Abstract
The σK checkpoint coordinates gene expression in the mother cell with signaling from the forespore during Bacillus subtilis sporulation. The signaling pathway involves SpoIVB, a serine peptidase produced in the forespore, which is believed to cross the innermost membrane surrounding the forespore and activate a complex of proteins, including BofA, SpoIVFA, and SpoIVFB, located in the outermost membrane surrounding the forespore. Activation of the complex allows proteolytic processing of pro-σK, and the resulting σK RNA polymerase transcribes genes in the mother cell. To investigate activation of the pro-σK processing complex, the level of SpoIVFA in extracts of sporulating cells was examined by Western blot analysis. The SpoIVFA level decreased when pro-σK processing began during sporulation. In extracts of a spoIVB mutant defective in forespore signaling, the SpoIVFA level failed to decrease normally and no processing of pro-σK was observed. Although these results are consistent with a model in which SpoIVFA inhibits processing until the SpoIVB-mediated signal is received from the forespore, we discovered that loss of SpoIVFA was insufficient to allow processing under certain conditions, including static incubation of the culture and continued shaking after the addition of inhibitors of oxidative phosphorylation or translation. Under these conditions, loss of SpoIVFA was independent of spoIVB. The inability to process pro-σK under these conditions was not due to loss of SpoIVFB, the putative processing enzyme, or to a requirement for ongoing synthesis of pro-σK. Rather, it was found that the requirements for shaking of the culture, for oxidative phosphorylation, and for translation could be bypassed by mutations that uncouple processing from dependence on forespore signaling. This suggests that ongoing translation is normally required for efficient pro-σK processing because synthesis of the SpoIVB signal protein is needed to activate the processing complex. When translation is blocked, synthesis of SpoIVB ceases, and the processing complex remains inactive despite the loss of SpoIVFA. Taken together, the results suggest that SpoIVB signaling activates the processing complex by performing another function in addition to causing loss of SpoIVFA or by causing loss of SpoIVFA in a different way than when translation is blocked. The results also demonstrate that the processing machinery can function in the absence of translation or an electrochemical gradient across membranes.
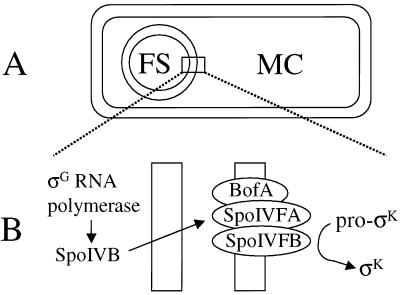
Communication between cells is an essential feature of many biological processes. During sporulation of the gram-positive bacterium Bacillus subtilis, two cell types are produced that communicate extensively in order to coordinate the developmental process (19, 20, 35). The two cell types are produced within a sporangium by asymmetric division, resulting in a larger mother cell and a smaller forespore. The membranes of the division septum migrate around the forespore and pinch it off as a protoplast within the mother cell (Fig. 1A). Upon completion of this step, a new sigma factor, σG, becomes active in the forespore. σG RNA polymerase transcribes the spoIVB gene, whose product somehow signals to the mother cell that it is time to produce active σK by proteolytic processing of inactive pro-σK (3, 4, 24). This signaling pathway has been called the σK checkpoint (Fig. 1B) because it delays mother-cell gene expression under the control of σK RNA polymerase for about 1 h (4). A breakdown in communication between the forespore and the mother cell can lead to premature production of σK, which causes a sporulation defect (4).
FIG. 1.
σK checkpoint. (A) Diagram of a sporangium in which the forespore (FS) has been pinched off as a protoplast within the mother cell (MC). (B) Expanded view of the two membranes separating the forespore and mother cell, depicting BofA, SpoIVFA, and SpoIVFB in the outermost membrane surrounding the forespore. The three proteins form a complex in which SpoIVFB is inactive prior to signaling from the forespore (4, 5, 16, 30, 31, 33). σG RNA polymerase transcribes spoIVB in the forespore (3), and SpoIVB is secreted into the space between the two membranes (38), where it activates the processing complex, leading to the production of σK in the mother cell.
Much is known about the components involved in the σK checkpoint, yet important questions remain. SpoIVB is a serine peptidase that is believed to cross the innermost membrane surrounding the forespore (14, 38). In the space between the two membranes surrounding the forespore, SpoIVB is thought to self-cleave, generating several species with slightly different N-terminal truncations (38). One or more of these species signals pro-σK processing, but the mechanism of signaling is unknown. For example, it is unclear whether further serine peptidase activity beyond the self-cleavage is required for signaling. SpoIVB has a PDZ domain likely to mediate protein-protein interactions (13, 27). These could include interactions between SpoIVB and other proteins which are important for signaling, but this is uncertain because the PDZ domain also plays a role in the autoproteolysis that activates the SpoIVB signaling function (13).
The target of SpoIVB signaling is a complex of three proteins, BofA, SpoIVFA, and SpoIVFB, that localize to the outermost membrane surrounding the forespore (5, 9, 16, 29, 31, 33, 37). These proteins are normally produced under the transcriptional control of σE RNA polymerase (5, 16, 31), which directs early gene expression in the mother cell. However, several insights have emerged from studies in which B. subtilis was engineered to produce different combinations of these proteins, as well as pro-σK, during growth. First, SpoIVFB alone is sufficient for pro-σK processing, suggesting that it is the protease that processes pro-σK (23, 30). Subsequent mutational analyses have supported the idea that SpoIVFB is a member of a large family of membrane metalloproteases (32, 40). Second, BofA and SpoIVFA together can inhibit processing by SpoIVFB (30). Third, BofA stabilizes SpoIVFA (28). These observations led to the model that SpoIVFA is an inhibitor of SpoIVFB protease activity during sporulation and that the role of BofA is to stabilize SpoIVFA (28).
We hypothesized that SpoIVB signaling from the forespore might target SpoIVFA, causing its level to decrease and relieving inhibition of SpoIVFB so that pro-σK processing would occur. We show that a decrease in the level of SpoIVFA in cell extracts coincides with the onset of pro-σK processing during sporulation and that in extracts of spoIVB mutant cells the SpoIVFA level fails to decrease normally. However, we also found that a decrease in the level of SpoIVFA was insufficient to allow pro-σK processing when shaking of the culture was discontinued or when shaking was continued after addition of inhibitors of oxidative phosphorylation or translation. Under these conditions, loss of SpoIVFA did not depend on SpoIVB-mediated forespore signaling. Although these conditions blocked further synthesis of pro-σK, this does not explain why processing was inhibited, because we confirmed a previous report that processing occurs posttranslationally (41). Moreover, we show that the dependence of pro-σK processing on shaking, oxidative phosphorylation, and translation can be relieved by mutations that bypass the need for forespore signaling.
