Abstract
Era is an essential GTPase in Escherichia coli, and Era has been implicated in a number of cellular functions. Homologues of Era have been identified in various bacteria and some eukaryotes. Using the era gene as bait in the yeast two-hybrid system to screen E. coli genomic libraries, we discovered that Era interacts with MazG, a protein of unknown function which is highly conserved among bacteria. The direct interaction between Era and MazG was also confirmed in vitro, being stronger in the presence of GDP than in the presence of GTPγS. MazG was characterized as a nucleoside triphosphate pyrophosphohydrolase which can hydrolyze all eight of the canonical ribo- and deoxynucleoside triphosphates to their respective monophosphates and PPi, with a preference for deoxynucleotides. A mazG deletion strain of E. coli was constructed by replacing the mazG gene with a kanamycin resistance gene. Unlike mutT, a gene for another conserved nucleotide triphosphate pyrophosphohydrolase that functions as a mutator gene, the mazG deletion did not result in a mutator phenotype in E. coli.
The era gene in Escherichia coli encodes an essential GTPase that binds GTP and GDP specifically and hydrolyzes GTP to GDP (1, 23). Era is associated with the cellular membrane (22). Era homologues are found in most bacteria, some of which have been shown to be essential for bacterial viability and able to cross-complement E. coli mutants defective in Era production (27, 32). These findings suggest that Era function is highly conserved in bacteria. Era homologues have also been identified in some higher eukaryotic organisms, including Antirrhinum, Caenorhabditis elegans, mice, and humans (7, 15). The Era homologue found in Antirrhinum has been shown to be essential for embryogenesis (15). It has been suggested that human Era plays a role in the regulation of apoptosis (3).
Era consists of an N-terminal GTP-binding domain and a conserved C-terminal domain that contains a putative KH domain (10) capable of binding RNA (13, 17, 24). The cellular function of Era still remains elusive. Partial loss of the Era GTPase activity or decreased synthesis of Era in E. coli resulted in elongated cells with multinucleoids and cell cycle arrest (9). It has been suggested that Era might play a role in the control of the cell cycle (8, 9, 12). It has also been shown that Era interacts with 16S rRNA and the 30S ribosomal subunit (13, 24, 29). Furthermore, 16S rRNA dimethyltransferase has been demonstrated to suppress a cold-sensitive mutant of Era E200K when expressed in a multicopy plasmid (19). An Era homologue in Streptococcus mutans was shown to modulate the GTP/GDP ratio in the cell (5). The reduction of Era production results in a lack of thermal induction of ppGpp pool levels and alters carbon metabolism in E. coli (18). Era has also been linked to the phosphoenolpyruvate-sugar phosphotransferase system, as an era temperature-sensitive mutation can be suppressed by disruption of pstN, encoding a nitrogen-related enzyme, IIA (IIAntr), a putative member of the phosphotransferase system (28).
GTP-binding proteins, known to function as molecular switches, change between an active (GTP-bound) and an inactive (GDP-bound) conformation. GTP-binding proteins can interact with GTPase activating proteins or guanine nucleotide release proteins to modulate the GTPase activity and also interact with several downstream target proteins to cause signal transduction. The interactions between the GTPase and its protein partners are important for its physiological function.
In order to identify potential proteins interacting with Era in E. coli we used the yeast two-hybrid system, with Era as bait. By screening E. coli genomic libraries, MazG was identified as a potential Era-interacting protein. The direct physical interaction between Era and MazG was confirmed by in vitro experiments. We purified MazG protein and characterized MazG as a novel nucleoside triphosphate pyrophosphohydrolase. In this study, we also explored the substrate specificity of MazG protein and its physiological role in E. coli.
MATERIALS AND METHODS
Reagents and enzymes.
Nucleotides, ampicillin, nalidixic acid, streptomycin, and rifampin were from Sigma. The restriction enzymes and DNA modifying enzymes used for cloning were from New England Biolabs. Pfu DNA polymerase was from Stratagene. The radioactive nucleotides used were from Amersham Pharmacia Biotech.
Strains and plasmids.
