Abstract
A new Salmonella enterica serovar Typhimurium strain has been constructed to facilitate tightly regulated gene expression. Arabinose-inducible and glucose-repressible expression of a T7 RNA polymerase gene that has been integrated with an adjacent araC-PBAD control element into the bacterial chromosome allows dynamic control of T7 promoter-driven RNA transcription.
We have designed a new Salmonella enterica serovar Typhimurium construct to emulate the Escherichia coli BL21(DE3) system (16, 17). The construct allows for improved control of gene expression, particularly in terms of decreasing induction-independent leaky expression (2). This Salmonella construct has stable, site-specific chromosomal integration of the araC-PBAD control element (6) in tandem with the T7 RNA polymerase gene (12). The arabinose araC-PBAD element allows for quite exact control of T7 RNA polymerase expression. The T7 RNA polymerase in turn transcribes genes under T7 promoter control, including those encoding external guide sequences (EGSs) (4). EGSs are oligoribonucleotides that are used to target a complementary mRNA for cleavage by RNase P in vitro or in vivo (4) in E. coli (5, 10). This new construct provides a general expression platform for tightly controlled T7 promoter-driven gene transcription in Salmonella and, in particular, allows for studies of EGSs in this bacterium.
Two plasmids were produced for the controlled expression of T7 RNA polymerase: one for chromosomal integration (pSBaadABADT7-1) and one for maintenance as a replicating plasmid (pACBADT7-1). Performance data for the latter are available on request. Our plasmid nomenclature employs the abbreviations BAD for the araC-PBAD control element (6) and T7-1 for T7 RNA polymerase (12). Figure 1 shows details of plasmid construction. The pSBaadABADT7-1 plasmid contains one copy of the araC-PBAD-T7pol genes, flanked by aadA sequences, in a pSB890 backbone (8).
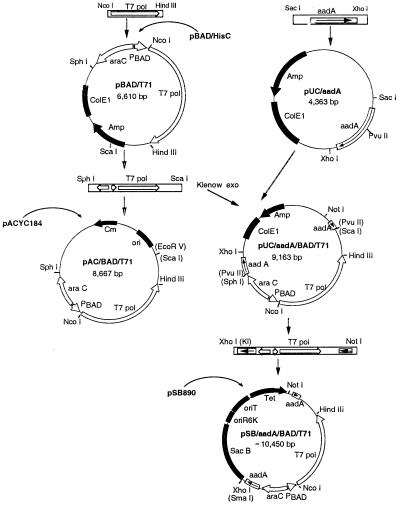
FIG. 1.
Schematic view of vector construction. Maps of the arabinose-inducible T7 RNA polymerase expression plasmids pSB/aadA/BADT7-1 and pAC/BADT7-1 and their construction intermediates are shown with relevant features. Products of the serial steps of construction are shown at the confluence of schematic arrows leading from that product's constituent parts: for example, pBAD/T7-1 results from the cloning of the NcoI-HindIII-delimited fragment containing T7pol into the pBAD/HisC plasmid. Plasmid names are in bold type, including those of vectors denoted by name alone (pBAD/HisC, pACYC184, and pSB890), which provided backbones for subsequent constructs. Cloned fragments used as inserts for construction steps are represented as ribbons that contain arrows denoting key functional components and their 5′-3′ orientations. The aadA gene in pUC/aadA is interrupted (by insertion into its PvuII site) during the construction of pUC/aadA/BADT7-1, and hence aadA is subsequently represented as two interrupted parts. Pertinent restriction sites are provided; restriction sites listed in parentheses [such as (SmaI) in pSB/aadA/BADT7-1] are destroyed by the cloning process. XhoI (Kl) is an XhoI site made blunt by the Klenow fragment.
The new Salmonella construct, SB300A#1, was produced from the Salmonella parent strain SB300 (8) by using homologous recombination techniques (7) in which E. coli SM10λpir (15) serves as a conjugation partner that donates pSBaadABADT7-1 to SB300. Key components of pSBaadABADT7-1 include tet, oriR6K (7, 11), sacB (7), aadA (7, 9), araC-PBAD (6), and the T7 RNA polymerase gene (12). The aadA gene, encoding a streptomycin adenylyltransferase which can mediate streptomycin and spectinomycin resistance, serves as the site of recombination (7, 9). Step-by-step details of our selection strategy for SB300A#1 are available on request. Briefly, we first selected for tetracycline resistance and then against sucrose sensitivity of the tet and sacB components, respectively, of pSBaadABADT7-1. This method is directly analogous to the approach described previously (7; Fig. 2 of this reference describes the genetics of the homologous recombination). Other selection maneuvers of potential utility for the pSBaadABADT7-1 system include screening for the loss of spectinomycin resistance (9) after homologous recombination into aadA (SB300A#1 loses the spectinomycin resistance exhibited by SB300 [data not shown]) and/or selecting for gain and subsequent loss (1) of tetracycline resistance.
