Abstract
The ruminant pathogen Mycoplasma agalactiae possesses a family of abundantly expressed variable surface lipoproteins called Vpmas. Phenotypic switches between Vpma members have previously been correlated with DNA rearrangements within a locus of vpma genes and are proposed to play an important role in disease pathogenesis. In this study, six vpma genes were characterized in the M. agalactiae type strain PG2. All vpma genes clustered within an 8-kb region and shared highly conserved 5′ untranslated regions, lipoprotein signal sequences, and short N-terminal sequences. Analyses of the vpma loci from consecutive clonal isolates showed that vpma DNA rearrangements were site specific and that cleavage and strand exchange occurred within a minimal region of 21 bp located within the 5′ untranslated region of all vpma genes. This process controlled expression of vpma genes by effectively linking the open reading frame (ORF) of a silent gene to a unique active promoter sequence within the locus. An ORF (xer1) immediately adjacent to one end of the vpma locus did not undergo rearrangement and had significant homology to a distinct subset of genes belonging to the λ integrase family of site-specific xer recombinases. It is proposed that xer1 codes for a site-specific recombinase that is not involved in chromosome dimer resolution but rather is responsible for the observed vpma-specific recombination in M. agalactiae.
Mycoplasmas belong to a group of cell wall-less pathogens that are widespread in nature and cause diseases that are generally chronic and difficult to eradicate in humans, animals, plants, and insects. Among free-living organisms, they possess one of the smallest genomes, which is mainly responsible for their parasitic lifestyle. Yet, despite having a limited coding capacity relative to other bacteria, mycoplasmas have developed quite an extensive repertoire of molecular mechanisms to generate surface protein variability at a high frequency either by phase or size variation (10, 28, 42). Many of these surface proteins belong to multigene families of lipoproteins that have the ability to switch the expression of individual components on and off and are suspected of playing a role in host cell adhesion. The ability of these proteins to switch expression between different forms is believed to be important in immune evasion and rapid adaptation in the host.
Mycoplasma agalactiae is the major etiological agent of the syndrome contagious agalactia, which primarily causes mastitis, arthritis, and keratoconjunctivitis in sheep and goats (4). Recently, a family of related but distinct surface lipoproteins, designated Vpmas, has been identified in this pathogen, and Vpma expression has been shown to undergo high-frequency variation (16). Vpmas are abundantly expressed in M. agalactiae and display an unusual processing of lipoproteins in which a peptidase II-like enzyme cleaves 2 amino acids (aa) (position −2) N terminal to the cysteine residue rather than at the cysteine to which the lipid anchor is attached. This atypical processing is also being increasingly observed with other mycoplasma lipoproteins such as P48 (position −2) from M. agalactiae (31) and MAA2 (position −1) from M. arthritidis (41). Detailed genetic analyses have suggested that the vpma system is composed of several genes, two of which have been sequenced (16). The 5′ untranslated regions and those encoding the signal peptide were found to be conserved within the vpma family and revealed a high identity to the equivalent regions in genes encoding a family of lipoproteins, called Vsps, in the phylogenetically related species M. bovis (3, 20, 23). Based on these data, the Vpmas of M. agalactiae were proposed to represent a system homologous to the Vsps in M. bovis (16). In addition, the two sequenced vpma genes were found to encode some of the epitopes that have been shown to be involved in Vsp host binding (32), suggesting that Vpmas could share a similar function. This, together with the fact that Vpmas are abundant and are released from their cell surfaces at levels as high as 59% relative to that present in the cells themselves per volume of medium (16), may emphasize the importance of Vpmas and Vpma switching in relation to disease progression. However, this issue has not yet been extensively examined as such studies require a precise characterization of the vpma system.
Control of vpma gene expression has been hypothesized to be linked to DNA rearrangements within the vpma locus, as vpma DNA restriction fragment length polymorphisms were found to correlate with switches in Vpma expression among clonal variants (16). DNA rearrangements associated with changes in gene expression have also been found for Vsps (20, 22). Two other well-described systems involving DNA rearrangements in mycoplasmas are the vsa and hsd systems in M. pulmonis (5, 35, 37, 38). Both systems generate variation in expression of a given product by exchanging the C-terminal region of the active gene with the open reading frame (ORF) of a silent gene by site-specific recombination. Each expressed gene possesses the same N-terminal sequence and active promoter. A third system, the vlh multigene family encoding surface lipoproteins in M. synoviae, involves gene conversion and the replacement of the C-terminal region of the expressed gene within an expression locus with another C-terminal sequence from a pool of vlh ORFs (25). The nature of the enzyme(s) responsible for this type of gene conversion is not known, but an as-yet-uncharacterized site-specific recombinase has been proposed to be involved in the DNA rearrangements observed for the vsa, hsd, vpma, and vsp systems.
This study describes the genetic organization of the entire vpma multigene locus in the PG2 type strain of M. agalactiae and the molecular characterization of the mechanism controlling Vpma expression. By isolation of PG2 clonal variants and characterization of their Vpma expression and vpma-specific DNA rearrangements, we show that a unique sequence containing the putative promoter for vpma expression is rearranged by site-specific recombination and linked to vpma ORFs to control their expression. An ORF unrelated to the vpma genes at one end of the locus and having homology to site-specific recombinases was found and is proposed to be responsible for the observed vpma-specific recombination in M. agalactiae. In addition, a pathogenicity island-like locus is described in a pathogenic mycoplasma species for the first time.
MATERIALS AND METHODS
Mycoplasma culture and derivation of clonal lineages.
Mycoplasmas were grown in standard medium, as described previously by Aluotto et al. (1), at 37°C. The mycoplasma clones used in this study were obtained from M. agalactiae type strain PG2 (39). The PG2 clones, 55-5 and 55-7, identified by using monoclonal antibody (MAb) 3B3 and the colony immunoblot technique, have been described previously (16). The same procedure was used to obtain the 3B3-negative clone, 55-5-10, from 55-5, and both 55-5-10-4 (3B3-positive) and 55-5-10-7 (3B3-negative) were derived from 55-5-10.
Oligonucleotides and plasmids.
All oligonucleotides listed in Table 1 were purchased from GibcoBRL, except A4F, which was modified at the 5′ end with digoxigenin (DIG) and synthesized by VBC-Genomics BIOSCIENCE Research-GmbH, Vienna, Austria.
TABLE 1.