From these results, we infer that synthesis of the SpoIVB signal protein is needed to activate the pro-σK processing machinery by a mechanism that involves more than just the loss of SpoIVFA observed when translation is blocked. Our results also show that processing of pro-σK can occur without ongoing translation and in the absence of a membrane potential when the need for forespore signaling is bypassed.
MATERIALS AND METHODS
Bacterial strains and sporulation.
B. subtilis strain OR758 (spoIVFB-gfp) (29) is indistinguishable from its parent, PY79 (39), with respect to pro-σK processing (data not shown) and sporulation (29). LK1 (spoIVBΔ::spc) was constructed by transforming PY79 with chromosomal DNA from RL1041 (spoIVBΔ::spc) (29) and selecting on Luria-Bertani (LB) agar (34) containing spectinomycin (100 μg/ml). BSL51 (spoIVFΔAB::cat) (25) served as a negative control to demonstrate the ability of our antibodies to detect SpoIVFA. OR745 (spoIVFΔB::spc) (29) and SC777 (bofB8 spoIIIGΔ1 cat) (4) have been described previously.
Sporulation was induced by resuspending growing cells in SM medium (11). The onset of sporulation (T0) is defined as the time of resuspension. Samples (0.5 ml) were microcentrifuged for 1 min, and cell pellets were stored at −70°C for preparation of whole-cell extracts. In parallel, samples were taken for determination of the optical density at 600 nm. The optical density of cultures treated with inhibitors decreased 10 to 20% at 60 to 90 min after addition of inhibitor, presumably due to cell lysis. Inhibitors (from Sigma) were added from the following stock solutions: carbonyl cyanide m-chlorophenylhydrazone (CCCP) at 20 mM in ethanol, sodium azide at 10% (wt/vol) in water, nigericin at 2 mM in ethanol, valinomycin at 2 mM in ethanol, chloramphenicol (Cm) at 34 mg/ml in ethanol, and kanamycin sulfate at 40 mg/ml in water. Control experiments showed that neither ethanol (used to dissolve several of the inhibitors) nor KCl (0.1 M) (required to observe inhibition by valinomycin) alone inhibited pro-σK processing.
Preparation of antibodies.
The antibodies used to detect pro-σK and σK have been described previously (24) and were used at a 1:10,000 dilution. A similar approach was used to produce polyclonal antibodies against recombinant green fluorescent protein (GFP) (Clontech) except TiterMax was used as the adjuvant. Serum was prepared (10) and used at a 1:5,000 dilution. To prepare antibodies against SpoIVFA, a fusion protein containing the C-terminal 116 amino acids of SpoIVFA fused to six histidine residues at the N terminus was produced in Escherichia coli and purified as described previously (29) with the following modifications. The cell extract was stirred for 2 h at room temperature, and the supernatant after centrifugation was stirred with 0.4 volume of nitrilotriacetic acid-agarose (Qiagen) (50% slurry equilibrated with buffer containing 40 mM sodium phosphate [pH 8.0], 400 mM NaCl, and 6 M guanidine-HCl) for 1 h at room temperature before packing into a column. The column was washed sequentially with buffer B (40 mM sodium phosphate, 400 mM NaCl, and 6 M urea) at pH 8.0, buffer B at pH 6.3, and buffer B at pH 5.9, and then eluted with buffer B at pH 4.6. After dialysis against phosphate-buffered saline (Gibco-BRL), the fusion protein was injected into rabbits as described for GFP, and the antiserum was used at a 1:3,000 dilution.
Western blot analysis.
Whole-cell extracts were prepared for Western blot analysis as described previously (29). Proteins in extracts from equivalent cell numbers (based on the optical density of the culture at the time of sample collection) were separated on sodium dodecyl sulfate-14% Prosieve polyacrylamide gels (FMC) with Tris-Tricine electrode buffer (0.1 M Tris, 0.1 M Tricine, 0.1% SDS) and electroblotted to Immobilon-P membranes (Millipore). Membranes were incubated in TBS (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl) with 5% nonfat dry milk for 1 to 2 h at room temperature with shaking to block nonspecific antibody binding and then incubated overnight at room temperature with shaking in antiserum diluted as described above into TBS with 2% nonfat dry milk. For immunodetection, membranes were incubated in TBS with 2% nonfat dry milk and a goat anti-rabbit immunoglobulin-horseradish peroxidase conjugate (Bio-Rad) at a 1:10,000 dilution for 1 h at room temperature with shaking, followed by chemiluminescent detection according to the manufacturer's instructions (ECL kit; Amersham). These instructions were also followed for experiments in which membranes were stripped and reprobed with a different antibody. Signals were quantified with a Kodak EDAS290 with 1D software.
Pulse-chase and immunoprecipitation.
Pulse-labeling of cells was performed as described previously (42). The chase typically involved addition of a 1,000-fold molar excess of unlabeled methionine, unless indicated otherwise. Immunoprecipitation was performed as described previously (42) with the following modifications. Polyclonal anti-pro-σK antibodies (2.5 μl, which was sufficient to quantitatively precipitate pro-σK and σK from a 1-ml culture lysate in a control experiment) were added to each lysate, and the mixture was incubated at 0°C for 2 h. Protein A-Sepharose CL-4B (Pharmacia) was used to bind antibody-antigen complexes as described previously (42), and samples were microcentrifuged at 1,000 rpm for 1 min to pellet the complexes. Immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described above. Pro-σK and σK bands were visualized by fluorography with En3Hance (NEN) as enhancing fluors and quantified with a Storm 820 PhosphorImager (Molecular Dynamics), with the background of each lane subtracted from the band intensity.
RESULTS
Level of SpoIVFA during sporulation.
