
MATERIALS AND METHODS
Bacterial strains and transformation.
E. coli strain DH10B was used for all cloning experiments and as the E. coli host for the chloramphenicol acetyltransferase (CAT) assays. M. tuberculosis H37Ra was used for the promoter mapping experiments. M. smegmatis mc2155 was used as the mycobacterial host for all the CAT assays. The E. coli cells were grown in Luria-Bertani (LB) medium. The mycobacterial cells were grown in modified Youmans and Karlson's medium (17) with 2% glycerol and 0.2% Tween 80. Kanamycin was added at 35 μg/ml where appropriate. The E. coli DH10B cells were transformed by the standard calcium chloride method (22). The M. smegmatis cells were transformed as described before (7). After transformation, the cells were plated on LB agar containing 0.5% glycerol with kanamycin (35 μg/ml), either alone or in combination with chloramphenicol (25 μg/ml).
RNA isolation, RT-PCR, and primer extension.
For RNA isolation, M. tuberculosis cells were grown for 15 days with intermittent shaking (≈1.0 optical density unit at 600 nm), harvested, and resuspended in Trizol reagent (Gibco-BRL). RNA was isolated as described previously (28). Primer extension was performed with Superscript II reverse transcriptase (Gibco-BRL) with appropriate primers (primer A for PA, B for PB1, and R for PR). Briefly, 2 μg of total RNA was mixed with 10 pmol of end-labeled specific primer, denatured at 95°C for 10 min, and quick chilled on ice immediately. After adding the reaction buffer, deoxynucleoside triphosphates (500 μM each), 10 mM dithiothreitol, and 10 U of pancreatic RNase inhibitor (Gibco-BRL), samples were incubated at 50°C for 2 min. The reaction was started with the addition of 200 U of Superscript II.
For reverse transcription (RT)-PCR, first-strand synthesis was performed with Superscript II reverse transcriptase and primer A as described above. Then 1/10th of the reaction was subjected to PCR with primers A and C with Taq polymerase, in two parts. For the first five cycles, the annealing was at 45°C, followed by 25 cycles with annealing at 55°C. The primer A sequence was 5′-TCGACCGGTTCGATCCGGTC-3′, that of primer B was 5′-CACCATGAATTCCTCGGTTCGTGTG-3′, that of primer C was 5′-CAGCCACGATCCGAATACTC-3′, and that of primer R was 5′-CGAAGCGAATTCGTATGCCGGACGTC-3′.
For induction by novobiocin, M. tuberculosis cells were grown for 15 days with intermittent shaking. Cultures were shifted to a water bath for continuous shaking. After allowing 24 h for adaptation, the cells were treated with 100 μg (final concentration) of novobiocin per ml. Aliquots were taken every 12 h, harvested, resuspended in Trizol reagent (Gibco-BRL), frozen in liquid nitrogen, and stored at −70°C. RNA was isolated as described previously (28).
DNA manipulation.
Putative promoter fragments were cloned at the BamHI site in the promoter selection shuttle vector pSD7 (7) for testing promoter strengths. pTUN1 and pTUN2 contain a 1.5-kb BamHI fragment from the region upstream of gyrB. pTUN3 and pTUN4 contain a 1.5-kb BamHI fragment including 100 bp upstream of gyrB and 1.4 kb of the gyrB gene itself. pTUN5 and pTUN6 contain a 900-bp BamHI fragment that includes 200 bp upstream of gyrA and 700 bp of the gyrA gene itself. All odd-numbered clones have the promoter elements in the correct orientation, while the even-numbered clones have them in the reverse orientation.
CAT assays and immunoblot analysis.
CAT assays were performed with exponentially growing M. smegmatis cells as described previously (28). For immunoblotting, 10 μg of the crude cell extract was resolved by 1% sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis and electroblotted onto a polyvinylidene difluoride membrane. The blots were probed with polyclonal antibodies (1:5,000) raised in rabbit against M. tuberculosis GyrA or GyrB. The blot was developed with secondary antibody conjugated with horseradish peroxidase (1:2,000; Sigma Chemicals). For GyrA, 3-amino-9-ethylcarbazole was used as the substrate, and for GyrB, the ECL-Plus system (Amersham Pharmacia Biotech) was used.
Sequence analysis.