Oligonucleotide sequences used in this study
| Name | Sequence (5′ to 3′)a |
|---|---|
| conF2 | cgcggatccAAGCTTAGTAAAATCAGTATAGAT |
| conF3 | CTGAACCATTAGGAGTAGTAAC |
| MAX2R | TTCTACCATATTGACTCCTATGT |
| Z3R | GACCTCCGCCCTGTGTGTCT |
| Z1F | cgcggatccCAAACAGATTCAACTCCGTCAAC |
| E1F | CCAAACTATTCAAGCCCAATTGAA |
| E1R | GTCAGATTCTGATTGTGCCGA |
| Z1R | aaactgcagTTATTCGTATTTAGGTAATAGTCTTC |
| E2F | GTCTGAAAATGATAAGAAATAAC |
| E2R | GCTCTAGTAAAAGGATTTGAACG |
| U1R | aaactgcagTCAACCTTAGATAAACCACCTAAC |
| U1F | cgcggatccGATAAAGAAGATAAGACAGGTGGTAG |
| UV2R | CTTGTGCCATTCTTTCAGGGG |
| UV1R | GGTTTTCCTGGTTGTTCTGTG |
| Y1R | gctctagaGATTAAACTTTTTTTACAGTAAATG |
| Y1F | ccggaattcAATGCAAACGCTGCAGAAAATG |
| YX1R | CTTYAAATTYTGGAGCTAAGCCTC |
| Y4R | TGACTGCCTTCTGCTGGAGT |
| X2R | TATTTTGACCTCTACCTTGTGTA |
| X1F | cgcggatccAAAGTAATGAAGGTCAATTACC |
| H4F | GAAACCCCAGAAGAAAGGGATG |
| X1R | aaactgcagGCTTAAGGATTTTTTAAAATGATG |
| W1R | aaactgcagAATAACTTTATCTAGTTCTATACC |
| H4R | CCTTAAAAATAGTGGTATAGAACTAGA |
| W3R | GTTGTTGCACTGTTACCATTACT |
| W1F | cgcggatccAATGGCGGAAATAGTAATGGTAAC |
| V2R | ATCTCAACGTATTATCGTTTTAC |
| V1R | gctctagattaTCCAGATGATGTTTCAACTTC |
| V1F | ccggaattcGTTGAGGAAGCAATTAAAACAGC |
| conR3 | GCAAATTAGTTGGTTTTTCAGC |
| conR2 | cgcggatccAAGCTTAATGATTTGTGCTATAAAA |
| A3F | AARTGYGGWGGWACWAMWRA |
| A4F-DIG | AARTGTGGTGGYACTAAAGA |
| 5′ leader1 | GGATAAATTTATGAAAAAATC |
| leader2 | ATGAAAAAATCAAADTTTKT |
Uppercase letters indicate target sequences, and lowercase letters indicate bases introduced to form restriction sites to aid in the cloning of subsequent PCR products.
The following plasmids contain DNA fragments from 55-5 (Fig. 1) in pUC18: p5H4.7 contains a 4.7-kbp HindIII fragment possessing ORF2′, xer1, vpmaZ, and vpmaU; p5H1.8 contains a 1.8-kbp HindIII fragment possessing the 5′ region of vpmaY and vpmaX; p5H3.0 contains a 2.9-kbp HindIII fragment possessing vpmaW and vpmaV; p5E2.2 contains a 2.2-kbp HindIII-EcoRI fragment from p5H4.7; p5E1.0 contains a 1.0-kbp EcoRI fragment from p5H4.7; p5E1.2 contains a 1.2-kbp EcoRI-HindIII fragment from p5H4.7; p5X0.26 contains a 262-bp XbaI fragment from p5H2.9; and p5X0.4 contains a 408-bp XbaI-HindIII fragment from p5H2.9. The 2.2-kbp XbaI fragment from p5H2.9, containing vpmaV and the 5′ region of vpmaW, was cloned into pBluescript II pKS. Plasmids pPCR9, pPCR75, and pPCR91 (16) contain PCR products (PCR9, PCR75, and PCR91, respectively) generated from A3F and 55-5 DNA (PCR9, 1.6 kbp) or 55-7 DNA (PCR75, 1.6 kbp; PCR91, 2.7 kbp).
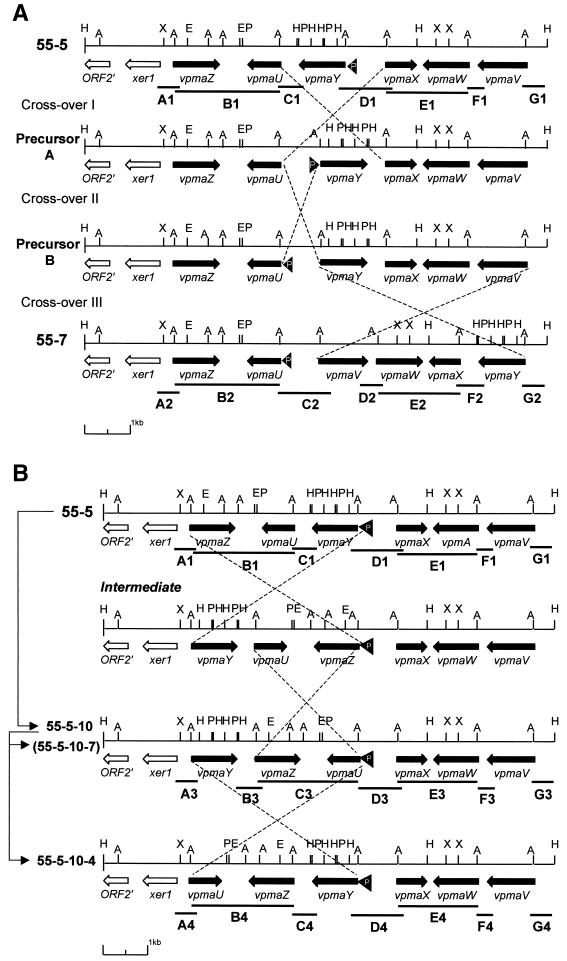
FIG. 1.
(A) Restriction map and arrangement of the ORFs of vpma genes within the vpma locus of 55-5. Black arrows represent vpma genes and white arrows indicate ORFs unrelated to vpma genes. Lines above vpma ORFs indicate the locations of the vpma-specific PCR probes. A, AlwI; As, AseI; E, EcoRI; H, HindIII; P, PstI; X, XbaI. The location of a tRNA-lys gene is indicated by a black arrowhead. (B) Southern blot analysis of restricted DNA (1 μg) from PG2 clones hybridized with A4F. The tree schematic above each lane outlines the derivation of PG2 clones; each number represents a different clone which was either MAb 3B3-positive (+) or 3B3-negative (−). Lanes 1, 55-7; lanes 2, 55-5; lanes 3, 55-5-10; lanes 4, 55-5-10-4; lanes 5, 55-5-10-7. The restriction enzyme used is indicated below each panel. SD, λHindIII DNA standard.
DNA manipulations, Southern blot analysis, and library construction.
Plasmid DNA was purified using AX100 columns from Macherey-Nagel. The genomic DNA isolation, DNA sequencing, and Southern blot technique have been described previously (16). Restriction endonucleases AlwI and AseI were obtained from New England BioLabs, and ClaI, EcoRI, HindIII, PstI, and XbaI were obtained from Promega. Restriction endonuclease digestions were performed as recommended by the manufacturers. Probes were hybridized with membranes overnight at the same temperatures that were used for the washing step. PCR-generated vpma gene-specific probes were washed from the membranes in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) for 1 h at 60°C (vpmaU, vpmaW, vpmaX, vpmaY, and vpmaZ) or 55°C (vpmaV). After hybridization, DIG-labeled A4F was washed from the membrane two times in 6× SSC-0.1% SDS for 10 min at 46°C. A library of HindIII-digested genomic DNA from M. agalactiae clone 55-5 was cloned into pUC18 (Pharmacia) and screened as previously described (16). DNA fragments were further cloned after restriction digestion either into pBluescript II pKS (Stratagene) or pUC18. PCR products were cloned into pGEM-Teasy (Promega). DNA inserts were sequenced as previously described (16)
PCR.