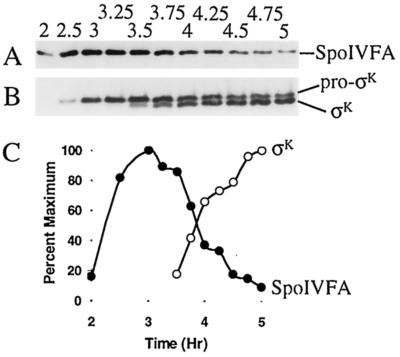
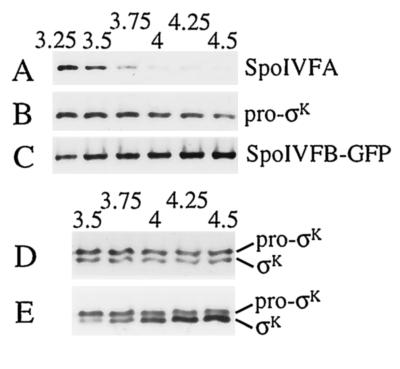
SpoIVFA is involved in the inhibition of pro-σK processing during sporulation (4, 5, 9, 28, 30, 33). One mechanism to relieve inhibition would be to decrease the level of SpoIVFA. To measure the level of SpoIVFA, we raised antibodies against a C-terminal part of the protein. These antibodies detected a polypeptide of the expected size for SpoIVFA (29) in extracts of sporulating B. subtilis OR758 cells (Fig. 2A). No signal was observed in extracts of a spoIVF mutant (data not shown). Quantification of the SpoIVFA level in extracts of OR758 cells is shown in Fig. 2C. SpoIVFA reached a maximum at hour 3 (T3) of sporulation, and the level decreased markedly between T3.5 and T4.
FIG. 2.
Levels of SpoIVFA, pro-σK, and σK during sporulation. (A) B. subtilis strain OR758 was induced to sporulate, samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against SpoIVFA. (B) The blot was stripped and reprobed with antibodies against pro-σK. (C) Quantification of the SpoIVFA (•) and σK (○) signal intensities from panels A and B, respectively.
To measure the levels of pro-σK and σK in these samples, the blot was stripped of antibodies and reprobed with antibodies against pro-σK, which also detect σK (Fig. 2B). The σK level increased markedly between T3.5 and T4, coincident with the decrease in SpoIVFA (Fig. 2C). We conclude that the onset of pro-σK processing during sporulation is accompanied by a decrease in the level of extractable SpoIVFA, consistent with the idea that SpoIVFA is involved in the inhibition of processing until a signal from the forespore relieves inhibition.
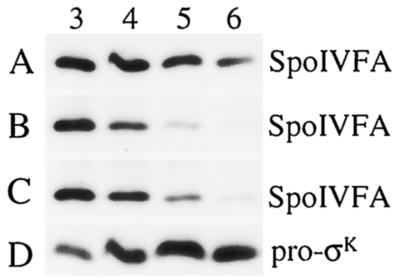
The signal that governs pro-σK processing in the mother cell involves SpoIVB (3), a serine peptidase produced in the forespore that may cross the innermost membrane surrounding the forespore (38) and influence SpoIVFA, which is located in the outermost membrane surrounding the forespore (29). Figure 3A shows the SpoIVFA level in extracts of a spoIVB mutant under sporulation conditions. The SpoIVFA level did not decrease markedly between T3 and T5, as it did in OR758 cell extracts (Fig. 2A). Rather, it remained easily detectable even at T6 (Fig. 3A). This difference was not due to sequestration of SpoIVFA in OR758 cells brought about by σK-dependent gene expression and morphological change because the SpoIVFA level decreased normally in extracts of a spoIVFB mutant (Fig. 3B), which fails to make σK (data not shown). Figure 3C shows wild-type cell extracts at the same time points for comparison. We conclude that the SpoIVB-mediated signal from the forespore is required for the SpoIVFA decrease that normally accompanies the onset of pro-σK processing, but neither SpoIVFB protease activity nor σK is needed for the decrease in the level of extractable SpoIVFA.
FIG. 3.
Levels of SpoIVFA and pro-σK in mutants. B. subtilis strains were induced to sporulate, samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against SpoIVFA. (A) LK1 (spoIVBΔ::spc). (B) OR745 (spoIVFΔB::spc). (C) PY79 (spo+). (D) The blot in panel A was stripped and reprobed with antibodies against pro-σK.
When the blot shown in Fig. 3A was stripped of antibodies and reprobed with anti-pro-σK antibodies, only pro-σK was observed (Fig. 3D), consistent with previous results showing that SpoIVB is necessary for processing of pro-σK to σK (24). These results are consistent with the model that SpoIVB causes the level of SpoIVFA to decrease, releasing SpoIVFB from inhibition so that it can cleave pro-σK.
Effect of shaking the culture.
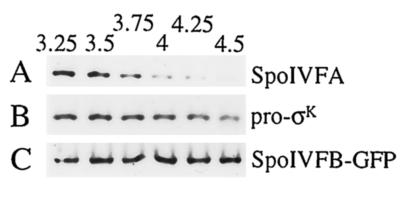
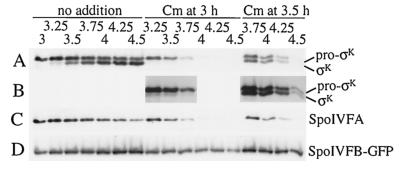
A goal of our laboratory has been to observe pro-σK processing in extracts of sporulating cells. Although some apparent processing can be observed, it does not appear to depend on SpoIVFB (L. Kroos, unpublished data), the protein believed to be responsible for processing in vivo (5, 23, 30, 32, 40). As a control to investigate why extracts of sporulating cells fail to process pro-σK to σK during static incubation, we tested the effect of static incubation on intact cells. Normally, the culture is shaken at 400 rpm to promote good aeration and efficient sporulation (11). We discovered that if shaking was discontinued after T3, the level of SpoIVFA in cell extracts still decreased (Fig. 4A), but pro-σK processing was not observed (Fig. 4B).
FIG. 4.
Effect of discontinued shaking on pro-σK processing and on the levels of SpoIVFA and SpoIVFB-GFP. A portion of the OR758 (spoIVFB-gfp) culture used in the experiment shown in Fig. 2 was removed from shaking at T3, and incubation was continued at 37°C without shaking. (A) Samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against SpoIVFA. (B) The blot was stripped and reprobed with antibodies against pro-σK. (C) The blot was again stripped and this time reprobed with antibodies against GFP to detect the SpoIVFB-GFP fusion protein.
One possible explanation of these results is that the putative SpoIVFB protease is not made or is unstable under these conditions. However, this was not the case. The B. subtilis strain OR758 used in our experiments has the gfp gene, encoding the GFP of Aequorea victoria fused to the 3′ end of spoIVFB (29). The SpoIVFB-GFP fusion protein is fully active for pro-σK processing (Fig. 2B and data not shown) and can be detected with antibodies against GFP. Figure 4C shows that SpoIVFB-GFP was present at T3.25, shortly after shaking was stopped, and its level remained constant at least until T4.5. Similar results were observed when shaking was continued after T3 (data not shown). Taken together, these results demonstrate that in the absence of shaking, the decrease in SpoIVFA is not sufficient to allow processing of pro-σK by SpoIVFB-GFP.