To develop a consensus for promoter elements for mycobacteria, 82 promoters for which the transcription start site had been experimentally defined were selected from the literature. Individual promoters were iteratively clustered into multiple groups. The final two groups of promoters included 80 of these promoters. The frequency of occurrence of different bases at individual positions was used to generate a consensus matrix. From this matrix, a simplified consensus was developed by selecting bases that were statistically overrepresented. Overrepresentation was determined by performing a χ2 test while taking into account the high GC content of mycobacterial genomes. Therefore, for instance, a 30% occurrence would not be considered significant for a G or C at a given position but would be considered significant for an A or T. The entire list of mycobacterial promoters and their analysis is available in the form of supplementary material upon request.
RESULTS
Mapping the transcription start site in the gyr locus.
The active DNA gyrase is composed of two subunits, GyrA and GyrB, products of separate genes, that form an A2B2 heterotetramer (20). The genomic arrangement of the genes that encode these two subunits varies greatly among different bacteria. For instance, in E. coli, while the gyrB gene is close to oriC, gyrA is almost at the diametrically opposite end (1). On the other hand, the genomes of many gram-positive organisms, including several mycobacterial species, have the two genes close to each other near oriC, with gyrA present downstream of gyrB in the vicinity of the chromosomal origin of replication (21). However, despite their proximity, in some organisms such as Bacillus subtilis, the two genes are transcribed independently (12), while in others, such as Borrelia burgdorferi (11) and M. smegmatis, they are part of a single dicistron (28).
The gyr genes in M. tuberculosis are located close to the origin of replication, with gyrB present 34 nucleotides upstream of gyrA (24). The short intergenic region is devoid of promoter or terminator-like features, implying that the genes are part of a single transcript, as in M. smegmatis (28). To identify the potential promoters upstream of gyrA and gyrB, primer extension analysis was performed with primers specific to gyrA and gyrB (Fig. 1). In contrast to the result with M. smegmatis, both reactions generated specific products, indicating that each gene is transcribed independently by its own promoter, located approximately 60 nucleotides upstream of the respective start codons. Interestingly, the putative promoter elements of the two genes were strikingly different (discussed later).
FIG. 1.
Primer extension analysis to map transcription start sites upstream of gyrB (B) and gyrA (C). (A) Schematic of the gyr operon in M. tuberculosis. Arrowheads represent the primers used for the analysis. The extension product corresponding to the transcription start site for each promoter is indicated. The sequencing lanes were used as markers. RNA was prepared from exponentially growing M. tuberculosis cells.
Promoter activity in E. coli and M. smegmatis.
To functionally test these promoters and to determine their relative strengths, fragments encompassing each promoter (Fig. 2) were cloned in the promoter selection vector pSD7 (7). All E. coli transformants were sensitive to chloramphenicol, and cell extracts from these transformants did not show any detectable CAT activity. On the other hand, the M. smegmatis transformants were resistant to chloramphenicol to different extents (compare pTUN3 and pTUN5 in Fig. 2). These results were paralleled by the specific CAT activity of these constructs, with the PB1 promoter fragment showing approximately 70-fold higher activity than PA (Fig. 2). However, in addition, there were some surprising results. First, while the fragment containing PA showed strict orientation-dependent expression (compare pTUN5 and pTUN6, Table 1), the fragment containing PB1 showed expression even in the reverse orientation, albeit at a lower level than in the correct orientation (compare pTUN3 and pTUN4, Fig. 2). The activity in the reverse orientation was about 13-fold weaker than that of PB1 and 5.5-fold stronger than that of PA. In addition, a fragment corresponding to a region upstream of PB1 (pTUN1 and pTUN2, Fig. 2) showed orientation-dependent expression comparable to that of PA (Fig. 2).
FIG. 2.
Functional analysis of putative promoters in M. smegmatis. The arrows denote the orientation of the clone. cfu denotes CFU obtained on plates containing either kanamycin alone (Knr) or with 12.5 or 25 μg of chloramphenicol (Chlr) per ml, as indicated. ∗, slow-growing colonies. ND, not determined.
TABLE 1.