Unless otherwise stated, PCR amplifications were performed using Taq DNA polymerase (Promega) as previously described (16). PCR or DIG labeling of vpma gene-specific probes by PCR was performed using the following primer sets and templates: U1F-U1R and p5E1.2 (for the vpmaU-specific probe); V1F-V1R and p5X2.2 (vpmaV); W1F-W1R and p5H3.0 (vpmaW); X1F-X1R and p5H1.8 (vpmaX); Y1F-Y1R and 55-5 genomic DNA (vpmaY); and Z1F-Z1R and p5H4.7 (vpmaZ). The PCR cycling conditions for these probes were as follows: 1 cycle of 94°C for 1 min and 28 cycles of 94°C for 20 s, 55°C (vpmaY), 60°C (vpmaW and vpmaX), 65°C (vpmaV), or 70°C (vpmaU) for 30 s, and 72°C for 10 s (vpmaV), 18 s (vpmaU, vpmaX, and vpmaY), or 54 s (vpmaW and vpmaZ), followed by 1 cycle at 72°C (performed in a Perkin-Elmer GeneAmp PCR System 2400 thermocycler). The PCR cycling conditions for the primer pairs with 55-5 DNA as a template were 1 cycle of 94°C for 1 min and 28 cycles of 94°C for 20 s, 55°C (E2F-E2R) or 60°C (E1F-E1R and H4F-H4R) for 30 s, and 72°C for 18 s, followed by 1 cycle at 72°C for 7 min.
Long-range (LR) PCR was conducted using the primer set conF2-conR2 at an annealing temperature of 60°C and the Expand Long Template PCR system from Roche according to the manufacturer's instructions.
The names of the PCR fragments and the primer sets and templates that were used for their generation, respectively, (see Fig. 4) are as follows: A2, Z3R-MAX2, and 55-7 LR PCR product (LRP); C2, UV2R, and 55-7 LRP; D2, V1F-W3R, and 55-7 LRP; F2, A3F, and 55-7 DNA; G2, conR2-Y4R, and 55-7 LRP; A3, YX1R-MAX2R, and 55-5-10 LRP; B3, Y1F-Z3R, and 55-5-10 LRP; D3, UV1R-X2R, and 55-5-10 LRP; F3, W3R-V2R, and 55-5-10 LRP; G3, UV1R-conR3, and 55-5-10 LRP; A4, MAX2R-UV1R, and 55-5-10-4 LRP; C4, Y1F-Z3R, and 55-5-10-4 LRP; D4, YX1R, and 55-5-10-4 LRP; F4, W3R-V2R, and 55-5-10-4 LRP; G4, UV1R-conR3, and 55-5-10-4 LRP. The LRP PCR templates were used at a final concentration of 0.1 ng/μl. The cycling conditions for these primer sets and templates were as follows: 1 cycle of 94°C for 3 min and 28 cycles of 94°C for 30 s, 55°C (G2), 57°C (A2, B3, D3, F3, G3, A4, C4, F4, and G4), 60°C (C2, D2, A3), or 63°C (D4) for 40 s, 72°C for 30 s (A2), 32 s (B3, F3, G3, A4, C4, F4, and G4), 33 s (D2), 38 s (G2), 44 s (A3), 1 min 30 s (C2 and D4), or 2 min (D3) (performed with a model PTC-100 thermocycler from MJ Research, Inc., and Taq DNA polymerase from Promega, except for A2, G2, and D4, for which enzyme and 1× buffer from GibcoBRL and a 1.5 mM final concentration of MgCl2 were used.) The PCR conditions for primer A3F have been described previously (16).
FIG. 4.
Transition states of vpma multigene loci of related M. agalactiae clones due to DNA inversions. (A) Transition states between 55-5 and 55-7 (the restriction map symbols are like those shown in Fig. 1A). The ends of the crossed broken lines indicate the locations of DNA recombinational crossover events (I, II, and III) between inverted identical sequences producing DNA inversions. Genotypes labeled Precursor A and Precursor B represent potential ancestors from which 55-5 and 55-7 could have arisen using a minimum of three crossover events. (B) Transitions between 55-5, 55-5-10, and 55-5-10-4. Arrowed lines at left show the lineage order of the clones. Clone 55-5-10-7 has a restriction map identical to that of 55-5-10. Lines labeled with a letter and number combination represent sequenced PCR products, except B1, E1, B2, E2, C3, E3, B4, and E4, which are AlwI-digested genomic DNA fragments identified by Southern blot using A3F or A4F. Each number following a letter represents a particular clone (1, 55-5; 2, 55-7; 3, 55-5-10; 4, 55-5-10-4). All remaining symbols are defined in the legend to Fig. 1A.
The PCR products (see Fig. 4) were cloned into the T-tailed pGEM-Teasy plasmid vector (Promega) and sequenced as previously described (16).
Northern blot analysis.
Total RNA was prepared and subjected to Northern blot analysis as previously described (15). The hybridization and washing conditions for the vpma gene-specific probes were the same as those for the Southern blot analysis. The control DNA consisted of unlabeled PCR products identical to the DIG-labeled vpma gene-specific PCR probes.
DNA analysis.
Blast and Advanced Blast (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/blast/blast.cgi]) were used to query amino acid or translated DNA sequences against all nonredundant sequences in the following databases: GenBank CDS translation, PDB, SwissProt, PIR, and PRF for protein and GenBank, EMBL, DDBJ, and PDB for DNA query sequences. DNA alignments were performed using Clustal W version 1.81 (European Bioinformatics Institute [http://www2.ebi.ac.uk/clustalw]). Identity and similarity values were calculated from the maximum length of the alignment, and the similarity values for protein alignments included the number of identical and conserved substitutions but excluded semiconserved substitutions. Translations and calculations of the molecular mass and pI of proteins were performed using the programs available at the Expert Protein Analysis System proteomics server (http://www.expasy.ch/).
Nucleotide sequence accession number. The sequence data described here have been submitted to the GenBank database under accession no. AF411984.
RESULTS AND DISCUSSION
vpma multigene locus.
In a previous study, a 1.8-kbp fragment and three adjacent HindIII fragments of approximately 0.3 kbp were sequenced from an M. agalactiae clonal variant (55-5) expressing a Vpma product of 39 kDa, revealing the presence of two vpma genes, vpmaX and vpmaY (16). However, Southern blot analysis suggested that the vpma multigene family is composed of at least four genes sharing a common 5′ region. The sequence of the entire vpma locus in clone 55-5 was obtained after the cloning of two additional HindIII fragments of 2.9 and 4.7 kbp (Fig. 1B) from the previously generated library, as described in Materials and Methods. The six HindIII fragments that carried the entire vpma gene repertoire all hybridized with either the common amino-terminal oligonucleotide probe, A4F, that recognizes all six vpma genes and/or to vpma-specific PCR probes (Fig. 1). The orientation of the HindIII fragments relative to each other was determined by amplification using primers located in adjacent fragments and was confirmed by sequencing.