Effect of inhibitors of oxidative phosphorylation.
Why is shaking of the culture needed for pro-σK processing? We hypothesized that good aeration of the culture permits energy generation via oxidative phosphorylation and that energy is needed to allow processing even when the SpoIVFA level is much diminished. To test this hypothesis, we added CCCP (5 μM), an H+ ionophore that uncouples oxidative phosphorylation, at T3 and then continued shaking the culture at 400 rpm. The effects on the SpoIVFA level (Fig. 5A), on pro-σK processing (Fig. 5B), and on the SpoIVFB-GFP level (Fig. 5C) were similar to those obtained when shaking was discontinued after T3 (Fig. 4). When CCCP was added at T3.5, after processing had begun, no further processing was observed (Fig. 5D), in contrast to the further increase in the σK level seen in a parallel untreated culture (Fig. 5E). Similar results were obtained with an inhibitor of oxidative phosphorylation, sodium azide (0.2%); an ionophore that dissipates the pH gradient, nigericin (1 μM); and an ionophore that dissipates the membrane potential, valinomycin (1 μM) in the presence of KCl (0.1 M) (data not shown). We conclude that oxidative phosphorylation is necessary for pro-σK processing to occur despite the virtual absence of SpoIVFA from cell extracts within 1 h after blockage of oxidative phosphorylation and the continued presence of the putative processing protease SpoIVFB-GFP.
FIG. 5.
Effect of an uncoupler of oxidative phosphorylation on pro-σK processing and on the levels of SpoIVFA and SpoIVFB-GFP. A portion of the OR758 (spoIVFB-gfp) culture used in the experiment shown in Fig. 2 was removed at T3, CCCP (5 μM) was added, and shaking was continued. (A) Samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against SpoIVFA. (B) The blot was stripped and reprobed with antibodies against pro-σK. (C) The blot was again stripped and this time reprobed with antibodies against GFP to detect the SpoIVFB-GFP fusion protein. (D) In a separate experiment, the same strain used in the experiment shown in Fig. 2 was induced to sporulate, a portion of the culture was removed at T3.5, CCCP (5 μM) was added, and shaking was continued. Samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against pro-σK. (E) A portion of the culture used in the experiment shown in panel D was left untreated, and samples were analyzed as in panel D.
Effect of protein synthesis inhibitors.
Why is oxidative phosphorylation needed for pro-σK processing? A clue came from comparing the pro-σK level in extracts of the spoIVB mutant (Fig. 3D) with that in extracts of wild-type OR758 (spoIVFB-gfp) cells that had been treated with inhibitors of oxidative phosphorylation (Fig. 5B and data not shown). Pro-σK continued to accumulate after T3 in the spoIVB mutant but not in OR758 cells treated with inhibitor. This suggested that the inhibitors of oxidative phosphorylation block the energy-intensive process of protein synthesis. To test whether protein synthesis is required for pro-σK processing, the translation inhibitor Cm (200 μg/ml) was added at T3 or T3.5 and shaking was continued. In the parallel untreated culture (Fig. 6A, no addition), a small amount of σK was observed at T3.25, considerably more was seen at T3.5, and by T4, σK was more abundant than pro-σK. Addition of Cm at T3 appeared to block processing (Fig. 6A, Cm at 3 h), but the interpretation was complicated by a decrease in the level of pro-σK, which was more rapid than when inhibitors of oxidative phosphorylation were added (Fig. 5B and data not shown) (see below).
FIG. 6.
Effect of a protein synthesis inhibitor on pro-σK processing and on the levels of SpoIVFA and SpoIVFB-GFP. (A) B. subtilis strain OR758 was induced to sporulate, and the culture was split at T3 into portions to which nothing was added or Cm (200 μg/ml) was added immediately (Cm at 3 h) or after 30 min (Cm at 3.5 h). Shaking was continuous except when the culture was split and Cm was added. Samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against pro-σK. (B) Eightfold-longer exposure of part of the blot shown in panel A. (C) The blot was stripped and reprobed with antibodies against SpoIVFA. (D) The same samples used to produce the blot shown in panels A to C were used to produce another blot that was probed with antibodies against GFP to detect the SpoIVFB-GFP fusion protein.
A longer exposure of the blot shown in Fig. 6A revealed that a small amount of σK was present 15 min after Cm addition, but thereafter the amount of σK relative to the amount of pro-σK did not rise (Fig. 6B, Cm at 3 h) as it did in the untreated culture (Fig. 6A, no addition). Clearly, Cm strongly inhibited pro-σK processing. Likewise, Cm appeared to block pro-σK processing shortly after its addition at T3.5 (Fig. 6A, Cm at 3.5 h), because the ratio of σK to pro-σK increased just slightly within 15 min after addition of the translation inhibitor, but thereafter the ratio did not increase (Fig. 6B, Cm at 3.5, shows a longer exposure) as it did in the untreated culture (Fig. 6A, no addition). We conclude that ongoing translation is required for efficient pro-σK processing.
Figure 6C shows that the level of SpoIVFA declined rapidly after addition of the protein synthesis inhibitor. In contrast, the SpoIVFB-GFP level remained fairly constant (Fig. 6D). These results demonstrate that the inhibition of pro-σK processing observed after Cm addition was not due to persistence of SpoIVFA or to loss of the proposed processing enzyme SpoIVFB-GFP. The entire experiment was also performed with another translation inhibitor, kanamycin (200 μg/ml), and similar results were observed (data not shown).
We suggested above that inhibitors of oxidative phosphorylation block the energy-intensive process of protein synthesis. However, the levels of pro-σK and σK declined much more rapidly after addition of translation inhibitors (Fig. 6A and data not shown) than after addition of oxidative phosphorylation inhibitors (Fig. 5B and 5D and data not shown). The inhibitors of oxidative phosphorylation either did not block translation completely or increased the stability of pro-σK and σK.
