Enterococci are gram-positive constituents of the normal human microflora, typically colonizing the intestinal tract and skin (34, 47). However, these organisms are capable of causing disease as opportunistic pathogens, mainly in immunocompromised patients (35, 58). The problem of enterococcal infection has been aggravated by the emergence in the past decade of multiple antibiotic resistance, which has converted some enterococci from being difficult to treat medically to being completely refractory to all antimicrobial regimens (22, 46).
Despite their pathogenicity, enterococci constitute a major component of the microflora of artisanal cheeses produced in Europe and are considered to play an important role in ripening and aroma development (20, 67). Additionally, enterococci are used as probiotics to improve the microbial balance of the intestine and to treat gastroenteritis in humans and animals (2, 43). These bacteria probably now represent the greatest risk to human health of any bacterial species currently used for these purposes (12, 21).
Additional general features of these organisms are hallmarks of their biology and may also contribute to their pathogenicity. Enterococci are distinguished for their ability to grow in 6.5% NaCl or at temperatures ranging from 10 to 45°C and to tolerate acidic and alkaline growth conditions.
These observations raise the question of how enterococcal physiology has evolved to allow the organisms to sense environmental changes and respond to the various stimuli with adaptive behavior. Monitoring and adapting to changing environmental conditions is the key function of bacterial signal transduction, which is generally carried out by the so-called two-component systems.
Two-component signal transduction systems have evolved to allow cells to monitor their environment and respond appropriately to a wide variety of stimuli, including nutrient deprivation, chemical changes, host-pathogen interaction, osmotic shock, and other stresses (6, 9, 28, 30, 57). While studies of enterococci have identified some general stress proteins (5, 18, 19, 24), a global view of the enterococcal signal transduction mechanisms has not been gained. The soon-to-be-completed sequences of the Enterococcus faecalis (strain V583) (The Institute for Genomic Research [TIGR]; www.tigr.org) and Enterococcus faecium (strain TX0016) (Human Genome Sequencing Center, Baylor College of Medicine) genomes will be invaluable tools to exploit in the effort to gain a better understanding of the physiology of these organisms and address the increasing threat they pose to human health.
OVERVIEW OF TWO-COMPONENT SYSTEMS IN E. FAECALIS STRAIN V583
Despite the great diversity in the types of signals detected and responses elicited, two-component systems utilize a common molecular mechanism that is based on the conversion of signal recognition into a chemical entity (50). The signal recognition function is carried out by a histidine protein kinase. Its autophosphorylation activity in response to a signal output is followed by phosphotransfer to a mated response regulator, thus activating its function. Response regulators are, for the most part, transcription regulators that, upon phosphorylation, activate and/or repress genes involved in the adaptive response. A recent survey of the genomic sequence databases has identified more than 500 members of the histidine kinase or response regulator families (26).
The length and amino acid sequence of the sensing domain of histidine kinases are highly variable from one protein to the other, reflecting the variety of signals to which these kinases respond. The sensing domain is coupled to a C-terminal cytoplasmic catalytic domain which, by contrast, consists of an invariant histidine residue and an ATP-binding domain showing high levels of sequence conservation. The response regulator contains a conserved amino-terminal regulatory domain that is phosphorylated on an aspartic acid residue by the histidine kinase to which it is paired. Phosphorylation of the regulatory domain activates the associated C-terminal or effector domain.
Following the original compilation of two-component systems in Escherichia coli based on the homology of the DNA-binding domain of the response regulators, two major families were identified, OmpR (class III) and NarL (class II) (45). Analysis of the two-component systems identified in the gram-positive organism Bacillus subtilis revealed that the same homology classes were obtained if the sequences surrounding the active-site histidine of the kinases were compared (14). This suggested that the catalytic domain of the kinases and both domains of the response regulators evolved as a unit from a common ancestor. Consistent with this conclusion was the observation that the gene order in the transcription unit in which the histidine kinase- and response regulator-coding genes reside is preserved within the classes (14).