Analysis of the entire contiguous sequence revealed that the locus contained a total of six vpma genes in both orientations, which were arbitrarily designated vpmaU to vpmaZ (Fig. 1A). As previously described for vpmaX and vpmaY (16), all vpma genes share a highly conserved 5′ untranslated region (92% identity over 71 nucleotides between all genes, with the sequences of vpmaV, vpmaW, and vpmaY being identical to each other) followed by a conserved sequence encoding a 25-aa lipoprotein leader ending in an identical acylation/peptidase II cleavage motif of AAKC. The leader sequences possess an overall nucleotide and amino acid identity of 76% (92% aa similarity), and this high identity continues for a short distance into the mature polypeptide, with the next 4 aa being identical between all genes, but drops to about 50% for the following 19 aa, after which no overall significant identity could be found. These same vpma 5′ untranslated and N-terminal-coding regions are also conserved in the equivalent regions of the vsp genes from M. bovis (3, 20, 23); however, beyond this, vsp genes have no significant similarity to any vpma genes in M. agalactiae.
The number of vpma genes in PG2 is quite low considering that there are at least 13 vsp genes in the homologous system in M. bovis (23). A number of efforts were made to find evidence of any additional vpma genes within the PG2 genome. First, an additional 2 kb of sequence was obtained at the 3′ end of the locus, which led to the identification of two ORFs (ORF3 and ORF4) with no relationship to vpma genes. Second, a 5-kbp XbaI fragment from 55-5 which hybridized to the vsp oligonucleotide probe, vspS-2 (identical in sequence to a region of the vspA leader from M. bovis), but not to the vpma common amino-terminal probe, A3F (16), was cloned and the appropriate region was sequenced. Analysis revealed the presence of a lipoprotein gene that showed a leader sequence similar to those of the vpma and vsp genes but with a different mature N-terminal sequence. Finally, two oligonucleotides, 5′leader1 and leader2 (Table 1), which would hybridize to all known vpma and vsp genes, revealed in Southern blot analysis a pattern identical to that obtained with A4F (data not shown). Thus, only six vpma genes are present in PG2 and all are grouped within a single cluster.
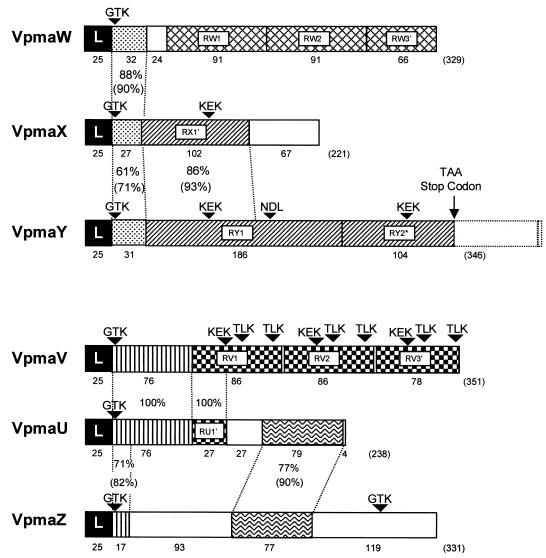
Based on their N-terminal sequences and other shared sequences, the vpma genes could be divided into two homology groups (Fig. 2). The first group (VpmaW, VpmaX, and VpmaY) shares a short region (between 27 and 33 aa) of high identity (Fig. 2). VpmaW is predominantly composed of 2.7 repeats of 91 aa. The vpmaY gene encodes two large repeats of 186 aa and is unusual as the second repeat is prematurely terminated due to the insertion of two nucleotides that introduce a TAA stop codon so that only 104 aa are translated. VpmaX also contains a truncated version of the vpmaY repeat, while the remainder of the ORF consists of a unique sequence. The second group (VpmaV, VpmaU, and VpmaZ) also shares various regions. VpmaV consists of 2.9 repeats of 86 aa and shares 76 of its N-terminal amino acids with VpmaU. VpmaU also contains a truncated version of the V repeat (31%) and shares 79 C-terminal amino acids with VpmaZ (Fig. 2).
FIG. 2.
Schematic of vpma ORFs. The ORFs are represented by boxes and begin with a homologous 25-aa leader sequence (L) followed by regions that have homology between genes or regions that are repeated (R, followed by gene designation, or R' for truncated repeats) within a gene. Homologous regions or repeated sequences are indicated with the same pattern, and unique sequences are not shaded. The percentages of amino acid identity between homologous regions between genes are indicated, and the percentages of similarity are in parentheses. The theoretical molecular mass (kDa), pI, and percentage of charged amino acids, respectively, calculated from the mature polypeptides for VpmaU, VpmaV, VpmaW, VpmaX, VpmaY, and VpmaZ are 23.2, 6.90, and 37%; 35.0, 9.09, and 34%; 33.1, 9.43, and 31%; 22.4, 6.33, and 42%; 35.2, 8.27, and 34%; and 34.2, 7.01, and 32%. Arrowheads indicate possible epitopes involved in host cytadhesion based on those determined for vsps from M. bovis (32).
The truncation in the second repeat of the vpmaY gene appears to be at an evolutionarily early stage since the remainder of the second repeat-coding sequence is still intact. Truncated repeats in other vpma genes (vpmaU, vpmaV, vpmaW, and vpmaX) may have simply arisen by deletion, since there was no evidence in all three reading frames that the repeat continued past the stop codon. The truncation of repeats in the C-terminal region may be a means by which unnecessary sequences are eliminated, while variability between genes is optimized by the creation of new carboxyl-terminal sequences such as those for the vpmaX and vpmaU gene products. The vpma genes encoding common amino-terminal sequences but different C-terminal sequences may also have derived from gene duplication followed by recombination to exchange the C-terminal portion of only one of the genes with that of an unrelated sequence. This phenomenon has been observed for the single vaa gene of M. hominis, in which subpopulations within the same strain possess different vaa alleles consisting of different numbers of a central repeat followed by different C-terminal sequences. This C-terminal variability has been proposed to confer cytadherence function depending on which sequence is present (44).
Regions of common sequence between vpma genes may also represent functionally important domains. Interestingly, vpma repeats are, in general, more conserved within a gene than between genes, particularly for the repeats of vpmaW and vpmaV, which each have 100% DNA identity within a gene. This may indicate that the number of these repeats periodically increases or decreases, a phenomenon which has been previously described for other variable surface lipoproteins belonging to gene families in mycoplasmas (2, 5, 43).
Four vpma genes, vpmaX, vpmaY, vpmaU, and vpmaZ, were found to be identical to four avg genes, avgA, avgB, avgC, and avgD, previously sequenced from the PG2 type strain of M. agalactiae by another group of researchers (12), with the exception of avgB, which only contains one repeat and an additional 4 aa at the C terminus. It is unclear from the available sequence (accession no. AF112467) whether avgB is prematurely truncated by mutation and still contains the coding sequence for a second repeat, as for vpmaY, or whether this second repeat has been completely deleted.
Only one pseudogene, which lacks a start codon and shares N-terminal coding sequence from the sixth amino acid of the leader sequence to 5 aa after the leader cysteine residue, was found downstream from, and in the same orientation to, vpmaZ. This homologous sequence shows an overall DNA identity of 69% (64% amino acid identity) with the other vpma genes. The leader sequence possesses an altered lipoprotein motif of ASKC and continues for an additional 5 aa (GDTKE) before terminating.
Identification of individual vpma gene expression within clones.