Saccharomyces cerevisiae strain PJ69-4A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4 gal80LYS2::GAL1-HIS3 GAL2-ADE2 met::GAL7-lacZ) was used for two-hybrid assays (16). Vectors pGAD-C1, pGAD-C2, pGAD-C3, and pGBD-C1 were described previously (16). The era gene and the various N-terminal and C-terminal deletion constructs of the era gene, as indicated in Fig. 2, were cloned into the EcoRI-BglII site of pGBD-C1 to create an in-frame translation fusion with the Gal4 DNA-binding domain. These plasmids were designated pGBD-Era, pGBD-EraΔ(1-50), pGBD-EraΔ(1-146), pGBD-EraΔ(151-301), and pGBD-EraΔ(233-301). The mutant era gene with a deletion of the G2 region was amplified by PCR from plasmid pJR302dE (26) and cloned into the EcoRI-BglII site of pGBD-C1, creating plasmid pGBD-EraΔ(40-49). The mazG gene and the various N-terminal and C-terminal deletion constructs of the mazG gene, as indicated in Fig. 2, were cloned into the EcoRI-PstI site of pGAD-C1 to create an in-frame translation fusion with the Gal4 activation domain. These plasmids were designated pGAD-MazG, pGAD-MazGΔ(1-57), pGAD-MazGΔ(1-87), pGAD-MazGΔ(1-123), pGAD-MazGΔ(1-149), pGAD-MazGΔ(200-263), and pGAD-MazGΔ(230-263). For the expression of Era, the era gene was cloned into the HindIII-EcoRI site of pET17b, creating plasmid pET17b-Era. For the expression of MBP-Era fusion protein, the era gene was cloned into the KpnI-XbaI site of pMAL-c2E (New England Biolabs), creating plasmid pMAL-Era. For the expression of MazG, the mazG gene was cloned into the NdeI-BamHI site of pET11a (New England Biolabs), creating plasmid pET11a-MazG. For the expression of MBP-MazG fusion protein, the mazG gene was cloned into the KpnI-BamHI site of pMAL-c2E, creating plasmid pMAL-MazG.
FIG. 2.
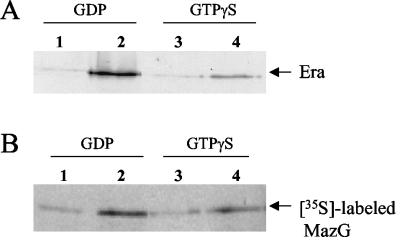
MazG and Era interaction in vitro. (A) Purified MBP-MazG fusion protein was mixed with Era in the presence of 20 μM GTPγS or GDP, and then the protein complexes were pulled down with amylose resin. The bound proteins on the resin were eluted, resolved on SDS-PAGE, blotted onto a polyvinylidene difluoride membrane, and detected with rabbit anti-Era antiserum. Lane 1, MBP, Era, and GDP; lane 2, MBP-MazG, Era, and GDP; lane 3, MBP, Era, and GTPγS; lane 4, MBP-MazG, Era, and GTPγS. (B) Purified MBP-Era fusion protein was mixed with in vitro-translated MazG labeled with [35S]Met in the presence of 20 μM GTPγS or GDP, and then the complexes were pulled down with amylose resin. The bound proteins on the resin were eluted and resolved on SDS-PAGE followed by autoradiography. Lane 1, MBP, [35S]MazG, and GDP; lane 2, MBP-Era, [35S]MazG, and GDP; lane 3, MBP, [35S]MazG, and GTPγS; lane 4, MBP-Era, [35S]MazG, and GTPγS.
An 865-bp 5′ upstream DNA fragment of mazG (from base −865 to −1; A of the initiation codon ATG is set as +1) and a 998-bp 3′ downstream fragment of mazG (from base +793 to +1790) were amplified from E. coli W3110 genomic DNA by PCR. The 865-bp upstream fragment was inserted into the EcoRI-SalI site of pUC19. The 998-bp downstream fragment was then cloned into the HindIII-SalI site of pUC19 harboring the 865-bp upstream fragment. The kanamycin-resistant gene fragment derived from SalI digestion of pUC7 was inserted into the SalI site of pUC19 harboring both the 865-bp upstream fragment and the 998-bp downstream fragment, creating plasmid pUCMDkana.
Construction of E. coli genomic two-hybrid libraries.
Genomic DNA was prepared from E. coli W3110 and partially digested with AciI, Hinp1I, and MspI, respectively. Resulting DNA fragments were separated by size on a gradient of 10 to 40% sucrose in STE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA). DNA fragments in the 500- to 2,000-bp size range were recovered by ethanol precipitation. Three different reading frame vectors, pGAD-C1, pGAD-C2, and pGAD-C3, were digested with ClaI and treated with calf intestinal alkaline phosphatase. Nine ligation reactions were performed (three vectors × three enzyme digests). Each ligation product was transformed into E. coli strain HB101 (Gibco-BRL) by electroporation. Approximately 106 transformant colonies from each transformation were used to form three E. coli genomic DNA plasmid libraries, each of which was constructed from one of the three vectors (14). The plasmid libraries were designated pGAD-EC1, pGAD-EC2, and pGAD-EC3, respectively.
Two-hybrid library screening.
Using the E. coli genomic DNA plasmid libraries pGAD-EC1, pGAD-EC2, and pGAD-EC3, two-hybrid screening was carried out according to the method of James et al. (16). Potential interactions in the yeast two-hybrid reporter strain PJ96-4A were initially selected and confirmed on synthetic dropout (SD) minimal medium (Clontech) lacking Trp, Leu, His, and adenine (Ade). The medium was supplemented with 1 mM 3-amino-1,2,4-triazole. To quantitate the protein-protein interaction, a spectrophotometric assay for β-galactosidase activity was performed using o-nitrophenyl-β-d-galactopyranoside (ONPG) as substrate.
Protein expression and purification.