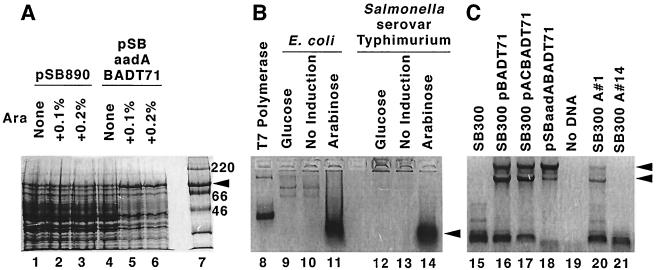
FIG. 2.
Assessment of intermediate construction steps. (A) Expression of presumptive T7 RNA polymerase after induction with arabinose. Protein expression is shown for crude cell extracts of E. coli which were transformed either with pSB890 (lanes 1 to 3) or with pSBaadABADT7-1 (lanes 4 to 6) and then exposed during growth in liquid culture to the indicated arabinose concentrations. Lane 7 provides molecular mass markers: a 97.4-kDa band is left of the arrowhead. For lanes 5 and 6, the arrowhead denotes the expression of a protein of appropriate size (∼99 kDa) for T7 RNA polymerase. This expression occurs after induction with arabinose in E. coli transformed with pSBaadABADT7-1 (lanes 4, 5, and 6) but not in pSB890 transformants (lanes 1, 2, and 3), which lack the araC control element and the T7 RNA polymerase gene. (B) In vitro enzymatic activity of T7 RNA polymerase that was recovered from crude cell extracts of bacterial transformants. T7 RNA polymerase activity was assessed by detection of RNA (denoted by the arrowhead) with an ethidium bromide agarose gel, resulting from transcription in vitro of DNA behind a T7 promoter. Purified recombinant T7 RNA polymerase was used as a positive control (lane 8). Other specimens are crude cell extracts of E. coli (lanes 9, 10, and 11) or Salmonella serovar Typhimurium (lanes 12, 13, and 14) transformants. Both E. coli and Salmonella serovar Typhimurium contain plasmids encoding T7 RNA polymerase and the araC control element and either were subjected in liquid culture to treatment with glucose (for repression) or arabinose (for induction) or were not induced, as shown. E. coli cells were transformed with pSBaadABADT7-1 and were used as donors for subsequent conjugation. Salmonella serovar Typhimurium cells were transformed with pBADT7, an intermediate plasmid in the construction of pSBaadABADT7-1 (Fig. 1). (C) Detection of the T7 RNA polymerase gene in bacterial transformants and conjugation products by using PCR amplification. Arrowheads indicate PCR products corresponding with the presence of the T7 RNA polymerase gene. PCR amplification of the T7 RNA polymerase gene from plasmid or genomic DNA is shown for various Salmonella serovar Typhimurium constructs. Lane 15 is a negative control, the parent Salmonella serovar Typhimurium strain (SB300) prior to manipulation. Lanes 16 and 17 are Salmonella serovar Typhimurium transformants containing the T7 RNA polymerase gene in the plasmids pBADT7-1 and pACBADT7-1 (Fig. 1). Lane 18 is a large-scale plasmid preparation of pSBaadABADT7-1 from an E. coli transformant. Lane 19 is the PCR mixture without DNA. Lanes 20 and 21 are Salmonella serovar Typhimurium strains recovered after the selection and screening steps described in the text. The strains in lanes 20 (denoted SB300 A#1) and 21 (denoted SB300 A#14) are with and without a PCR-detectable T7 RNA polymerase gene, respectively. Both SB300A#1 and SB300A#14 were used for the in vivo experiment described in the legend for Fig. 3; SB300A#1 was used for the experiments reported in the legend for Fig. 4.