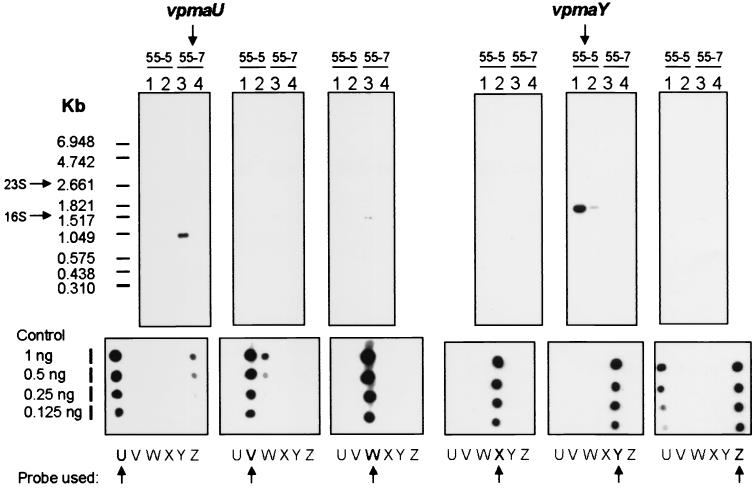
In a previous study, two clonal variants, 55-5 and 55-7, were derived from the same PG2 strain and were shown to express Vpma39 and Vpma34, respectively (16). These two related but distinct Vpma products were not previously assigned to any one particular vpma gene, as all vpma genes had not been identified. After the entire vpma locus had been sequenced, gene assignments were still difficult to make because the N-terminal sequence of Vpma39 (14 aa) was identical to the predicted N-termini of VpmaY, VpmaX, and VpmaW, and the N-terminal sequence of Vpma34 (14 aa) was identical to those of VpmaU, and VpmaV. To resolve this problem, Northern blot analyses were performed (Fig. 3) using vpma gene-specific PCR probes (the probe positions relative to each vpma gene are shown in Fig. 1). The results established that the Vpma39 protein, expressed by 55-5, was encoded by the vpmaY gene. If vpma genes used a promoter located similarly to that of the expressed vsp genes of M. bovis (see below and Fig. 5), then the 1.60-kb vpmaY transcript would include the untranslated remainder of the second vpmaY repeat and would terminate very close to the start of the downstream gene, vpmaU. If this termination were to occur within the 5′ region of the downstream gene, vpmaU in this case, then this may affect the transcript size or even stability of vpmaY in other backgrounds with different configurations of vpma genes directly downstream from expressed vpmaY genes. The Vpma34 protein expressed by 55-7 was found to be encoded by a 1.05-kb vpmaU gene transcript (Fig. 3). This transcript has the potential to encode the entire vpmaU ORF, with termination occurring between vpmaU and vpmaZ, approximately 100 bases immediately downstream from the vpmaU stop codon. These results indicate that vpma gene transcripts are monocistronic, with only one vpma gene being expressed at any one time in each clonal isolate. Therefore, we conclude that control in vpma gene expression is at the transcriptional level and suggest that a single active promoter is present within the locus.
FIG. 3.
Northern blot analysis of replica loadings of total RNA from clones 55-5 and 55-7 (lanes 1 and 3, 2 μg; lanes 2 and 4, 0.5 μg) hybridized with each vpma gene-specific probe. The vpma probe used for each replica is indicated by an arrow below each panel showing dot blotted unlabeled PCR probe DNA (underneath the panels showing the results of Northern blot analysis) diluted in a twofold series.
FIG. 5.
DNA alignment of 5′ untranslated regions ending at the start codon of six vpma genes and vspA from M. bovis (21). A putative Shine-Dalgarno (SD) sequence, the start point of transcription (+1), and the promoter element (−10) are indicated by lines above the sequence based on vspA. An inverted repeat is identified by half arrows. The box indicated with broken lines shows the minimum region where DNA cleavage must occur during recombination between vpma genes. The boxes indicated with solid lines show regions of complete identity between vspA and vpmaY sequences upstream from the minimum region of recombination. Asterisks represent identity between all sequences, and dashes indicate gaps introduced to optimize the alignment.
Since MAbs were used to obtain the M. agalactiae clones investigated, it is not surprising that the collection expressed only a limited range of vpma genes from the locus, namely, vpmaU and vpmaY. Preliminary data obtained by using polyclonal antibodies specific to each vpma gene product in both colony and Western blot analyses have also provided evidence for the expression of VpmaV, VpmaW, VpmaX, and VpmaZ in PG2.
Recombination within the vpma multigene locus is site specific and links a single promoter-like element to individual vpma ORFs to control vpma gene expression.
As hypothesized in an earlier paper (16), switches in Vpma expression are due to the occurrence of specific DNA rearrangements within the vpma locus. In order to locate the exact sequences involved in these events, a detailed genetic analysis of the vpma locus was performed using the two sibling clones, 55-5 and 55-7, and three clonal variants derived from 55-5 (55-5-10, 55-5-10-4, and 55-5-10-7), each expressing either Vpma39 (MAb-positive) or Vpma34 (MAb-negative) (Fig. 1B). For this purpose, a strategy based on the sequence of clone 55-5 was devised that combined genomic DNA restriction digestion with AlwI or AseI and Southern blot analysis to identify DNA fragments containing single vpma genes or up to two vpma genes orientated convergently (Fig. 1). This was possible because the enzymes cleaved within the conserved 5′ regions of each vpma gene. Southern blot analysis could then identify the AlwI or AseI fragment that hybridized with the common N-terminal probe, A4F. Interestingly, an identical fragment banding pattern for both restriction enzymes was observed for all five clones tested (55-7, 55-5, 55-5-10, 55-5-10-4, and 55-5-10-7 [Fig. 1B]). In contrast, different restriction fragment length polymorphisms were observed when HindIII was used, while in all clones, the vpma genes were clustered on the same ClaI fragment (Fig. 1B). This indicated that the intergenic regions, rather than the vpma coding sequences themselves, were involved in the apparent vpma DNA rearrangements between clones.