Era was purified from the E. coli strain BL21(DE3)ndk::Cmr (20) harboring the expression plasmid pET17b-Era as described previously (31). MBP-MazG and MBP-Era were expressed in E. coli DH5α and purified with amylose resin (New England Biolabs) according to the manufacturer's instructions followed by Q-Sepharose column chromatography. MazG was purified from the E. coli BL21 strain harboring the expression plasmid pET11a-MazG by ammonium sulfate fractionation (30 to 60%) followed by gel filtration on a Sephadex G-100 column and Q-Sepharose column chromatography.
In vitro protein-protein interaction assay.
Purified MBP-MazG (5 μg) and Era (1 μg) were mixed in reaction buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT]) at 4°C for 4 h in the presence of 20 μM GDP or GTPγS. Amylose resin was washed with reaction buffer containing 20 μM GDP or GTPγS. An equal volume of amylose resin (50% [vol/vol]) was added into the protein mixture, and the mixture was then incubated for 2 h at 4°C with gentle mixing. The resin was washed three times with reaction buffer containing 20 μM GDP or GTPγS using ultrafree-MC centrifugal filters (Millipore), and the bound proteins were eluted with 10 mM maltose in reaction buffer. The eluted proteins were assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot with anti-Era antiserum.
In vitro translation of MazG was performed in an E. coli cell-free system with the plasmid pET11a-MazG. MazG protein was labeled with [35S]Met. Purified MBP-Era (5 μg) and radiolabeled MazG protein (6 μl of a 30-μl translation reaction) were mixed in reaction buffer in the presence of 20 μM GDP or GTPγS, and then the mixture was incubated at 4°C for 4 h. MBP pull-down assay on amylose resin was performed as mentioned above. The eluted proteins were subjected to SDS-PAGE followed by autoradiography.
Thin-layer chromatography.
The hydrolysis reaction was carried out in 20 μl of reaction buffer containing α-32P-labeled nucleoside triphosphates and the appropriate amount of protein at 37°C for 30 min. The stop solution (2% SDS, 20 mM EDTA) was added to terminate the reaction. Using the terminated reaction mixture (1 μl), thin-layer chromatography was carried out as described previously (14).
Detection of pyrophosphate.
Nucleotide triphosphate hydrolysis was performed as mentioned above with [γ-32P]GTP as substrate for MazG. The samples were spotted onto a Whatman 3MM paper. The unlabeled monophosphate and pyrophosphate were spotted alongside as markers. Paper chromatography was carried out as described by Schwemmle and Staeheli (30).
Substrate specificity of MazG protein.
A standard colorimetric assay was used to measure the nucleoside triphosphate pyrophosphohydrolase activity of MazG, with nonradioactive nucleoside triphosphates as substrates. The reaction was carried out in 50 μl of reaction buffer containing 4 mM nucleoside triphosphates, 0.5 U of yeast inorganic pyrophosphatase (Sigma), and 2 μg of MazG protein at 37°C for 15 min and then terminated by the addition of 50 μl of a mixture of four parts 20% Norit A (Sigma) and one part 7% perchloric acid. After centrifugation, Pi in the supernatant was measured by the method of Ames and Dubin (4). Pi detected in control assays without MazG was subtracted as background.
Deletion of the mazG gene.
The plasmid pUCMDkana was digested with EcoRI and HindIII. A 3.2-kb DNA fragment containing the 865-bp upstream fragment, the 1.4-kb kanamycin-resistant gene from pUC7, and the 998-bp downstream fragment was recovered and transformed into the E. coli strain BW25113 harboring a Red recombinase expression helper plasmid pKD46 (Ampr) (11). The transformants resistant to both ampicillin and kanamycin at 30°C were isolated. They were colony purified once on the plate with kanamycin at 42°C to remove the helper plasmid. Replacement of the chromosomal mazG gene with the kanamycin-resistant gene was further confirmed by PCR. The mazG deletion strain was named BW25113MD.
Assay for mutator phenotype.
The mazG deletion strain, BW25113MD, and its parent strain, BW25113, were grown in Luria-Bertani (LB) medium at 37°C to near stationary phase. Mutation frequencies were then determined using selective plates containing 100 μg of rifampin per ml, 30 μg of nalidixic acid per ml, or 150 μg of streptomycin per ml. The total viable cell numbers were determined with nonselective LB plates. Mutation frequencies were obtained by using the averages of five separate determinations.
RESULTS AND DISCUSSION
Interaction between MazG and Era in the yeast two-hybrid system.
To identify E. coli proteins that interact with Era, we constructed three E. coli genomic libraries and screened the libraries using Era as bait in the yeast two-hybrid system. Potential interactions were selected in the yeast two-hybrid reporter strain PJ69-4A (16). A library screen was performed as described in Materials and Methods. Library plasmids, which enabled PJ69-4A cells to grow on SD minimal medium lacking Trp, Leu, His, and Ade only in the presence of plasmid pGBD-Era, were sequenced and subjected to a BLAST search of the E. coli genome database. Twelve positive library plasmids containing the various fragments from the mazG gene were identified. The MazG protein consists of 263 amino acid residues. Eight positive plasmids contained a fragment from residues 58 to 263, three from residues 124 to 263, and one from residues 88 to 263. Sequence analysis revealed that all of the mazG sequences in these plasmids were in the same reading frame as the GAL4 transcriptional activation domain.