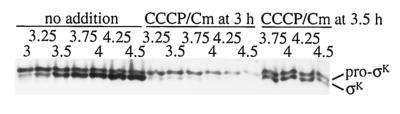
To distinguish between these possibilities, we added both the oxidative phosphorylation inhibitor CCCP (5 μM) and the translation inhibitor Cm (200 μg/ml) at T3 or T3.5 and continued shaking the cultures. In this experiment, a small amount of σK had already accumulated at T3 in the parallel untreated culture (Fig. 7, no addition). The combination of inhibitors not only blocked pro-σK processing, as expected from the preceding experiments, but allowed pro-σK and σK to persist longer (Fig. 7, CCCP/Cm at 3 h or at 3.5 h) than when Cm alone was added (Fig. 6A, Cm at 3 h or at 3.5 h). Similar results were obtained when other inhibitors of oxidative phosphorylation, including sodium azide (0.2%) and nigericin (1 μM), were added in combination with Cm (data not shown). These results indicate that inhibition of oxidative phosphorylation increases the stability of pro-σK and σK in sporulating cells. We infer that degradation of pro-σK and σK may involve one or more ATP-dependent proteases.
FIG. 7.
Effect of a combination of inhibitors on pro-σK and σK levels. B. subtilis strain OR758 was induced to sporulate, and the culture was split at T3 into portions to which nothing was added or CCCP (5 μM) and Cm (200 μg/ml) were added immediately (CCCP/Cm at 3 h) or after 30 min (CCCP/Cm at 3.5 h). Shaking was continuous except when the culture was split and inhibitors were added. Samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against pro-σK.
Effect of inhibitors on SpoIVFA level in a spoIVB mutant.
As shown in Fig. 8, the loss of SpoIVFA observed when translation or oxidative phosphorylation was inhibited did not require SpoIVB. Treatment of a spoIVB mutant with Cm, CCCP, azide, or nigericin at T3 resulted in decreases in SpoIVFA with similar kinetics as for strain OR758 (Fig. 5A and 6C and data not shown). These results demonstrate that sporulating cells possess a powerful SpoIVB-independent mechanism(s) that can cause loss of SpoIVFA when translation or oxidative phosphorylation is inhibited. Yet pro-σK processing was not observed (Fig. 5 and 6 and data not shown).
FIG. 8.
Effect of inhibitors on the level of SpoIVFA in a spoIVB mutant. B. subtilis strain LK1 (spoIVBΔ::spc) was induced to sporulate, and the culture was split at T3 into portions to which nothing was added or CCCP (5 μM), sodium azide (0.2%), nigericin (1 μM), or Cm (200 μg/ml) was added. Shaking was continuous except when the culture was split and inhibitors were added. Samples were collected at the indicated times (hours) after sporulation was induced, and whole-cell extracts were subjected to Western blot analysis with antibodies against SpoIVFA.
Pulse-chase analysis of pro-σK processing.
We considered the possibility that ongoing translation is necessary for efficient pro-σK processing because processing occurs cotranslationally. According to this model, full-length pro-σK is not processed but is degraded, and processing occurs while nascent pro-σK is being synthesized. A prediction of this model is that [35S]methionine-labeled pro-σK should not be processed to σK during a chase period with unlabeled methionine. A previous pulse-chase experiment suggested that labeled pro-σK can be chased to σK (41), arguing against cotranslational processing, but in light of the results presented here, we were concerned about the adequacy of the chase (i.e., the possibility of continued incorporation of label during the chase period) in the previous experiment.
We repeated the previous experiment, labeling cells with [35S]methionine for 5 min at T3 and then chasing with a 1,000-fold molar excess of unlabeled methionine (Fig. 9A, lanes 1 to 3). As reported previously (41), pro-σK was processed to σK during the chase period. Quantification of the signals revealed little change in the total amount of pro-σK plus σK (data not shown). In contrast, Western blot analysis of whole-cell extracts of samples collected in parallel showed a greater than twofold increase in the total amount of pro-σK plus σK by 60 min after the pulse (data not shown). Taken together, these results indicate that synthesis of unlabeled pro-σK continued during the chase period and suggest that synthesis of labeled pro-σK was prevented by the 1,000-fold molar excess of unlabeled methionine.
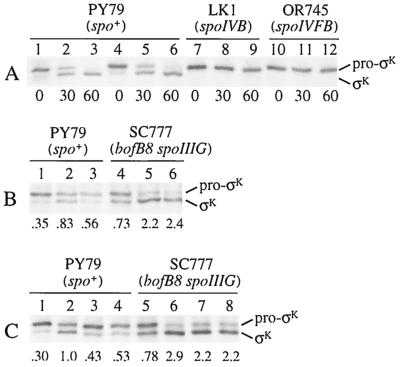
FIG. 9.
Pulse-chase analysis of pro-σK processing. (A) The indicated B. subtilis strains were induced to sporulate, pulse-labeled with [35S]methionine at T3, and chased with a 1,000-fold molar excess of unlabeled methionine (lanes 1 to 3 and 7 to 12) or chased by collecting the cells by centrifugation and resuspending them in fresh medium with a 10,000-fold molar excess of unlabeled methionine (lanes 4 to 6). Numbers below the lanes indicate the length of the chase period in minutes. (B) The indicated strains were induced to sporulate, pulse-labeled with [35S]methionine at T3 (lanes 1 and 4), and chased with a 1,000-fold molar excess of unlabeled methionine for 30 min in the absence (lanes 2 and 5) or presence (lanes 3 and 6) of Cm (200 μg/ml). The numbers below the lanes indicate the σK/pro-σK ratio based on quantification of the [35S]methionine-labeled protein bands. (C) The indicated strains were induced to sporulate, pulse-labeled with [35S]methionine at T3 (lanes 1 and 5), and chased with a 1,000-fold molar excess of unlabeled methionine for 30 min with shaking in the absence (lanes 2 and 6) or presence (lanes 3 and 7) of 5 μM CCCP or for 30 min without shaking (lanes 4 and 8). The numbers below the lanes indicate the σK/pro-σK ratio.
To be certain that continued synthesis of labeled pro-σK did not occur during the chase period, cells were collected by centrifugation immediately after the 5-min pulse-labeling and resuspended in fresh medium containing a 10,000-fold molar excess (relative to the concentration of labeled methionine used during the pulse) of unlabeled methionine. Similar results were observed (Fig. 9A, lanes 4 to 6). When spoIVB or spoIVFB mutant cells were pulse-labeled at T3 and chased with a 1,000-fold molar excess of unlabeled methionine, pro-σK persisted and was not processed to σK (Fig. 9A, lanes 7 to 12). We conclude that pro-σK is processed to σK posttranslationally.