The sequencing of the genome of E. faecalis strain V583 revealed 17 two-component systems based on homology searches that were carried out using the region surrounding the phosphorylatable histidine of the histidine kinases and the DNA-binding domain of the response regulators from B. subtilis. These two-component systems possess linked genes for a sensor histidine kinase and a response regulator (Tables 1 and 2). An orphan response regulator without a linked sensor kinase has also been identified (Table 2). Since the available sequence covers the complete genome, the two-component systems presented here should represent the full complement of the systems encoded by the E. faecalis V583 genome (a compilation of the amino acid sequences of the two-component systems described in this review is available at www.scripps.edu/mem/cb/links).
TABLE 1.
Histidine kinases identified on the E. faecalis V583 genome
| Groupa | Locusb | Alignmentc | Size (amino acids) | Coordinatesd | Similarity (% of identical residues/% of conserved residues)e | Reference(s) |
|---|---|---|---|---|---|---|
| I | EF2219 (HK01) | LQSQINPHFLYNTLEYIR | 570 | 2133921 | YesM (34/54) | 14 |
| EF3197 (HK02) | LQAQVNPHFFFNAINTIS | 589 | 3070495 | LytS (52/71) | 7 | |
| II | EF2912 (HK03) | HRLAREIHDSVSQQLFAAM | 367 | 2790823 | YvqE (45/68); VraS (48/69) | 14, 39 |
| IIIA | EF1704 (HK04) | DFVSNVSHELKTPVTSLLG | 591 | 1649393 | PhoR (41/57) | 60 |
| EF3290 (HK05) | ELITNVSHDIRTPLTSIIG | 364 | 3171476 | |||
| EF1261 (HK06) | QFMADASHERMTPLTTING | 489 | 1227473 | YclK (49/65) | 14 | |
| EF1194 (HK07) | EFVSNVSHELRTPLTSMRS | 609 | 1160481 | YycG (54/74) | 15 | |
| EF1863 (HK08) | DFFKGASHELKTPLASLKI | 439 | 1810225 | VncS (37/64) | 53 | |
| EF0927 (HK09) | DYIDSWVHEIKVPLAAITL | 341 | 889562 | |||
| EF1051 (HK10) | QFVEDVSHELKTPIAAVSV | 502 | 1016027 | LisK (58/72) | 10 | |
| EF2298 (HK11) | YFFAAASHELKTPIAAVSV | 447 | 2218126 | VanSB (100) | 13 | |
| EF0570 (HK12) | NLLRAVSHDLRTPLTVISG | 856 | 527240 | KdpD (38/61) | 66 | |
| EF0373 (HK13) | ELIANISHDLKTPITSIIG | 493 | 342321 | |||
| IV | EF1209 (HK14) | SALQSQSHEFMNKMHVIYG | 392 | 1174587 | YufL (28/47); DcuS (30/53) | 14, 69 |
| EF1820 (HK15) | EELAMFRHDYKNLLYSLYS | 447 | 1766071 | FsrC (100) | 56 | |
| EF1335 (HK16) | TNVLKLKHDLKNQYLTILG | 436 | 1306490 | |||
| EF1632 (HK17) | VAIREIHHRVKNNLQSVVS | 477 | 1588528 |
Grouping is based on the homology of the kinase region surrounding the phosphorylatable histidine (14, 45).
Listed are the locus numbers according to the TIGR database as of 20 June 2002. Shown in parentheses are our working numbers.
Shown is the region surrounding the phosphorylatable histidine (boldface).
Listed are the coordinates as of 20 June 2002. Numbers are subject to change upon completion of the genome sequencing.
Percentages refer to the catalytic domain only, spanning the region from the first residue shown in the third column to the stop codon.
TABLE 2.