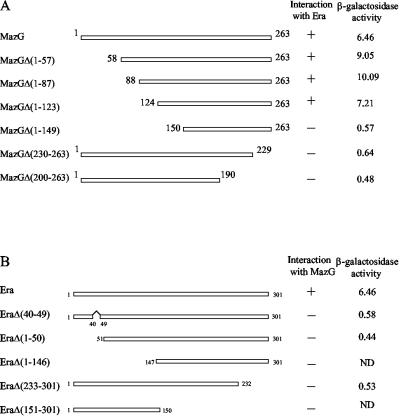
Two-hybrid assays were further performed to unambiguously demonstrate that Era indeed interacts with MazG, excluding any effects from genes downstream of the mazG gene. For this purpose, the full-length mazG gene was cloned into the pGAD-C1 vector and cotransformed with pGBD-Era or pGBD-C1 vector into PJ69-4A yeast cells. In order to localize the interaction regions between Era and MazG proteins, a series of N- and C-terminal deletions of MazG were also constructed in pGAD-C1 and cotransformed with pGBD-Era or pGBD-C1 vector into PJ69-4A cells, as shown in Fig. 1A. The cotransformants harboring pGAD-MazG, pGAD-MazGΔ(1-57), pGAD-MazGΔ(1-87), or pGAD-MazGΔ(1-123) with pGBD-Era were able to grow on SD minimal medium lacking Trp, Leu, His, and Ade, while the cotransformants with the pGBD-C1 vector were not. All of these cotransformants with pGBD-Era displayed a significant increase in β-galactosidase activity (Fig. 1A). Similar results were obtained in the reciprocal two-hybrid assays with the era gene inserted in the pGAD-C1 vector and mazG fragments inserted in the pGBD-C1 vector (data not shown). These data confirmed the initial two-hybrid library screen results, demonstrating that full-length MazG, MazGΔ(1-57), MazGΔ(1-87), and MazGΔ(1-123) can interact with Era. Further deletion from the N terminus of MazG [MazGΔ(1-149)] and deletions from the C terminus of MazG [MazGΔ(230-263) and MazGΔ(200-263)] (Fig. 1A) disrupted the two-hybrid interaction with Era. These results indicate that the C-terminal domain consisting of 140 amino acid residues of MazG is indispensable in the interaction with Era in the two-hybrid system.
FIG. 1.
MazG and Era interaction in the yeast two-hybrid system. (A) The MazG C-terminal region is required for the interaction with Era. All mazG truncation mutations were constructed in pGAD-C1 as described in Materials and Methods. Numbers refer to the amino acid positions in MazG. Protein-protein interactions were tested in yeast PJ69-4A cells in the presence of pGBD-Era on SD minimal medium plates lacking Trp, Leu, His, and Ade and supplemented with 1 mM 3-amino-1,2,4-triazole. (B) Both the N- and C-terminal regions of Era were essential for the interaction with MazG. All era mutations were constructed in pGBD-C1. Protein-protein interactions were tested in yeast PJ69-4A cells in the presence of pGAD-MazG on the same SD minimal medium plates as mentioned above. +, visible colonies formed in 5 days; −, no visible colonies formed in 5 days; ND, not detected. β-Galactosidase activity (Miller units) was measured using ONPG as substrate as described in Materials and Methods. The results shown are the averages of three independent experiments.
As shown in Fig. 1B, a series of truncation mutations from the N and C termini of Era and a G2 region deletion mutation of Era were constructed in pGBD-C1 and cotransformed with pGAD-MazG into PJ69-4A cells. All of these cotransformed yeast cells were unable to grow on SD minimal medium in the absence of Trp, Leu, His, and Ade, indicating that these Era mutants were unable to interact with MazG. Therefore, both the N- and C-terminal regions of Era appear to be essential for the interaction with MazG. It is possible that both termini of Era contain residues involved in the interaction with MazG, or the whole structure of Era may be required for binding to MazG.
Interaction of MazG with Era in vitro.
In vitro experiments were performed to confirm the interaction between MazG and Era. MBP-MazG was mixed with Era, and then amylose resin was added to the mixture. Proteins binding to the amylose resin were eluted with maltose and were then subjected to SDS-PAGE. By Western blot analysis using the anti-Era antiserum as shown in Fig. 2A, it was found that Era coeluted with MBP-MazG in the presence of GDP (Fig. 2A, lane 2) or GTPγS (Fig. 2A, lane 4), but not with MBP protein (Fig. 2A, lanes 1 and 3). These results were reproducible, indicating that Era can form a complex with MBP-MazG by interacting with MazG. The complex formation seems to be stronger in the presence of GDP (Fig. 2A, lane 2) than in the presence of GTPγS (Fig. 2A, lane 4). In the absence of either nucleotide, the Era binding affinity to MazG was about at the same level as in the presence of GTPγS (data not shown).