We next addressed the possibility that ongoing translation is needed to synthesize the SpoIVB signaling protein in the forespore. This would explain our results if SpoIVB signaling is needed to activate pro-σK processing despite the loss of SpoIVFA upon translational arrest. The normal dependence of processing on SpoIVB can be bypassed by bof (which stands for bypass of forespore) mutations in SpoIVFA or BofA (4, 5, 16, 31). We repeated the pulse-chase experiment, including Cm to block translation during the chase period, and compared the wild-type strain with a bofB8 spoIIIG double mutant in which the spoIIIG mutation blocks production of sufficient SpoIVB for signaling (3) and the bofB8 mutation in spoIVFA nevertheless permits processing (4).
Figure 9B shows that Cm inhibited processing of pro-σK to σK during the first 30 min of the chase period in wild-type cells. Quantification of the signals showed that the ratio of σK to pro-σK increased from 0.35 (lane 1) to 0.83 (lane 2) during the 30-min chase in the absence of Cm, but in the presence of Cm it increased only to 0.56 (lane 3). In contrast, Cm did not inhibit processing in the bypass mutant (Fig. 9B, lanes 4 to 6). The ratio of σK to pro-σK after the pulse at T3 was higher in the bypass mutant (lane 4) than in wild-type cells (lane 1) because processing was not delayed by dependence on forespore signaling. During the chase, the ratio of σK to pro-σK in the bypass mutant increased more than twofold in the presence of Cm (lane 6) as well as in its absence (lane 5). A similar result was observed for a bofA mutant (data not shown). These results suggest that the need for ongoing translation to observe efficient processing in wild-type cells reflects the need to synthesize SpoIVB.
Can a bypass mutation also alleviate the need for oxidative phosphorylation and shaking of the culture in order to observe processing? Figure 9C shows that CCCP addition (lane 3) and static incubation of the culture (lane 4) inhibited pro-σK processing to a similar extent as Cm addition (Fig. 9B, lane 3) during the first 30 min of the chase period in wild-type cells but had little effect on processing in the bypass mutant (Fig. 9C, lanes 5 to 8). Taken together, the results of our pulse-chase analysis indicate that processing of pro-σK to σK occurs posttranslationally and its dependence on shaking, oxidative phosphorylation, and translation can be relieved by a mutation that bypasses the need for forespore signaling.
DISCUSSION
Our results provide several new insights into the signaling pathway that leads to activation of pro-σK processing. First, the SpoIVB-mediated signal from the forespore causes a decrease in the level of extractable SpoIVFA that coincides with the onset of pro-σK processing during sporulation. Second, loss of SpoIVFA brought about by static incubation of the culture or inhibition of oxidative phosphorylation or translation does not result in processing of pro-σK, even though the putative processing protease, SpoIVFB-GFP, is present. Third, this inhibition of processing can be overcome by mutations that bypass the dependence of processing on forespore signaling. We conclude that forespore signaling is necessary to activate the pro-σK processing complex despite loss of SpoIVFA under conditions that inhibit translation. We discuss two possible explanations for this novel finding below.
One possibility is that although SpoIVB-mediated signaling normally causes loss of SpoIVFA from cell extracts, it also performs another function necessary to activate the pro-σK processing complex. What might that function be? Rudner and Losick (33) showed recently that SpoIVFA interacts with both BofA and SpoIVFB, possibly enabling BofA to inhibit SpoIVFB. If this model is correct, our results might be indicating that BofA can inhibit SpoIVFB protease activity after SpoIVFA is lost from the complex. If so, we would predict that BofA, like SpoIVFB-GFP, would persist when shaking of the culture is stopped or when oxidative phosphorylation or translation is inhibited, rather than diminishing like SpoIVFA (Fig. 4, 5, and 6). According to this model, the primary function of SpoIVB signaling is to relieve BofA inhibition of the SpoIVFB protease, and SpoIVFA is lost as a consequence of this activation event.
A second possible explanation of our results is that SpoIVB signaling causes loss of SpoIVFA in a way that activates the SpoIVFB protease, but the conditions that we discovered cause loss of SpoIVFA (i.e., static incubation and inhibition of oxidative phosphorylation or translation), do so in a way that fails to activate the SpoIVFB protease. The loss of SpoIVFA observed when translation or oxidative phosphorylation is inhibited does not require SpoIVB (Fig. 8). The mechanism of SpoIVFA loss under these conditions is unknown.
Some insight into the mechanisms of SpoIVFA and pro-σK/σK loss after addition of inhibitors can be gleaned from our data. The kinetics of SpoIVFA loss were similar after treatment of sporulating cells with inhibitors of oxidative phosphorylation (Fig. 5A and data not shown) and translation (Fig. 6C and data not shown). In contrast, the levels of pro-σK and σK declined much more rapidly after addition of translation inhibitors (Fig. 6A and data not shown) than after addition of oxidative phosphorylation inhibitors (Fig. 5B and 5D and data not shown), and we showed that the effect of the oxidative phosphorylation inhibitors was dominant, allowing pro-σK and σK to persist longer when a combination of the two types of inhibitors was added (Fig. 7) than when translation inhibitor alone was added (Fig. 6A).
Oxidative phosphorylation generates ATP (and, indirectly, GTP), needed for translation. Certain proteases also require ATP (8). Blocking ATP synthesis with inhibitors of oxidative phosphorylation is expected to block not only translation but also ATP-dependent mechanisms of proteolysis. The stabilizing effect of oxidative phosphorylation inhibitors on pro-σK and σK in the presence of a translation inhibitor (Fig. 7 and data not shown) suggests that degradation of pro-σK and σK may involve an ATP-dependent protease(s). FtsH is a membrane-bound ATP- and Zn2+-dependent protease that might be involved in pro-σK and σK turnover. In E. coli, FtsH degrades the heat shock transcription factor σ32 (12, 36). Also, the SpoVM protein of B. subtilis can inhibit FtsH-mediated degradation of E. coli σ32 in vitro, and certain mutations in the B. subtilis ftsH gene can partially suppress the sporulation defect of certain spoVM mutants, leading to speculation that SpoVM might antagonize FtsH-dependent degradation of sigma factors during sporulation (2).
FtsH has also been implicated in the turnover of SpoIVFA in B. subtilis. A null mutation in ftsH enhances SpoIVFA accumulation in cells engineered to produce SpoIVFA during growth (28). However, our results showing that the SpoIVFA level declines as rapidly after treatment of sporulating cells with inhibitors of oxidative phosphorylation (Fig. 5A and data not shown) as after treatment with translation inhibitors (Fig. 6C and data not shown) suggest that degradation of SpoIVFA under these conditions can occur by an ATP-independent mechanism, so FtsH is not involved.