Representative promoters from each class
| Promoter | Species | Gene | Sequence
|
% Match | ||||
|---|---|---|---|---|---|---|---|---|
| −35 | Spacer (nt) | −10 | Spacer | +1 | ||||
| SigA | M. smegmatis | ace | TTGACT | 16 | TATATT | 6 | G | 92 |
| M. tuberculosis | rrnA P3 | TTGACT | 18 | TAGACT | 6 | G | 92 | |
| L5 | Pleft | TTGACA | 18 | CATTCT | 6 | A | 83 | |
| M. fortuitum | rrnA P3 | TTGACA | 18 | TAAGCT | 6 | G | 83 | |
| M. leprae | 16S rRNA | TTGACT | 16 | ATTAAT | 7 | G | 83 | |
| M. paratuberculosis | PAN | TCGACA | 17 | TACACT | 7 | A | 83 | |
| M. phlei | rrnA PCL1 | TTGACG | 18 | TAGACT | 6 | G | 83 | |
| M. smegmatis | rrnA P3 | TTGACA | 18 | TAAGCT | 6 | G | 83 | |
| M. smegmatis | furA | TTGACT | TAGCCT | 83 | ||||
| M. tuberculosis | furA | TTGACT | TATTGT | 83 | ||||
| SigGC | M. paratuberculosis | pAJB303 | TGGCGT | 16 | CGGCAC | 7 | T | 92 |
| M. paratuberculosis | pAJB73 | TGCCGC | 20 | CTCCAG | 7 | T | 83 | |
| M. paratuberculosis | pAJB86 | TGACGT | 17 | CGGTCC | 6 | T | 83 | |
| M. paratuberculosis | pAJB300 | TGACGC | 17 | CAGCCG | 7 | A | 83 | |
| M. bovis | mpb70 | TACCGA | 19 | CATCAG | 6 | G | 75 | |
| M. paratuberculosis | pAJB301 | TCCAGT | 20 | CTGGCC | 7 | T | 67 | |
| M. tuberculosis | 85A | TACACG | 17 | CGCCTG | 7 | A | 58 | |
| M. paratuberculosis | pAJB305 | TGTTGG | 17 | TGGTTG | 7 | T | 50 | |
| M. tuberculosis | katG PC | TTCGCG | 14 | CACAGC | 7 | C | 50 | |
| M. tuberculosis | cpn60 | TGCTCA | 17 | GGCGGC | 7 | A | 50 | |
| M. tuberculosis | gyrA | CGACGC | 17 | CCCGCA | 7 | G | 50 | |
The transcription start corresponding to the promoter in the reverse orientation (PR) was mapped with RNA isolated from M. tuberculosis (Fig. 3). The position of the transcription start site implies that PR substantially overlaps PB1. Repeated attempts to precisely locate the upstream promoter activity (PB2) by primer extension failed, probably due to its weak nature. To further substantiate the results from the functional assay, primer extension analysis was carried out with RNA isolated from M. smegmatis cells transformed with appropriate constructs. The transcription start sites for PB1 and PR mapped to the same position as obtained with RNA from M. tuberculosis (not shown). However, not surprisingly, due to its weak activity, the transcription start site corresponding to PA could not be detected in M. smegmatis.
FIG. 3.
Primer extension to map transcription start site corresponding to the reverse promoter. The extension product corresponding to the transcription start site for each promoter is indicated. The sequencing lanes were used as markers. RNA was prepared from exponentially growing M. tuberculosis cells.
Organization of gyr genes in M. tuberculosis.
The presence of a promoter specific to gyrA itself raised the possibility that the two gyr genes of M. tuberculosis were transcribed independently, unlike the dicistronic arrangement in M. smegmatis. On the other hand, PB1 was 70-fold stronger than PA, at least when tested in M. smegmatis, indicating that it could be the primary initiation site for both genes. To ascertain whether the gyr genes were part of a single dicistron, RT-PCR was performed with M. tuberculosis RNA and primers specific to gyrA and gyrB. These showed specific amplification of a 240-bp product encompassing the intergenic region, proving that at least the primary transcript was dicistronic.
Autoregulation of DNA gyrase.