The araC-PBAD and T7 RNA polymerase components (not previously used in pSB890-based systems) were assessed in several ways. Portions (totaling ∼250 bases at the 5′ end [data not shown]) of the T7 RNA polymerase gene were sequenced, and all were as per the reported sequence (12). An appropriately sized protein (∼99 kDa) for T7 RNA polymerase was produced, in an arabinose-inducible manner, in E. coli cells that had been transformed with pSBaadABADT7-1 (Fig. 2A). In all experiments using metabolic regulators, a 0.2% concentration of arabinose or glucose in Luria-Bertani medium was used unless otherwise specified. Culture incubation conditions were as previously described (10). We will supply complete information regarding experimental methods on request.
The arabinose-inducible T7 RNA polymerase expression product recovered from 2 μl of crude cell extracts of either E. coli or Salmonella serovar Typhimurium transformants was shown to have functional enzymatic activity in the transcription in vitro of 2 μg of the DNA template from plasmid pBSC5 (3) under T7 promoter control (Fig. 2B). PCR analysis was used to demonstrate the new acquisition of the T7 RNA polymerase gene by Salmonella strain SB300 after either transformation of cells (13) with relevant plasmids or selection of cells based on the stable integration of the gene into the chromosome of SB300 (Fig. 2C). For PCRs, we used T7NcoF (ACCATGGGGAACACGATTAACATCGC) and T7HIIIR (CCGAAGCTTACGCGAACGCGAAG) as primers and 25 cycles of 94, 37, and 72°C. A total of four stable integration candidates (two are shown in Fig. 2C) were screened via PCR, with two candidates (including SB300A#1) being positive and two (including SB300A#14) being negative.
The control in vivo and the activity of the integrated T7 RNA polymerase in SB300A#1 were assessed by using Northern blotting to detect a T7 promoter-driven RNA strand, transcribed from the plasmid pUC19 M1:AEF2HH (C. Guerrier-Takada, unpublished data), which encodes the M1 RNA component of RNase P (14) fused with a “three-quarter” form EGS (4). In cell extracts of cultures (grown for 1 h with or without induction) containing similar amounts of constitutively expressed RNA, E. coli BL21(DE3) exhibits leaky basal expression of a T7 promoter-driven RNA even without induction with IPTG (isopropyl-β-d-thiogalactopyranoside) (Fig. 3). By contrast, in the new Salmonella serovar Typhimurium construct SB300A#1, leaky expression of the T7 promoter-driven RNA is not detected. Expression is seen in SB300A#1 after induction with arabinose (Fig. 3) but not in SB300A#14, which lacks integrated T7 RNA polymerase (Fig. 2 and 3).
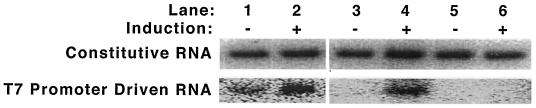
FIG. 3.
Basal and induced gene expression in E. coli BL21(DE3) and the new Salmonella serovar Typhimurium construct. Northern blots of constitutive and induced RNA transcription are shown for two systems: E. coli lac-based induction with IPTG (lanes 1 and 2) and the new Salmonella serovar Typhimurium system, ara-based induction with arabinose (lanes 3 to 6). The Salmonella serovar Typhimurium in lanes 3 and 4 (SB300A#1) has a PCR-detectable T7 RNA polymerase gene in genomic DNA, whereas the Salmonella serovar Typhimurium in lanes 5 and 6 (SB300A#14) lacks an integrated T7 RNA polymerase gene (Fig. 2C). Note the leaky transcription of T7 promoter-driven RNA even without induction in the E. coli lac system, in contrast to the off-on expression in the new Salmonella serovar Typhimurium ara system. Levels of constitutive RNA expression (assessed here by Northern blotting for the M1 RNA component of RNase P) are grossly similar in all samples, which were total RNA extracts from bacteria pelleted from liquid cultures.