In order to further confirm Era-MazG complex formation, MBP-Era and 35S-labeled MazG produced in a cell-free system were mixed and protein complexes were trapped by amylose resin as mentioned above. The proteins eluted from the resin with maltose were subjected to SDS-PAGE, and then 35S-labeled MazG was detected by autoradiography. It was found that 35S-labeled MazG could form a complex with MBP-Era in the presence of GDP (Fig. 2B, lane 2) or GTPγS (Fig. 2B, lane 4), but not with MBP (Fig. 2B, lanes 1 and 3). It appeared that more 35S-labeled MazG was trapped on the resin with MBP-Era in the presence of GDP (Fig. 2B, lane 2) than in the presence of GTPγS (Fig. 2B, lane 4). The results of the in vitro experiments were consistent with the results from the yeast two-hybrid system that Era is able to interact with MazG and furthermore suggested that GDP-bound Era may bind more strongly to MazG than to GTP-bound Era.
Expression and purification of MazG.
After E. coli BL21 harboring pET11a-MazG was induced with isopropyl-β-d-thiogalactopyranoside (IPTG), MazG was produced to about 30% of the total protein and formed a major band with an approximate molecular mass of 30 kDa on the SDS-PAGE gel (not shown). MazG was purified by gel filtration followed by Q-Sepharose column chromatography to near homogeneity. When analyzed by gel filtration, the purified MazG was eluted at around 60 kDa, indicating that MazG exists as a dimer (data not shown).
MazG has no effect on the GTPase activity of Era.
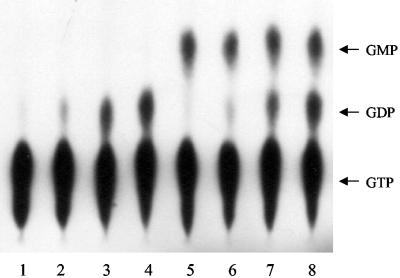
In order to examine whether the interaction between Era and MazG has any effect on the GTPase activity of Era, Era was incubated with MazG in the presence of [α-32P]GTP at 37°C. Nucleotide analysis was performed by polyethyleneimine thin-layer chromatography. It was found that MazG had no significant effect on the GTPase activity of Era, while, surprisingly, MazG could efficiently convert GTP to GMP, suggesting that MazG may have a nucleoside triphosphate pyrophosphohydrolase activity against GTP. Era also had no effect on the GTP hydrolysis activity of MazG (Fig. 3). As the GDP-bound Era has a stronger binding affinity to MazG than GTP-bound Era, GDP was added into the reaction mixture in 5 to 10 times molar excess of GTP to examine the effect of GDP on GTP hydrolysis. It was found that the GTPase activity of Era was significantly inhibited with higher GDP concentrations but was not affected by adding MazG into the reaction mixture (data not shown), indicating that the interaction between Era and MazG does not modulate their individual GTP hydrolysis activities.
FIG. 3.
Effect of MazG on the GTPase activity of Era. MazG was incubated with increasing concentrations of Era and 100 μM GTP with 5 μCi of [α-32P]GTP in 20 μl of reaction buffer at 37°C for 30 min as described in Materials and Methods. Lanes 1 to 4, 0, 1, 5, and 10 μM Era, respectively, without MazG; lanes 5 to 8, 0, 1, 5, and 10 μM Era, respectively, with 0.5 μg of MazG. The hydrolysis products were assayed by polyethyleneimine-cellulose thin-layer chromatography. The positions of GTP, GDP, and GMP are indicated.
Properties of MazG.
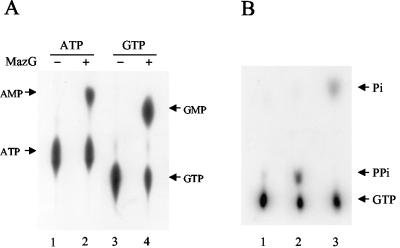
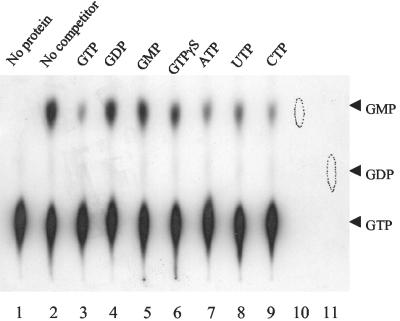
We further attempted to characterize the nucleoside triphosphate pyrophosphohydrolase activity by incubating the purified MazG with [α-32P]ATP and [α-32P]GTP at 37°C, respectively. Nucleotide analysis was performed by polyethyleneimine-cellulose thin-layer chromatography. It was revealed that MazG not only converted GTP to GMP (Fig. 4A, lane 4) but also converted ATP to AMP (Fig. 4A, lane 2). When [γ-32P]GTP was used as the substrate for MazG and the reaction products were separated by paper chromatography and visualized by autoradiography, a strong radioactive signal was observed at the position of the pyrophosphate marker (Fig. 4B, lane 2). When yeast inorganic pyrophosphatase was added into the reaction mixture, the radioactive pyrophosphate spot disappeared with the concomitant appearance of a new spot corresponding to the position of the inorganic phosphate marker (Fig. 4B, lane 3). These results indicate that MazG has nucleoside triphosphate pyrophosphohydrolase activity hydrolyzing GTP to GMP and PPi. When the GTP hydrolysis activity of MazG was tested in the presence of the unlabeled nucleotides as competitors (200-fold excess), GTP, ATP, CTP, and UTP effectively competed with GTP to block the reaction. On the other hand, GDP, GMP, and GTPγS had no significant influence on the hydrolysis of GTP (Fig. 5). It was also found that dGTP, dATP, dCTP, and dTTP effectively competed with GTP to block the reaction (data not shown).