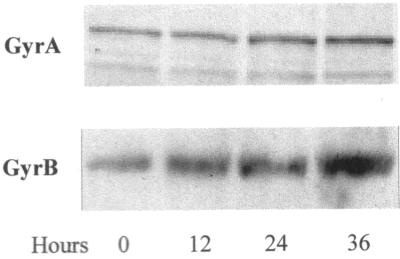
The presence of multiple promoters appeared to suggest complex regulation of the gyr genes in M. tuberculosis. However, because it is a slow-growing organism, we expected it to be more tolerant of changes in topology than faster-growing species, probably producing a more subtle response spread over a longer duration. In accordance with this expectation, when global relaxation was induced in M. tuberculosis cells by novobiocin treatment (9), there was little change in the steady-state level of either GyrA or GyrB up to 12 h. It should be noted that much shorter durations are sufficient to induce relaxation-stimulated transcription in E. coli and M. smegmatis (15, 28). However, treatment for longer durations results in a time-dependent increase in both GyrA and GyrB, as in M. smegmatis, except over a long time period (Fig. 4). This induction is reflected at the level of transcription from the PB1 promoter (Fig. 5). Thus, the phenomenon of relaxation-stimulated transcription is conserved in M. tuberculosis as well, although the kinetics of induction are significantly slower. Concomitant to this induction, there was a decrease in transcription from the divergently organized PR. In addition, a small yet reproducible decrease in transcription from PA, the gyrA-specific promoter, was observed.
FIG. 4.
Increase in DNA gyrase protein level in response to novobiocin. Western blot analysis of GyrA and GyrB with polyclonal antibodies raised against the individual proteins. Protein extracts were prepared after treatment of cells for the indicated durations with 100 μg of novobiocin per ml.
FIG. 5.
Induction of the gyr transcript in response to novobiocin. Primer extension analysis was used to assess changes in the levels of transcript from PA, PB1, and PR. RNA was prepared from cells before (−) and after (+) 12 h of treatment with 100 μg of novobiocin/ml. The primers illustrated in Fig. 1 and 3 were used for primer extension analysis.
DISCUSSION
A comparison of the expression of DNA gyrase in M. smegmatis and M. tuberculosis reveals an amalgamation of conserved and divergent features. The genomic arrangement of the gyr locus is substantially conserved between the two mycobacterial species, and the primary transcript is dicistronic in both species. In addition, the primary promoter in M. tuberculosis, PB1, is located upstream of the gyrB gene at a position similar to that of the M. smegmatis gyr promoter. Furthermore, the promoter region per se for PB1 shows extensive conservation with Pgyr, the promoter driving the gyr genes of M. smegmatis (Fig. 6), indicating that they are evolutionarily related.
FIG. 6.
Promoters in M. smegmatis and M. tuberculosis. (A) Comparison of the gyr locus in M. smegmatis and M. tuberculosis. (B) Putative promoter elements of the gyr promoters in the two species. The putative elements and transcription start sites are in bold uppercase letters. The sequence conservation between the primary promoters in the two species is also shown.
Apart from the primary promoter, the gyr locus in M. tuberculosis employs at least two other promoters (Fig. 6). These additional promoters are weak and probably play a regulatory role. PA is 70-fold weaker than PB1 in exponentially growing M. smegmatis. Therefore, it is unlikely to contribute greatly to the steady-state levels of the GyrA protein. On the other hand, it is possible that it employs an M. tuberculosis-specific σ factor or regulatory protein that is absent in M. smegmatis. Moreover, PA may be induced under specific conditions which require only the production of excess GyrA. For instance, there is at least one report of induction of GyrA alone in E. coli in response to treatment with GyrA inhibitors (18).
The other weak promoter, PR, is divergently oriented and almost completely overlaps PB1. Therefore, the binding of RNA polymerase to one of them would prevent binding in the opposite orientation. It should be noted that there are no identifiable coding sequences upstream of gyrB that PR could be involved in transcribing. Thus, the function of PR is also likely to be purely regulatory. Overlapping, mutually exclusive promoters are one mechanism for regulating gene expression (16). For instance, recruitment of the polymerase to PR would decrease expression of DNA gyrase by reducing transcription initiation. In the converse scenario, as in relaxation of the template, PR is repressed and PB1 gets induced to almost the same extent.