To further define the sequences involved in vpma recombination, a strategy was devised to fast-track the analysis of only the vpma intergenic regions rather than sequencing the entire vpma locus from each chosen M. agalactiae clone. Clones 55-7, 55-5, 55-5-10, 55-5-10-4, and 55-5-10-7 were subjected to LR PCR using primers at each end of the vpma locus shown in Fig. 1A, including the two extreme HindIII sites. An approximately 10-kbp LR PCR fragment was obtained for each clone and was used to map the location and orientation of the vpma genes by using Southern blot analysis with gene-specific probes and single and double digestions of a combination of the following enzymes: EcoRI, HindIII, PstI, and XbaI (Fig. 4). For the LR PCR fragment from 55-7, partial digestion with AseI and Southern blot using two separate internal oligonucleotides immediately downstream from each LR primer was used as a second method for ordering the vpma genes and for restriction map confirmation. The vpma intergenic regions for each locus were then amplified by gene-specific primers designed at each end of every vpma gene, and the PCR product was cloned and sequenced. The location of each intergenic PCR fragment is indicated in Fig. 4, except for B1, B2, C3, B4, E1, E2, E3, and E4, whose locations were determined by Southern blot analysis. This analysis showed that the vpma gene organization of clone 55-7 differs from that of its sibling, 55-5. Interestingly, the 5′ untranslated sequence of the expressed vpmaY gene of 55-5, which contains the promoter element discussed below, is located upstream from the expressed vpmaU gene of 55-7 rather than the upstream sequence of the silent vpmaU gene of 55-5. This suggests that recombinational crossover events must have occurred in the parental population so that a minimum of three crossover events (I, II, and III [Fig. 4A]) are theoretically required to generate both vpma genotypes of 55-7 and 55-5 from a common ancestor. It is possible that crossover III, shown in Fig. 4A, may have taken place before crossover II, which would then have resulted in an intermediate, with the vpmaV gene possessing the promoter element discussed below. Likewise, two recombinational crossover events must have taken place to produce the necessary DNA inversions to obtain the 55-5-10 vpma genotype from 55-5, while only one would be required to produce 55-5-10-4 from 55-5-10 (Fig. 4B). The sibling clone to 55-5-10-4, 55-5-10-7, retained the genotype of the parent, 55-5-10 (Fig. 4B). All vpma genes, except vpmaW, were documented to have been involved in vpma rearrangement. The results indicate that the strand exchange must have taken place within the conserved 5′ untranslated regions of the genes. The exchange between vpmaY and vpmaV extends this possible region halfway into the leader sequence, since their sequences are identical up to this point. Based on the recombination events involving the vpmaZ gene, the minimal region in which the recombinational strand exchange could have taken place was identified as a 21-bp region within the 5′ untranslated region between nucleotides 51 and 71 upstream from vpma start codons (Fig. 5). It is possible, but unlikely, that this minimal sequence could be extended by 6 bases at the 3′ end, where vpmaZ possesses three nucleotide differences relative to all other vpma genes, if the cleavage of the DNA were to occur on either side of these 6 bases. Cleavage involving heterogeneous sites for recombination between vsaA and vsaF in M. pulmonis, where there are two base differences within the proposed 6-base cleavage site in vsaF relative to the same region in vsaA, has been documented. After recombination between vsaA and vsaF, two forms of each gene were observed with one or the other 6-base sequence (35). No evidence was observed of the three base differences in vpmaZ, immediately 3′ to the proposed minimal region involved in vpma DNA recombination, being transferred to either of the two genes, vpmaY or vpmaU, after crossover with vpmaZ (Fig. 4B), although direct sequencing of the A3, A4, B3, C4, D3, and D4 PCR products was not done. Also, no sequence similarity was observed between the vpma sequences undergoing recombination and those of vsa and hsd of M. pulmonis (35, 37, 38).
Recombinational crossover events for all PG2 clones analyzed were also only observed to occur between two divergently or convergently oriented 21-bp minimal regions. This would be necessary, as recombination between two sites in the same orientation would lead to deletion of the interspanning DNA between them. It is possible that this process does occur at a low frequency, since some clones, upon amplification of their vpma loci, produced smaller-sized PCR fragments in a lower proportion to the expected full-length fragment (data not shown).
Recently, it has been documented that the region involved in DNA inversions between two vsp genes in M. bovis (vspA and vspO) includes a 34-bp region between (and including) nucleotides 38 and 71 upstream from the vspA start codon (20). This region includes the equivalent 21-bp region we have observed to be involved in DNA inversions between vpma genes (Fig. 5), indicating that these two gene families share a highly conserved mechanism for site-specific recombination in the two related mycoplasma species.
To understand which 5′ sequences are necessary for transcribing vpma genes, it was necessary to identify which vpma gene was expressed in each related clone that was analyzed at the molecular level. It was already known that clone 55-5 expressed vpmaY and that clone 55-7 expressed vpmaU. Since specific rabbit antisera had previously been raised to both VpmaY and VpmaU (16), they were used to confirm that the MAb-negative clones 55-5-10 and 55-5-10-7 expressed VpmaU and that 55-5-10-4 (MAb-positive) expressed VpmaY (data not shown). All vpmaU and vpmaY genes that were actively expressed in these clones were also found to possess a 5′ untranslated sequence identical to that of the vpmaY gene in 55-5, located upstream from the minimal region involved in vpma DNA recombination. This region should contain the active promoter element that drives vpma gene expression, and this sequence is highly conserved with the equivalent region of the expressed vspA gene of M. bovis (Fig. 5) but it is not present in silent vpma genes and published vsp genes (3, 20, 21, 23). Very recently, the start point for transcription of the expressed gene, vspA, was found to be located 192 bp upstream from the initiation codon (Fig. 5), and it was proposed that this start point possesses a σ70-like −10 consensus sequence promoter element (21). Since this promoter region of expressed vsp genes is highly conserved with that of expressed vpma genes, it is most likely that vpmaY and other expressed vpma genes use the same start point for transcription. Overall, these data imply that the 5′ untranslated region, specific to expressed genes only, provides the promoter required to initiate transcription of vpma genes and is rearranged and linked with a silent gene via site-specific DNA inversion during switches of expression from one vpma gene to another within the locus.
A possible recombinase candidate in M. agalactiae is related to the Xer site-specific recombinases.
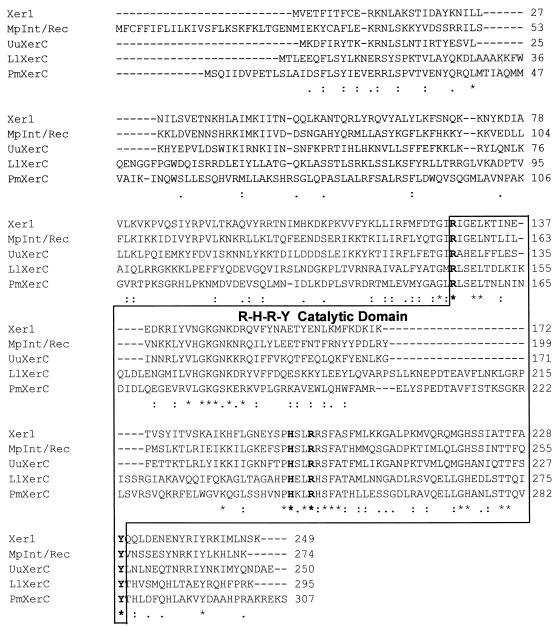
In the course of cloning and sequencing the vpma locus, two ORFs unrelated to vpma genes were found immediately adjacent and in the opposite orientation to vpmaZ (Fig. 1A). ORF2′ is a partial ORF and has homology to a conserved hypothetical protein with various proposed functions in bacteria. More importantly, the complete ORF immediately adjacent to vpmaZ has homology to Xer recombinases included in the databanks mentioned above (xer1 [Fig. 1A]). The highest-scoring alignments were obtained with the XerC homologues in Ureaplasma urealyticum (accession no. AAF30630 [14]) and M. pulmonis CAC13704 (8), which have overall identities of 38% (68% similarity) and 42% (69% similarity), respectively, to xer1. The Xer recombinases belong to a large tyrosine recombinase family called the λ integrase family that shares an invariant tetrad of amino acid residues involved in catalysis, R-H-R-Y (11, 26). Within this group, the Xer proteins form a distinct subfamily based on their C-terminal regions (11). Xer1 in 55-5 also possesses the R-H-R-Y tetrad (Fig. 6) and numerous other distinctive residues in common with Xer proteins (26). The Xer site-specific recombination system has been best studied in Escherichia coli and consists of two homologous recombinases, XerC and XerD, which act on specific chromosomal sequences called dif sites to initiate DNA strand exchange to resolve chromosome dimers that have arisen from homologous recombination before chromosome segregation and cell division (7, 9, 36).