Response regulators identified on the E. faecalis V583 genome
| Groupa | Locusb | Family | Gene orderc | Size (amino acids) | Coordinatesd | Similarity (% of identical residues/ % of conserved residues) | Reference(s) |
|---|---|---|---|---|---|---|---|
| I | EF2218 (RR01) | AraC | HK-RR | 493 | 2132197 | YesN (41/66)e | 14 |
| EF3196 (RR02) | Lyt | HK-RR | 242 | 3068745 | 49 | ||
| II | EF2911 (RR03) | NarL | HK-RR | 210 | 2789745 | YvqC (62/78); VraR(62/78) | 14, 39 |
| IIIA | EF1703 (RR04) | OmpR | RR-HK | 236 | 1648686 | PhoP (54/75) | 60 |
| EF3289 (RR05) | OmpR | RR-HK | 229 | 3170710 | BacR (41/59) | ||
| EF1260 (RR06) | OmpR | RR-HK | 239 | 1226754 | YclJ (60/72) | ||
| EF1193 (RR07) | OmpR | RR-HK | 234 | 1159770 | YycF (70/79) | 15 | |
| EF1864 (RR08) | OmpR | RR-HK | 217 | 1810875 | YncR (62/79) | 53 | |
| EF0926 (RR09) | OmpR | RR-HK | 224 | 888874 | |||
| EF1050 (RR10) | OmpR | RR-HK | 228 | 1015330 | LisR (68/77) | 10 | |
| EF2299 (RR11) | OmpR | RR-HK | 220 | 2218788 | VanRB (100) | 13 | |
| EF0571 (RR12) | OmpR | HK-RR | 229 | 529822 | KdpE (41/60) | 66 | |
| EF0372 (RR13) | OmpR | HK-RR | 230 | 340843 | SrrA (37/61) | 68 | |
| EF3329 (RR18) | OmpR | Orphan | 240 | 3215076 | |||
| IV | EF1210 (RR14) | Other-A | HK-RR | 230 | 1175746 | YufM (32/57); DcuR (31/55) | 14, 69 |
| EF1822 (RR15) | Agr | RR-P-HK | 243 | 1767616 | FsrA (100) | 49, 56 | |
| EF1336 (RR16) | Agr | RR-HK | 234 | 1307194 | 49 | ||
| EF1633 (RR17) | RR-HK | 190 | 1589101 |
Grouping is based on the homology of the corresponding kinase region surrounding the phosphorylatable histidine (Table 1) (14, 45).
Listed are the locus numbers according to the TIGR database as of 20 June 2002. Shown in parentheses are our working numbers.
HK, histidine kinase; RR, response regulator; RR-P-HK, response regulator-peptide-histidine kinase.
Listed are the coordinates as of 20 June 2002. Numbers are subject to change upon completion of the genome sequencing.
The percentages were calculated for proteins lacking the linker domain.
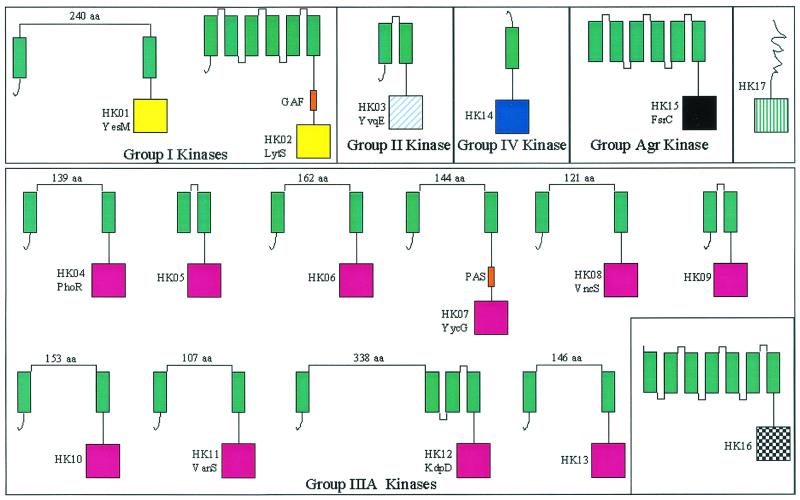
The two-component systems were grouped based on the conservation of amino acids surrounding the phosphorylatable histidine residue and on the structural homology of the DNA-binding domain of the response regulator according to the classification scheme proposed by Fabret et al. (14). Application of this classification scheme (Table 1) revealed that E. faecalis strain V583 has two histidine kinases that are members of the group I family of kinases and one group II kinase. Ten histidine kinases belong to the IIIA class. This class of histidine kinases appears to be the most abundant class of kinases in the gram-positive pathogens whose genomes have been published to date (17, 40, 63). A single group IV histidine kinase was found. The three remaining histidine kinases do not appear to fit into the current classification scheme based on the residues around the conserved histidine. These include the recently described FsrC kinase (56) and two histidine kinases, HK16 and HK17, whose function remains unclear based on sequence similarity.