Although FtsH does not appear to be involved in the loss of SpoIVFA in sporulating cells treated with inhibitors of oxidative phosphorylation or translation, we cannot exclude the involvement of FtsH in untreated sporulating cells, because the mechanism of SpoIVFA loss caused by SpoIVB signaling is unknown and, as noted above, it could be different. A simple model would be that the serine peptidase activity of SpoIVB (14, 38) acts directly on SpoIVFA, leading to degradation of SpoIVFA. Perhaps a product of this specific degradation pathway is necessary to activate the SpoIVFB protease. Alternatively, SpoIVB might cause sequestration rather than degradation of SpoIVFA.
In addition to its signaling role in the σK checkpoint, SpoIVB has a second distinct function required for heat-resistant spore formation, which may involve a role in synthesis of the germ cell wall deposited between the membranes surrounding the forespore (26). We showed that the SpoIVFA decrease was not due to sequestration brought about by σK-dependent gene expression and morphological change (Fig. 3B), but it remains possible that SpoIVB causes sequestration of SpoIVFA (e.g., related to the postulated role of SpoIVB in germ cell wall synthesis). Interestingly, Rudner and Losick (33) noted a region of SpoIVFA with similarity to proteins involved in peptidoglycan remodeling and proposed that SpoIVFA interacts with peptidoglycan between the membranes surrounding the forespore. Perhaps SpoIVB directly or indirectly causes linkage of SpoIVFA to peptidoglycan, resulting in activation of the pro-σK processing complex and loss of SpoIVFA from cell extracts.
Further studies are needed to determine the mechanism of SpoIVFA loss caused by SpoIVB signaling and how it differs from that caused by static incubation or treatment with inhibitors of oxidative phosphorylation or translation. The significance of such studies hinges on whether the first explanation of our results, offered above, is correct, because in this model, SpoIVFA loss is a mere consequence of activation of the processing complex, not a cause of activation. Instead, BofA is the key to regulation of the complex. Rudner and Losick (33) favor this model because they showed that a functional GFP-SpoIVFA fusion protein, which accumulates in the absence of BofA better than native SpoIVFA, did not impair sporulation or pro-σK processing. They inferred that the instability of SpoIVFA is not critical to activation of the SpoIVFB protease. However, as Rudner and Losick (33) pointed out, it is conceivable that a small amount of the GFP-SpoIVFA fusion protein was destroyed (or linked to peptidoglycan) during sporulation in their experiments. This leaves open the possibility that loss of SpoIVFA plays a role in the regulation of pro-σK processing. Rudner and Losick (33) believed this was unlikely because GFP-SpoIVFA failed to inhibit pro-σK processing even in a strain that made 5- to 10-fold less SpoIVFB. However, this only strengthens the argument if a molecule of GFP-SpoIVFA not present in a complex initially can replace a molecule of GFP-SpoIVFA lost from a complex upon its activation.
If BofA is the key to regulation of pro-σK processing, how does SpoIVB signaling overcome BofA-mediated inhibition of the SpoIVFB protease? It is possible that BofA is a direct target of SpoIVB protease activity (14, 38). Alternatively, a protein-protein interaction via SpoIVB's PDZ domain (13, 27) might somehow relieve BofA inhibition of SpoIVFB processing activity.
In bof mutant cells, the SpoIVFB protease is not inhibited, so there would be no need for translation to produce SpoIVB, and our results show that processing occurs in the presence of Cm in bof mutants (Fig. 9B and data not shown). Moreover, the processing inhibition caused by static incubation and inhibitors of oxidative phosphorylation was relieved by a bypass mutation in a similar fashion (Fig. 9C). These results demonstrate that processing of pro-σK does not require ongoing translation or an electrochemical gradient across membranes if the need for forespore signaling is bypassed mutationally. These insights are guiding our efforts to reconstitute pro-σK processing in vitro.
We favor the model that ongoing translation is normally required for efficient pro-σK processing because synthesis of the SpoIVB signal protein is needed to activate the processing complex. The bof mutant used in the experiments shown in Fig. 9B and C also contained a mutation in the spoIIIG gene that encodes σG, which is necessary to direct sufficient expression of the spoIVB gene in order for SpoIVB to carry out its signaling function in the σK checkpoint (3, 7). Indeed, transcription of spoIVB is the only role of σG that is essential for pro-σK processing (6). This is one reason we propose that the need for ongoing translation in wild-type cells reflects the need to synthesize SpoIVB. Another reason is that the active SpoIVB species may be unstable (13, 14, 38).
SpoIVB self-cleaves, generating several species with slightly different N-terminal truncations. One or more of these was proposed to be active in signaling pro-σK processing (38). However, these intermediate (44 to 46 kDa) forms of SpoIVB are subject to secondary proteolysis by one or more unidentified enzymes that may inactivate SpoIVB signaling function. Because secondary proteolysis began almost immediately after SpoIVB autoproteolysis during sporulation, it was proposed that active SpoIVB intermediates have a short half-life (38). If this model is correct, it might explain the rapid cessation of processing that we observed after adding inhibitors of oxidative phosphorylation or translation at T3.5 (Fig. 5D and 6A and data not shown), when some cells in the population had already begun processing. However, it is important to note that sporulation is somewhat asynchronous in a population of cells, so our data do not address the kinetics of signaling or processing in single cells. For example, the rapid cessation of processing that we observed could reflect the need to attain a threshold level of an active SpoIVB intermediate in a cell, which rapidly signals complete (or nearly complete) processing of all the pro-σK in that cell. According to this model, the inhibitors of oxidative phosphorylation or translation prevent any more cells in the population from attaining the threshold level of SpoIVB needed to initiate rapid signaling and processing.
Interestingly, the addition of Cm or rifampin (an inhibitor of transcription) has been shown to arrest pro-σE processing (17). Pro-σE processing requires the synthesis of SpoIIR to activate the apparent protease SpoIIGA (15, 18, 22). Hence, the inhibition of pro-σE processing by Cm or rifampin may reflect inhibition of SpoIIR synthesis.
Understanding the molecular mechanism governing pro-σK processing is important because it is a model for a type of signaling pathway used broadly in living organisms. SpoIVFB appears to be a signal-transducing membrane metalloprotease representative of a large family of such proteases present in eubacteria, archaea, and eukaryotes (21, 32, 40). Regulated intramembrane proteolysis has been proposed to be a common theme in processes as diverse as bacterial sporulation and mating, Notch signaling in animal development, human lipid metabolism, the response to unfolded proteins in the endoplasmic reticulum of mammalian cells, and processing of the amyloid precursor protein implicated in Alzheimer's disease (1). The insights gained from this study are guiding our efforts to reconstitute pro-σK processing and its regulation in vitro.