FIG. 6.
Amino acid alignment of Xer1 from M. agalactiae type strain PG2 with XerC recombinases from M. pulmonis (MpInt/Rec), U. urealyticum (UuXerC), Proteus mirabilis (PmXerC), and Lactobacillus leichmannii (LlXerC). The boxed C-terminal sequences form the catalytic domain that includes the four amino acids (R, H, R, and Y, shown in boldface type) that are invariant for the λ integrase family of recombinases. Dashes indicate gaps introduced to optimize alignment, and asterisks indicate positions which have a single fully conserved residue. Colons and dots indicate that one residue of the strong and weak groups (as defined by ClustalW, version 1.8) is fully conserved.
Homologues of XerC and XerD have been identified in a number of gram-negative and gram-positive bacteria (11, 26) but are absent in the circular genomes of M. genitalium and M. pneumoniae (18), which is hypothesized to be due to a deficiency in homologous recombination in these organisms which would render the Xer system obsolete (29). Although dif, XerC, and XerD are dispensable, their removal from E. coli (6, 19), B. subtilis (except XerC) (33), or Haemophilus influenzae (24) produces a subpopulation of filamentous cells that contain aberrant nucleoids. The absence of an Xer system may also explain why some Mycoplasma species have been observed to form filamentous branching forms (13, 34). Since the Xer system is unnecessary in mycoplasmas, it is proposed that the Xer homologue found in M. agalactiae and located immediately adjacent to the vpma multigene locus is involved in the observed vpma-specific DNA recombination that controls vpma gene expression rather than chromosome segregation.
Quite strikingly, the annotated integrase/recombinase from M. pulmonis mentioned above (accession no. CAC13704), which is the only recombinase found in this organism, is located adjacent (3.216 kb) to the vsa multigene locus. Therefore, it is also possible that this recombinase functions to control Vsa expression by DNA inversions generated by site-specific recombination.
It would be expected that the Xer-like recombinases of M. agalactiae and M. pulmonis would function in a fashion similar to that of the Xer site-specific recombinases of E. coli but that they would be different in one very important aspect: in mycoplasmas, the reaction would require only one recombinase rather than two different recombinases. Interestingly, the region that provides DNA-binding specificity to XerC and XerD in E. coli and has sequence conservation among Xer proteins (26, 40) is not conserved in either Xer1 or the integrase/recombinase from M. pulmonis, which may explain why vpma and vsa genes do not use the same dif sequence. Additionally, the gene for the conserved hypothetical protein in M. pulmonis, which is the homologue to ORF2′ in M. agalactiae, is situated at a distance of approximately 200 kbp from the integrase/recombinase gene. Thus, the organization of these two genes in M. pulmonis is not conserved relative to ORF2′ and xer1 in M. agalactiae.
The presence of the vpma pseudogene mentioned above suggests that the vpma locus is likely to be prone to instability. Whether this is the result of aberrant recombinase activity at nonpermissive recombination sites or simply of homologous recombination is not clear. Also, analysis of 32 other M. agalactiae strains and field isolates for their vpma gene contents, using the probes specific for vpmaU to vpmaZ from PG2, revealed that the vpma repertoires varied among strains. Approximately 50% of the strains were missing certain vpma genes and/or had additional new genes (unpublished data). Again, the question arises concerning whether an active recombinase aids and abets this evolutionary process for vpma loci or whether it is due to gene duplication, RecA-dependent homologous recombination, and/or lateral transfer of vpma (or even vsp) genes.
The vpma multigene family, the xer1 recombinase gene, and an adjacent tRNA-lys gene are typical elements of a pathogenicity island.
Adjacent to the vpma locus is a tRNA-lys (anticodon CTT) gene (Fig. 1A) located between xer1 and ORF2′ (Fig. 1A) which appears to be related more to genes from Streptomyces lividans and Brucella abortus than those from mycoplasmas (M. genitalium, M. pneumoniae, and M. capricolum). The tRNA-lys (CTT) gene is missing from M. pulmonis, but its absence might be explained by the fact that tRNA-lys (TTT) can read both AAA and AAG codons. It is important to note that vpma genes use an above-average percentage of AAA (12.7%) and AAG (3.7%) lysine codons compared with the average percentages used by all translated genes for the genomes of M. genitalium, M. pneumoniae, and M. pulmonis (averages of 7.01, 4.73, and 9.79%, respectively, for AAA and 2.44, 3.83, and 1.20%, respectively, for AAG), except for the average percentage of AAG codon usage in M. pneumoniae. The xer1 gene also exhibits high usage of the AAG lysine codon (4.4%). Keeping in mind that the characterized gene products of two vpma genes, vpmaY and vpmaU, are each abundantly expressed in their respective clones and represent the most abundant proteins in their membrane fractions (16), the presence of the adjacent tRNA-lys (CTT) gene and the codon bias may play an important regulatory role in vpma and xer1 gene expression, similar to that played by leuX in the expression of the FimB recombinase (30) and numerous other proteins, including type 1 fimbriae (27), in E. coli.
Several loci, each consisting of genes encoding a recombinase and factors involved in virulence, are usually associated with a tRNA gene(s) and form mobility units in bacteria that have been termed pathogenicity islands (17). It is interesting that these same elements are present within the vpma locus (Fig. 1A). This poses the question of whether vpma loci represent mobile elements important for intraspecies, or even interspecies (vsps of M. bovis), evolution and whether Vpmas per se or switches in Vpma phenotype contribute to virulence in M. agalactiae.
Acknowledgments
We thank Evelyne Sagné for her technical help.
This work was supported by grant P14725-GEN from the Austrian Science Fund (Fonds zur Förderung der wissenschaftlichen Forschung to C. Citti and R. Rosengarten).