FIG. 4.
Nucleoside triphosphate pyrophosphohydrolase activity of MazG. (A) Conversion of ATP to AMP and of GTP to GMP by MazG. Reactions were performed in 20 μl of reaction buffer containing 10 μM ATP with 1 μCi of [α-32P]ATP (lanes 1 and 2) or 10 μM GTP with 1 μCi of [α-32P]GTP (lanes 3 and 4) at 37°C for 30 min. Lanes 1 and 3, controls without MazG; lanes 2 and 4, samples with 1 μg of MazG. The hydrolysis products were assayed by polyethyleneimine-cellulose thin-layer chromatography. The positions of ATP, AMP, GTP, and GMP are indicated. (B) Production of pyrophosphate in the MazG-catalyzed hydrolysis of GTP. Reactions were performed in 20 μl of reaction buffer containing 10 μM GTP with 1 μCi of [γ-32P]GTP at 37°C for 15 min. Samples were analyzed by paper chromatography as described in Materials and Methods. Lane 1, without MazG protein; lane 2, with 0.5 μg of MazG; lane 3, the sample in lane 2 was treated with 0.5 U of yeast inorganic pyrophosphatase at 37°C for 10 min. The positions of GTP, monophosphate, and pyrophosphate are indicated.
FIG. 5.
GTP hydrolysis activity of MazG in the presence of nucleotide competitors. The GTP hydrolysis activity assays were carried out in 20 μl of reaction buffer containing 10 μM GTP with 1 μCi of [α-32P]GTP and 0.5 μg of MazG at 37°C for 30 min. The hydrolysis products were assayed by polyethyleneimine-cellulose thin-layer chromatography. Lane 1, without MazG protein; lane 2, without any nucleotide competitor; lanes 3 to 9, with 2 mM nucleotide competitors as indicated. Circles in lanes 10 and 11 correspond to unlabeled GMP and GDP, respectively. The positions of GTP, GDP, and GMP are indicated.
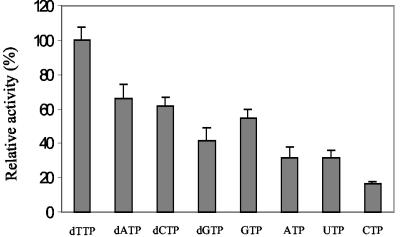
Next, the activity of MazG toward each of the eight canonical ribo- and deoxynucleoside triphosphates was determined by the colorimetric assay as described in Materials and Methods. MazG hydrolyzed all eight of the canonical ribo- and deoxynucleoside triphosphates to their respective monophosphates and PPi. Pi was not detected unless inorganic pyrophosphatase was added to the reaction mixtures. At a fixed concentration of 4 mM, the most preferred substrate for MazG was found to be dTTP, followed by dATP and dCTP (Fig. 6). It seems that MazG has higher activity towards deoxynucleoside triphosphates than ribonucleoside triphosphates, indicating that MazG recognizes not only the nucleotide base but also the sugar group.
FIG. 6.
Substrate specificity of MazG. The hydrolysis of various substrates was detected with a standard colorimetric assay as described in Materials and Methods. The hydrolysis activity with each substrate was depicted relative to the activity with dTTP, which was taken as 100 percent. Each value is the mean from three separate experiments.
Similar to other nucleoside triphosphate pyrophosphohydrolases, the nucleotide hydrolysis by MazG requires divalent cation Mg2+ or Mn2+ (5 mM). The addition of EDTA effectively blocked its activity. The effect of pH on the nucleotide hydrolysis activity of MazG was analyzed under various pH conditions. The nucleotide hydrolysis activity of MazG has an optimum at pH 9.5 (not shown).
What is the role of MazG?
There are 52 protein sequences homologous to E. coli MazG, two of them from Archaea, and the rest from bacteria. Sequence alignments of MazG proteins from E. coli, Yersinia pestis, Vibrio cholerae, Pasteurella multocida, Haemophilus influenzae, Caulobacter crescentus, Agrobacterium tumefaciens, and Thermotoga maritima are shown in Fig. 7. MazG is highly conserved and the sequence similarities to E. coli MazG in these bacteria are present throughout the sequence.
FIG. 7.