Acknowledgments
We thank O. Resnekov, D. Rudner, and R. Losick for sending bacterial strains and for communicating results prior to publication. We thank K. Carr for help with quantification of Western blot signals.
This research was supported by the Michigan Agricultural Experiment Station and by grants GM43585 (to L.K.) and GM26916 (to S.F.-M.) from the National Institutes of Health.
REFERENCES
- 1.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 2.Cutting, S., M. Anderson, E. Lysenko, A. Page, T. Tomoyasu, K. Tatematsu, T. Tatsuta, L. Kroos, and T. Ogura. 1997. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J. Bacteriol. 179:5534-5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutting, S., A. Driks, R. Schmidt, B. Kunkel, and R. Losick. 1991. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis. Genes Dev. 5:456-466. [DOI] [PubMed] [Google Scholar]
- 4.Cutting, S., V. Oke, A. Driks, R. Losick, S. Lu, and L. Kroos. 1990. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell 62:239-250. [DOI] [PubMed] [Google Scholar]
- 5.Cutting, S., S. Roels, and R. Losick. 1991. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J. Mol. Biol. 221:1237-1256. [DOI] [PubMed] [Google Scholar]
- 6.Gomez, M., S. Cutting, and P. Stragier. 1995. Transcription of spoIVB is the only role of σG that is essential for pro-σK processing during spore formation in Bacillus subtilis. J. Bacteriol. 177:4825-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez, M., and S. M. Cutting. 1996. Expression of the Bacillus subtilis spoIVB gene is under dual σF/σG control. Microbiology 142:3453-3457. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 9.Green, D., and S. Cutting. 2000. Membrane topology of the Bacillus subtilis pro-σK processing complex. J. Bacteriol. 182:278-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harlow, E., and D. Lane. 1988. Antibodies. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England.
- 12.Herman, C., D. Thevenet, R. D'Ari, and P. Bouloc. 1995. Degradation of σ32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 92:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoa, N. T., J. A. Brannigan, and S. M. Cutting. 2001. The PDZ domain of the SpoIVB serine peptidase facilitates multiple functions. J. Bacteriol. 183:4364-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoa, N. T., J. A. Brannigan, and S. M. Cutting. 2002. The Bacillus subtilis signaling protein SpoIVB defines a new family of serine peptidases. J. Bacteriol. 184:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmeister, A. E. M., A. Londono-Vallejo, E. Harry, P. Stragier, and R. Losick. 1995. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 83:219-226. [DOI] [PubMed] [Google Scholar]
- 16.Ireton, K., and A. Grossman. 1992. Interaction among mutations that cause altered timing of gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 174:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonas, R. M., S. C. Holt, and W. G. Haldenwang. 1990. Effects of antibiotics on synthesis and persistence of σE in sporulating Bacillus subtilis. J. Bacteriol. 172:4616-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karow, M. L., P. Glaser, and P. Piggot. 1995. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroos, L., and Y. T. Yu. 2000. Regulation of sigma factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 3:553-560. [DOI] [PubMed] [Google Scholar]
- 20.Kroos, L., B. Zhang, H. Ichikawa, and Y.-T. N. Yu. 1999. Control of σ factor activity during Bacillus subtilis sporulation. Mol. Microbiol. 31:1285-1294. [DOI] [PubMed] [Google Scholar]
- 21.Lewis, A., and P. Thomas. 1999. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Protein Sci. 8:439-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Londono-Vallejo, J. A., and P. Stragier. 1995. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 9:503-508. [DOI] [PubMed] [Google Scholar]
- 23.Lu, S., S. Cutting, and L. Kroos. 1995. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor pro-σK in the absence of other sporulation gene products. J. Bacteriol. 177:1082-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, S., R. Halberg, and L. Kroos. 1990. Processing of the mother-cell σ factor, σK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc. Natl. Acad. Sci. USA 87:9722-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, S., and L. Kroos. 1994. Overproducing the Bacillus subtilis mother-cell sigma factor precursor, pro-σK, uncouples σK-dependent gene expression from dependence on intercompartmental communication. J. Bacteriol. 176:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oke, V., M. Shchepetov, and S. Cutting. 1997. SpoIVB has two distinct functions during spore formation in Bacillus subtilis. Mol. Microbiol. 23:223-230. [DOI] [PubMed] [Google Scholar]
- 27.Pallen, M. J., and C. P. Ponting. 1997. PDZ domains in bacterial proteins. Mol. Microbiol. 26:411-413. [DOI] [PubMed] [Google Scholar]
- 28.Resnekov, O. 1999. Role of the sporulation protein BofA in regulating activation of the Bacillus subtilis developmental transcription factor σK. J. Bacteriol. 181:5384-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resnekov, O., S. Alper, and R. Losick. 1996. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells 1:529-542. [DOI] [PubMed] [Google Scholar]
- 30.Resnekov, O., and R. Losick. 1998. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 95:3162-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricca, E., S. Cutting, and R. Losick. 1992. Characterization of bofA, a gene involved in intercompartmental regulation of pro-σK processing during sporulation in Bacillus subtilis. J. Bacteriol. 174:3177-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudner, D., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA 96:14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudner, D. Z., and R. Losick. 2002. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 16:1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 36.Tomoyasu, T., J. Gamer, B. Bukau, M. Kanemori, H. Mori, A. J. Rutman, A. B. Oppenheim, T. Yura, K. Yamanaka, H. Niki, S. Hiraga, and T. Ogura. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 14:2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varcamonti, M., R. Marasco, M. De Felice, and M. Sacco. 1997. Membrane topology analysis of the Bacillus subtilis BofA protein involved in pro-σK processing. Microbiology 143:1053-1058. [DOI] [PubMed] [Google Scholar]
- 38.Wakeley, P. R., R. Dorazi, N. T. Hoa, J. R. Bowyer, and S. M. Cutting. 2000. Proteolysis of SpolVB is a critical determinant in signalling of pro-σK processing in Bacillus subtilis. Mol. Microbiol. 36:1336-1348. [DOI] [PubMed] [Google Scholar]
- 39.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
- 40.Yu, Y.-T. N., and L. Kroos. 2000. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 182:3305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, B., A. Hofmeister, and L. Kroos. 1998. The pro-sequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J. Bacteriol. 180:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, B., and L. Kroos. 1997. A feedback loop regulates the switch from one sigma factor to the next in the cascade controlling Bacillus subtilis mother cell gene expression. J. Bacteriol. 179:6138-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]