REFERENCES
- 1.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of Mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 2.Behrens, A., M. Heller, H. Kirchhoff, D. Yogev, and R. Rosengarten. 1994. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect. Immun. 62:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beier, T., H. Hotzel, I. Lysnyansky, C. Grajetzki, M. Heller, B. Rabeling, D. Yogev, and K. Sachse. 1998. Intraspecies polymorphism of vsp genes and expression profiles of variable surface protein antigens (Vsps) in field isolates of Mycoplasma bovis. Vet. Microbiol. 63:189-203. [DOI] [PubMed] [Google Scholar]
- 4.Bergonier, D., X. Berthelot, and F. Poumarat. 1997. Contagious agalactia of small ruminants: current knowledge concerning epidemiology, diagnosis and control. Rev. Sci. Tech. Off. Int. Epizoot. 16:848-873. [DOI] [PubMed] [Google Scholar]
- 5.Bhugra, B., L. L. Voelker, N. Zou, H. Yu, and K. Dybvig. 1995. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol. Microbiol. 18:703-714. [DOI] [PubMed] [Google Scholar]
- 6.Blakely, G., S. Colloms, G. May, M. Burke, and D. J. Sherratt. 1991. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 3:789-798. [PubMed] [Google Scholar]
- 7.Blakely, G. W., A. O. Davidson, and D. J. Sherratt. 1997. Binding and cleavage of nicked substrates by site-specific recombinases XerC and XerD. J. Mol. Biol. 265:30-39. [DOI] [PubMed] [Google Scholar]
- 8.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. C. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colloms, S. D., R. McCulloch, K. Grant, L. Neilson, and D. J. Sherratt. 1996. Xer-mediated site-specific recombination in vitro. EMBO J. 15:1172-1181. [PMC free article] [PubMed] [Google Scholar]
- 10.Dybvig, K., and L. L. Voelker. 1996. Molecular biology of mycoplasmas. Annu. Rev. Microbiol. 50:25-57. [DOI] [PubMed] [Google Scholar]
- 11.Esposito, E., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flitman-Tene, R., S. Levisohn, I. Lysnyansky, E. Rapoport, and D. Yogev. 2000. A chromosomal region of Mycoplasma agalactiae containing vsp-related genes undergoes in vivo rearrangement in naturally infected animals. FEMS Microbiol. Lett. 191:205-212. [DOI] [PubMed] [Google Scholar]
- 13.Girón, J. A., M. Lange, and J. B. Baseman. 1996. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect. Immun. 64:197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 15.Glew, M. D., P. F. Markham, G. F. Browning, and I. D. Walker. 1995. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum S6. Microbiology 141:3005-3014. [DOI] [PubMed] [Google Scholar]
- 16.Glew, M. D., L. Papazisi, F. Poumarat, D. Bergonier, R. Rosengarten, and C. Citti. 2000. Characterization of a multigene family undergoing high-frequency DNA rearrangements and coding for abundant variable surface proteins in Mycoplasma agalactiae. Infect. Immun. 68:4539-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker, J., G. Blum-Oehler, I. Mühldorfer, and H. Tschäpe. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 18.Himmelreich, R., H. Plagens, H. Hilbert, B. Reiner, and R. Herrmann. 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuempel, P. L., J. M. Henson, L. Dircks, M. Tecklenburg, and D. F. Lim. 1991. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 3:799-811. [PubMed] [Google Scholar]
- 20.Lysnyansky, I., Y. Ron, K. Sachse, and D. Yogev. 2001. Intrachromosomal recombination within the vsp locus of Mycoplasma bovis generates a chimeric variable surface lipoprotein antigen. Infect. Immun. 69:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lysnyansky, I., Y. Ron, and D. Yogev. 2001. Juxtaposition of an active promoter to vsp genes via site-specific DNA inversions generates antigenic variation in Mycoplasma bovis. J. Bacteriol. 183:5698-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lysnyansky, I., R. Rosengarten, and D. Yogev. 1996. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J. Bacteriol. 178:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lysnyansky, I., K. Sachse, R. Rosenbusch, S. Levisohn, and D. Yogev. 1999. The vsp locus of Mycoplasma bovis: gene organization and structural features. J. Bacteriol. 181:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neilson, L., G. Blakely, and D. J. Sherratt. 1999. Site-specific recombination at dif by Haemophilus influenzae XerC. Mol. Microbiol. 31:915-926. [DOI] [PubMed] [Google Scholar]
- 25.Noormohammadi, A. H., P. F. Markham, A. Kanci, K. G. Whithear, and G. F. Browning. 2001. A novel mechanism for control of antigenic variation in the haemagglutinin gene family of Mycoplasma synoviae. Mol. Microbiol. 35:911-923. [DOI] [PubMed] [Google Scholar]
- 26.Nunes-Düby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piechaczek, K., U. Dobrindt, A. Schierhorn, G. S. Fischer, M. Hecker, and J. Hacker. 2000. Influence of pathogenicity islands and the minor leuX-encoded tRNA5Leu on the proteome pattern of the uropathogenic Escherichia coli strain 536. Int. J. Med. Microbiol. 290:75-84. [DOI] [PubMed] [Google Scholar]
- 28.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Recchia, G. D., and D. J. Sherratt. 1999. Conservation of xer site-specific recombination genes in bacteria. Mol. Microbiol. 34:1146-1148. [DOI] [PubMed] [Google Scholar]
- 30.Ritter, A., D. L. Gally, P. B. Olsen, U. Dobrindt, A. Friedrich, P. Klemm, and J. Hacker. 1997. The Pai-associated leuX specific tRNA5LEU affects type 1 fimbriation in pathogenic Escherichia coli by control of FimB recombinase expression. Mol. Microbiol. 25:871-882. [DOI] [PubMed] [Google Scholar]
- 31.Rosati, S., S. Pozzi, P. Robino, B. Montinaro, A. Conti, M. Fadda, and M. Pittau. 1999. P48 major surface antigen of Mycoplasma agalactiae is homologous to a malp product of Mycoplasma fermentans and belongs to a selected family of bacterial lipoproteins. Infect. Immun. 67:6213-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachse, K., J. H. Helbig, I. Lysnyansky, C. Grajetzki, W. Müller, E. Jacobs, and D. Yogev. 2000. Epitope mapping of immunogenic and adhesive structures in repetitive domains of Mycoplasma bovis variable surface lipoproteins. Infect. Immun. 68:680-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sciochetti, S. A., P. J. Piggot, D. J. Sherratt, and G. Blakely. 1999. The ripX locus of Bacillus subtilis encodes a site-specific recombinase involved in proper chromosome partitioning. J. Bacteriol. 181:6053-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto, S., and M. Miyata. 1998. Cell reproduction and morphological changes in Mycoplasma capricolum. J. Bacteriol. 180:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen, X., J. Gumulak, H. Yu, C. T. French, N. Zou, and K. Dybvig. 2000. Gene rearrangements in the vsa locus of Mycoplasma pulmonis. J. Bacteriol. 182:2900-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherratt, D. J., L. K. Arciszewska, G. Blakely, S. Colloms, K. Grant, N. Leslie, and R. McCulloch. 1995. Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond. Biol. Sci. 347:37-42. [DOI] [PubMed] [Google Scholar]
- 37.Simmons, W. L., C. Zuhua, J. I. Glass, J. W. Simecka, G. H. Cassell, and H. L. Watson. 1996. Sequence analysis of the chromosomal region around and within the V-1-encoding gene of Mycoplasma pulmonis: evidence for DNA inversion as a mechanism for V-1 variation. Infect. Immun. 64:472-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sitaraman, R., and K. Dybvig. 1997. The hsd loci of Mycoplasma pulmonis: organization, rearrangements and expression of genes. Mol. Microbiol. 26:109-120. [DOI] [PubMed] [Google Scholar]
- 39.Solsona, M., M. Lambert, and F. Poumarat. 1996. Genomic, protein homogeneity and antigenic variability of Mycoplasma agalactiae. Vet. Microbiol. 50:45-58. [DOI] [PubMed] [Google Scholar]
- 40.Subramanya, H. S., L. K. Arciszewska, R. A. Baker, L. E. Bird, D. J. Sherratt, and D. B. Wigley. 1997. Crystal structure of the site-specific recombinase, XerD. EMBO J. 16:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Washburn, L. R., K. E. Weaver, E. J. Weaver, W. Donelan, and S. Al-Sheboul. 1998. Molecular characterization of Mycoplasma arthritidis variable surface protein MAA2. Infect. Immun. 66:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wise, K. S. 1993. Adaptive surface variation in mycoplasmas. Trends Microbiol. 1:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yogev, D., R. Rosengarten, R. Watson-McKown, and K. S. Wise. 1991. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 10:4069-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Q., and K. S. Wise. 1996. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect. Immun. 64:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]