Another point of interest was that all these promoters showed no detectable activity in E. coli while showing a wide range of activity in mycobacteria. The identification of promoters that function only in mycobacteria raised the possibility of defining features that are specific to mycobacterial transcription initiation. Sequence analysis revealed that the putative promoter elements of PA do not follow the E. coli σ70 promoter consensus (Fig. 6). On the other hand, Pgyr from M. smegmatis along with PB1 and PR from M. tuberculosis show moderate resemblance to the E. coli consensus (Fig. 6).
To develop a general consensus matrix for promoter elements for mycobacterial promoters, we analyzed 82 mycobacterial promoters for which the transcription start site had been experimentally defined. A majority of these promoters approximate the E. coli consensus to various extents. However, a subset of promoters, including PA, have extremely GC-rich −10 and −35 regions. Therefore, we clustered the promoters iteratively into two classes. The consensus elements (Fig. 7) for the two classes of promoters are shown, along with representative members (Table 1). The complete consensus matrices as well as the classification of all promoters are included as supplementary material. These two classes encompass 80 of the 82 promoters used in the analysis. The only promoters that were excluded were two extremely weak promoters identified in M. paratuberculosis (2).
FIG. 7.
Comparison of promoter consensus between E. coli and mycobacteria. The most frequently occurring bases at each position are listed. The number in the subscript denotes the frequency of occurrence. R denotes A or G, S denotes C or G, M denotes A or C, and W denotes A or T. The base numbering reflects the numbering used in the consensus matrix in the supplementary material (available on request).
The major class includes 69 promoters that show considerable resemblance to the E. coli σ70 consensus (10). Since all residues known to be involved in base-specific contact of the promoter are conserved between E. coli and mycobacterial σ70 (3, 4), these promoters are probably recognized by SigA, the principal sigma factor in mycobacteria. However, it is not clear what additional features of these promoters make them functional in mycobacteria while still, by and large, being nonfunctional in E. coli.
On the other hand, the second class of 11 promoters represent a completely mycobacterium-specific consensus distinct from any promoter consensus reported in any organism so far. However, further work is required to identify which of the 13 sigma factors in M. tuberculosis recognizes this class of promoters (the putative SigGC). Since the representative promoter of this class (gyrA promoter from M. tuberculosis, PA) is also recognized in M. smegmatis, the completion of the M. smegmatis genome sequence would provide some clues to SigGC. It is noteworthy that in both classes of promoters, there is a correlation between the strength of a promoter and how closely it approximates the consensus. For instance, among the putative SigA-driven promoters, those that show the closest match to the consensus include some of the strongest mycobacterial promoters (S16, ace, and rrnA promoters in Table 1). In addition, among the putative SigGC-driven promoters, the promoter that most closely resembles the consensus is the strongest promoter (pAJB303 in Table 1) identified in the random promoter screen by Bannantine et al. (2). Furthermore, the six promoters from their study that fall into this class show a correlation between their strength and the extent to which they approximate the consensus (Table 1). This is reminiscent of the E. coli σ70 paradigm, for which there is a similar correlation and the consensus promoter actually shows maximal activity (10).
Finally, as discussed above, M. tuberculosis appears to respond to relaxation of the genome by altering the utilization of promoters leading to increased synthesis of DNA gyrase. Therefore, the phenomenon of relaxation-stimulated transcription appears to be conserved in M. tuberculosis. However, the kinetics of response is extremely slow compared to that in E. coli or M. smegmatis. E. coli shows maximal response in as little as 5 min after treatment with novobiocin (15), while M. smegmatis takes 3 h (28). In contrast, M. tuberculosis takes over 24 h to reach a peak. However, independent of the time taken, all species appear to increase the level of protein by about threefold. Papavinasasundaram and coworkers reported a slow response in the induction of RecA in mycobacteria (19). Thus, the kinetics of the response seems to parallel the rate of growth and metabolism of each organism.
Acknowledgments
We thank U. H. Manjunatha for providing polyclonal antibodies against M. tuberculosis GyrA and M. smegmatis GyrB, A. K. Tyagi for pSD7, and other members of the laboratory for discussions.