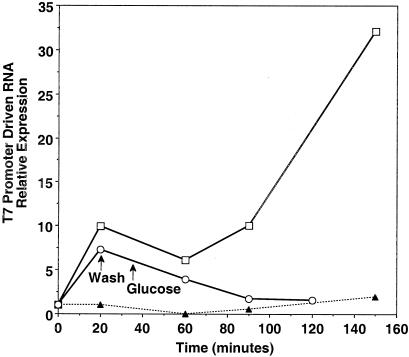
The kinetics of T7 promoter-driven RNA expression in SB300A#1 are shown in Fig. 4. The results are normalized to constitutively expressed 5S RNA. Induced RNA expression is detected within 20 min of arabinose induction of SB300A#1 liquid cultures. This RNA shows an increased level of expression from 1 to 2.5 h after induction, and this level decreases by 19 h after induction. This induced expression can be modulated, as shown when glucose was added to a liquid culture of SB300A#1 bacteria, which was pelleted and washed after 20 min of induction with arabinose. Arabinose-induced T7 promoter-driven RNA expression was repressed to the background levels seen in uninduced bacteria within 60 to 90 min of the addition of glucose. Resuspension of the same arabinose-induced cultures in glucose-free medium does not inhibit expression of T7 promoter-driven RNA at 90 min (data not shown).
FIG. 4.
Kinetics of gene expression control. Quantitation of on and off gene expression after induction with arabinose and repression with glucose. Northern blot signals of induced T7 promoter-driven RNA expression, normalized to constitutive RNA expression, are shown over time. Specimens are total RNA extracts of Salmonella serovar Typhimurium bacteria pelleted from liquid cultures at various times after the addition of the inducing agent arabinose (□), with arabinose induction followed by washing and by the addition of glucose (○), or without the addition of arabinose (▴).
The dynamic response in gene expression observed with this system was not guaranteed. While the araC control element does exhibit a clear distinction between induction and repression, the T7 RNA polymerase expressed under araC control must subsequently transcribe the gene of interest. The induced T7 RNA polymerase appears to have net effects on transcription which peak and then decrease rapidly. Furthermore, the reporter RNA we measure as an output of our control system likely has a short survival half-life in the cell. Together, these features prevent the system from becoming overly damped. That is, despite a series of required steps between the addition of arabinose (or glucose) and the expression (or repression) of the gene product of interest, the transition between on and off expression states remains clear.
Acknowledgments
We thank Matthew Kelly and Alessandra Pancetti for their expert advice on conjugation and selection steps.
This research was conducted while J.M. was a Pfizer postdoctoral fellow. This work was supported by a National Institutes of Health research fellow training program (NIH T32 AI07210-17), a Yale Children's Health Research Center Scholar Award (to J.M.), and National Institutes of Health grant GM19422 (to S.A.).
REFERENCES
- 1.Bochner, B. R., H.-C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doherty, A. J., B. A. Connolly, and A. F. Worrall. 1993. Overproduction of the toxic protein, bovine pancreatic DNase I, in Escherichia coli using a tightly controlled T7-promoter-based vector. Gene 136:337-340. [DOI] [PubMed] [Google Scholar]
- 3.Gopalan, V., A. D. Baxevanis, D. Landsman, and S. Altman. 1997. Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J. Mol. Biol. 267:818-829. [DOI] [PubMed] [Google Scholar]
- 4.Guerrier-Takada, C., and S. Altman. 2000. Inactivation of gene expression using ribonuclease P and external guide sequences. Methods Enzymol. 313:442-456. [DOI] [PubMed] [Google Scholar]
- 5.Guerrier-Takada, C., Y. Li, and S. Altman. 1995. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc. Natl. Acad. Sci. USA 92:11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 8.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 9.Leung, K. Y., S. R. Ruschkowski, and B. B. Finlay. 1992. Isolation and characterization of the aadA aminoglycoside-resistance gene from Salmonella choleraesuis. Mol. Microbiol. 6:2453-2460. [DOI] [PubMed] [Google Scholar]
- 10.McKinney, J., C. Guerrier-Takada, D. Wesolowski, and S. Altman. 2001. Inhibition of Escherichia coli viability by external guide sequences complementary to two essential genes. Proc. Natl. Acad. Sci. USA 98:6605-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffatt, B. A., J. J. Dunn, and F. W. Studier. 1984. Nucleotide sequence of the gene for bacteriophage T7 RNA polymerase. J. Mol. Biol. 173:265-269. [DOI] [PubMed] [Google Scholar]
- 13.O'Callaghan, D., and A. Charbit. 1990. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol. Gen. Genet. 223:156-158. [DOI] [PubMed] [Google Scholar]
- 14.Reed, R. E., M. F. Baer, C. Guerrier-Takada, H. Donis-Keller, and S. Altman. 1982. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell 30:627-636. [DOI] [PubMed] [Google Scholar]
- 15.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 16.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 17.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]