Sequence alignments of MazG proteins. Sequence alignments of eight bacterial MazG proteins are shown. The ClustalW program was used for alignment analysis. Identical residues among the eight different proteins are shown in black boxes, and similar residues are shown in gray boxes. Gaps (indicated by dashes) were introduced to optimize the alignment. The sequences are from E. coli (GenBank accession no. P33646), Y. pestis (NP_406840), V. cholerae (NP_232079), P. multocida (NP_246222), H. influenzae (NP_438621), C. crescentus (NP_420555), A. tumefaciens (NP_354459), and T. maritima (NP_228721). The numbers correspond to amino acid residue numbers.
In E. coli, the mazG gene is located downstream of mazEF, which is a chromosomal “addiction module” proposed to be responsible for programmed cell death (2). The relationship between mazG and mazEF is still unknown. In order to discover if mazG is an essential gene for E. coli, we have constructed a mazG deletion strain by replacing the mazG gene with a kanamycin-resistant gene as described in Materials and Methods. The mazG deletion strain was able to form colonies under several growth conditions such as low (15°C) and high (37 and 42°C) temperatures on LB medium or M9 medium, indicating that the mazG function is not required under normal growth conditions.
Some other E. coli proteins, such as MutT and Orf17, also catalyze the hydrolysis of ribo- and deoxynucleoside triphosphates, yielding inorganic pyrophosphate and nucleoside monophosphates (6, 25). However, they have different and more substrate specificities than MazG. dATP is the preferred substrate for Orf17, and dGTP is the preferred substrate for MutT, while MazG does not have a strong substrate specificity among the eight canonical nucleoside triphosphates. Moreover, there is no significant amino acid sequence similarity between MazG and MutT or Orf17. It has been shown that MutT hydrolyzes 8-oxo-dGTP, the mutagenic form of dGTP, thus preventing AT-to-CG mutations (21). Orf17 is of unknown function and does not have antimutator properties (25). In order to find if MazG is also involved in preventing mutations, the mutation frequencies of the mazG deletion strain and its parent strain were determined on LB plates containing different antibiotics. As shown in Table 1, frequencies of spontaneous mutation to rifampin, nalidixic acid, and streptomycin resistance did not significantly increase in the mazG deletion strain, indicating that MazG does not play a role as an antimutator.
TABLE 1.
Spontaneous mutation frequencies of the mazG deletion strain
| Strain | No. of drug-resistant mutants/108 cellsa
|
||
|---|---|---|---|
| Rifampin | Nalidixic acid | Streptomycin | |
| BW25113 | 7.2 ± 1.4 | 1.6 ± 0.5 | 0.2 ± 0.1 |
| BW25113MD | 10.4 ± 3.4 | 0.2 ± 0.1 | 0.9 ± 0.5 |
Numbers represent the average (± standard deviation) drug-resistant cell numbers from five separate experiments.
Although our two-hybrid system and in vitro experiments demonstrated their physical interaction, the significance of the interaction between MazG and Era remains elusive at present. It is possible that another protein factor(s) may be needed for their functional interaction. It is interesting that GDP-bound Era binds more tightly to MazG than GTP-bound Era, suggesting that Era may play a role as a molecular switch in regulating the function associated with MazG. Further studies on the function of MazG may provide insights into our understanding of the physiological role of MazG and the functional significance of its interaction with Era.
Acknowledgments
We thank Sangita Phadtare for her assistance in manuscript editing and Mangjing Pan for technical assistance in the in vitro translation.
REFERENCES
- 1.Ahnn, J., P. E. March, H. E. Takiff, and M. Inouye. 1986. A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc. Natl. Acad. Sci. USA 83:8849-8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama, T., J. Gohda, S. Shibata, Y. Nomura, S. Azuma, Y. Ohmori, S. Sugano, H. Arai, T. Yamamoto, and J. Inoue. 2001. Mammalian homologue of E. coli Ras-like GTPase (ERA) is a possible apoptosis regulator with RNA binding activity. Genes Cells 6:987-1001. [DOI] [PubMed] [Google Scholar]
- 4.Ames, B. N., and D. T. Dubin. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acids. J. Biol. Chem. 235:769-775. [PubMed] [Google Scholar]
- 5.Baev, D., R. England, and H. K. Kuramitsu. 1999. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect. Immun. 67:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatnagar, S. K., L. C. Bullions, and M. J. Bessman. 1991. Characterization of the mutT nucleoside triphosphatase of Escherichia coli. J. Biol. Chem. 266:9050-9054. [PubMed] [Google Scholar]
- 7.Britton, R. A., S. M. Chen, D. Wallis, T. Koeuth, B. S. Powell, L. G. Shaffer, D. Largaespada, N. A. Jenkins, N. G. Copeland, D. L. Court, and J. R. Lupski. 2000. Isolation and preliminary characterization of the human and mouse homologues of the bacterial cell cycle gene era. Genomics 67:78-82. [DOI] [PubMed] [Google Scholar]