REFERENCES
- 1.Bachmann, B. J. 1987. Linkage map of Escherichia coli K-12, edition 7, p. 807-877. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 2.Bannantine, J. P., R. G. Barletta, C. O. Thoen, and R. E. Andrews, Jr. 1997. Identification of Mycobacterium paratuberculosis gene expression signals. Microbiology 143:921-928. [DOI] [PubMed] [Google Scholar]
- 3.Bashyam, M. D., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.DasGupta, S. K., M. D. Bashyam, and A. K. Tyagi. 1993. Cloning and assessment of mycobacterial promoters by using a plasmid shuttle vector. J. Bacteriol. 175:5186-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DasGupta, S. K., S. Jain, D. Kaushal, and A. K. Tyagi. 1998. Expression systems for study of mycobacterial gene regulation and development of recombinant BCG vaccines. Biochem. Biophys. Res. Commun. 246:797-804. [DOI] [PubMed] [Google Scholar]
- 8.Dorman, C. J. 1995. 1995 Flemming Lecture: DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology 141:1271-1280. [DOI] [PubMed] [Google Scholar]
- 9.Drlica, K., and M. Snyder. 1978. Superhelical Escherichia coli DNA: relaxation by coumermycin. J. Mol. Biol. 120:145-154. [DOI] [PubMed] [Google Scholar]
- 10.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight, S. W., and D. S. Samuels. 1999. Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J. 18:4875-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampe, M. F., and K. F. Bott. 1985. Genetic and physical organization of the cloned gyrA and gyrB genes of Bacillus subtilis. J. Bacteriol. 162:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menzel, R., and M. Gellert. 1987. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc. Natl. Acad. Sci. USA 84:4185-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menzel, R., and M. Gellert. 1983. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell 34:105-113. [DOI] [PubMed] [Google Scholar]
- 16.Nagaraja, V. 1993. Control of transcription initiation. J. Biosci. 18:13-25. [Google Scholar]
- 17.Nagaraja, V., and K. P. Gopinathan. 1980. Requirement for calcium ions in mycobacteriophage I3 DNA injection and propagation. Arch. Microbiol. 124:249-254. [DOI] [PubMed] [Google Scholar]
- 18.Neumann, S., and A. Quinones. 1997. Discoordinate gene expression of gyrA and gyrB in response to DNA gyrase inhibition in Escherichia coli. J. Basic Microbiol. 37:53-69. [DOI] [PubMed] [Google Scholar]
- 19.Papavinasasundaram, K. G., C. Anderson, P. C. Brooks, N. A. Thomas, F. Movahedzadeh, P. J. Jenner, M. J. Colston, and E. O. Davis. 2001. Slow induction of RecA by DNA damage in Mycobacterium tuberculosis. Microbiology 147:3271-3279. [DOI] [PubMed] [Google Scholar]
- 20.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 21.Salazar, L., H. Fsihi, E. de Rossi, G. Riccardi, C. Rios, S. T. Cole, and H. E. Takiff. 1996. Organization of the origins of replication of the chromosomes of Mycobacterium smegmatis, Mycobacterium leprae and Mycobacterium tuberculosis and isolation of a functional origin from M. smegmatis. Mol. Microbiol. 20:283-293. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Straney, R., R. Krah, and R. Menzel. 1994. Mutations in the −10 TATAAT sequence of the gyrA promoter affect both promoter strength and sensitivity to DNA supercoiling. J. Bacteriol. 176:5999-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takiff, H. E., L. Salazar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs, Jr., and A. Telenti. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiara, A. S., and E. Cundliffe. 1989. Interplay of novobiocin-resistant and -sensitive DNA gyrase activities in self-protection of the novobiocin producer Streptomyces sphaeroides. Gene 81:65-72. [DOI] [PubMed] [Google Scholar]
- 26.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unniraman, S., and V. Nagaraja. 2001. Axial distortion as a sensor of supercoil changes: a molecular model for the homeostatic regulation of DNA gyrase. J. Genet. 80:119-124. [DOI] [PubMed] [Google Scholar]
- 28.Unniraman, S., and V. Nagaraja. 1999. Regulation of DNA gyrase operon in Mycobacterium smegmatis: a distinct mechanism of relaxation stimulated transcription. Genes Cells 4:697-706. [DOI] [PubMed] [Google Scholar]
- 29.Zechiedrich, E. L., A. B. Khodursky, S. Bachellier, R. Schneider, D. Chen, D. M. Lilley, and N. R. Cozzarelli. 2000. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275:8103-8113. [DOI] [PubMed] [Google Scholar]