- 8.Britton, R. A., B. S. Powell, D. L. Court, and J. R. Lupski. 1997. Characterization of mutations affecting the Escherichia coli essential GTPase era that suppress two temperature-sensitive dnaG alleles. J. Bacteriol. 179:4575-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britton, R. A., B. S. Powell, S. Dasgupta, Q. Sun, W. Margolin, J. R. Lupski, and D. L. Court. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 27:739-750. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X., D. L. Court, and X. Ji. 1999. Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc. Natl. Acad. Sci. USA 96:8396-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gollop, N., and P. E. March. 1991. A GTP-binding protein (Era) has an essential role in growth rate and cell cycle control in Escherichia coli. J. Bacteriol. 173:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hang, J. Q., T. I. Meier, and G. Zhao. 2001. Analysis of the interaction of 16S rRNA and cytoplasmic membrane with the C-terminal part of the Streptococcus pneumoniae Era GTPase. Eur. J. Biochem. 268:5570-5577. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, J., and M. Inouye. 2001. An essential GTPase, der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritima. J. Biol. Chem. 276:31415-31421. [DOI] [PubMed] [Google Scholar]
- 15.Ingram, G. C., R. Simon, R. Carpenter, and E. S. Coen. 1998. The Antirrhinum ERG gene encodes a protein related to bacterial small GTPases and is required for embryonic viability. Curr. Biol. 8:1079-1082. [DOI] [PubMed] [Google Scholar]
- 16.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnstone, B. H., A. A. Handler, D. K. Chao, V. Nguyen, M. Smith, S. Y. Ryu, E. L. Simons, P. E. Anderson, and R. W. Simons. 1999. The widely conserved Era G-protein contains an RNA-binding domain required for Era function in vivo. Mol. Microbiol. 33:1118-1131. [DOI] [PubMed] [Google Scholar]
- 18.Lerner, C. G., and M. Inouye. 1991. Pleiotropic changes resulting from depletion of Era, an essential GTP-binding protein in Escherichia coli. Mol. Microbiol. 5:951-957. [DOI] [PubMed] [Google Scholar]
- 19.Lu, Q., and M. Inouye. 1998. The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of era, an essential RAS-like GTP-binding protein in Escherichia coli. J. Bacteriol. 180:5243-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, Q., X. Zhang, N. Almaula, C. K. Mathews, and M. Inouye. 1995. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J. Mol. Biol. 254:337-341. [DOI] [PubMed] [Google Scholar]
- 21.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355:273-275. [DOI] [PubMed] [Google Scholar]
- 22.March, P. E. 1992. Membrane-associated GTPases in bacteria. Mol. Microbiol. 6:1253-1257. [DOI] [PubMed] [Google Scholar]
- 23.March, P. E., C. G. Lerner, J. Ahnn, X. Cui, and M. Inouye. 1988. The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene 2:539-544. [PubMed] [Google Scholar]
- 24.Meier, T. I., R. B. Peery, S. R. Jaskunas, and G. Zhao. 1999. 16S rRNA is bound to era of Streptococcus pneumoniae. J. Bacteriol. 181:5242-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Handley, S. F., D. N. Frick, L. C. Bullions, A. S. Mildvan, and M. J. Bessman. 1996. Escherichia coli orf17 codes for a nucleoside triphosphate pyrophosphohydrolase member of the MutT family of proteins. Cloning, purification, and characterization of the enzyme. J. Biol. Chem. 271:24649-24654. [DOI] [PubMed] [Google Scholar]
- 26.Pillutla, R. C., J. Ahnn, and M. Inouye. 1996. Deletion of the putative effector region of Era, an essential GTP-binding protein in Escherichia coli, causes a dominant-negative phenotype. FEMS Microbiol. Lett. 143:47-55. [DOI] [PubMed] [Google Scholar]
- 27.Pillutla, R. C., J. D. Sharer, P. S. Gulati, E. Wu, Y. Yamashita, C. G. Lerner, M. Inouye, and P. E. March. 1995. Cross-species complementation of the indispensable Escherichia coli era gene highlights amino acid regions essential for activity. J. Bacteriol. 177:2194-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell, B. S., D. L. Court, T. Inada, Y. Nakamura, V. Michotey, X. Cui, A. Reizer, M. H. Saier, Jr., and J. Reizer. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 270:4822-4839. [DOI] [PubMed] [Google Scholar]
- 29.Sayed, A., S. Matsuyama, and M. Inouye. 1999. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 264:51-54. [DOI] [PubMed] [Google Scholar]
- 30.Schwemmle, M., and P. Staeheli. 1994. The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem. 269:11299-11305. [PubMed] [Google Scholar]
- 31.Sood, P., C. G. Lerner, T. Shimamoto, Q. Lu, and M. Inouye. 1994. Characterization of the autophosphorylation of Era, an essential Escherichia coli GTPase. Mol. Microbiol. 12:201-208. [DOI] [PubMed] [Google Scholar]
- 32.Zuber, M., T. A. Hoover, M. T. Dertzbaugh, and D. L. Court. 1997. A Francisella tularensis DNA clone complements Escherichia coli defective for the production of Era, an essential Ras-like GTP-binding protein. Gene 189:31-34. [DOI] [PubMed] [Google Scholar]