With the exception of a single histidine kinase (HK17), all of the sensor histidine kinases in E. faecalis strain V583 are predicted to be membrane localized (Fig. 1). This is consistent with the observation that localization of the sensor kinase to the membrane of the bacterial cell appears to be a general feature of most two-component regulatory systems (36).
FIG. 1.
Schematic structure of the 17 histidine kinases identified in the E. faecalis V583 genome. The groups were determined from the homology of the residues surrounding the phosphorylated histidine in the catalytic domain according to the classification of Fabret et al. (14) and from the homology of the DNA-binding domain of response regulators. The color coding is as previously presented by Fabret et al. (14). Green rectangles, transmembrane domains; squares, histidine phosphotransferase domains; orange rectangles, additional domains (PAS and GAF).
Classification of the response regulators in two-component systems was performed based on the relatedness of their output domains (14, 45) (Table 2). In most cases, examination of the output domains of the response regulators is predictive of the class of sensor kinases with which the regulators interact. This stems from the coevolution predicted to take place between sensor kinases and their cognate regulators to maintain the specificity of the phosphotransfer (14, 31).
Eighteen response regulator genes were identified in the E. faecalis V583 genome by using BLASTP analysis. Using the PFAM database at TIGR, we examined the output domain of each response regulator. This analysis revealed similarity to at least three families of DNA-binding proteins, including the OmpR (33), NarL (1), and AraC (65) protein families. For B. subtilis, the only other gram-positive organism for which an analysis of this kind has been performed (14), it has been demonstrated that response regulators grouped in the NarL family were always paired with class II sensor histidine kinases and that members of the OmpR family were consistently coupled with class IIIA kinases. Observations made in our analysis of the E. faecalis two-component systems were consistent with this. Members of the AraC family were not identified in the B. subtilis study (14), but subsequent evaluation of the genome identified the response regulator YesN as being a member of this family (65). While the precise function of YesN is unknown, it possesses an unusual structure for a response regulator in that the region that links the receiver and output domains (linker domain) is predicted to be large (∼150 amino acid residues). By comparison, the linker region of NarL, a protein for which the crystal structure has been solved, is only 23 residues in length (1). A YesN homologue, designated RR01, was identified in E. faecalis (Table 2) and is similar to yesN relatives in the Streptococcus pyogenes (17) and Streptococcus pneumoniae (41, 63) genomes. Analysis of ClustalW alignments of these closely related proteins revealed extensive similarity over both the receiver and output domains but little similarity within the linker region (data not shown). What role, if any, this large linker region plays in the function of these proteins awaits further elucidation; however, its variability and length suggest that it may serve solely to tether the two domains together.
Our analysis of the E. faecalis response regulator output domains identified 11 members of the OmpR family, 1 member of the NarL family, and 1 member of the AraC family. The output domains of the remaining response regulators did not fall into any of the established protein families (Table 2).
INFERRED BIOLOGICAL FUNCTION BASED ON SEQUENCE SIMILARITY
Both members of each two-component system identified in the E. faecalis genome were queried against protein and nucleotide sequence databases by using the BLAST programs. Chromosomal regions immediately surrounding the kinase and response regulator genes were also analyzed, as some two-component systems are adjacent to genes that they control or are involved in their function.
The most significant matches observed are summarized in Tables 1 and 2. The percentages of identity and similarity reported indicate that, among the 17 two-component systems identified in E. faecalis, two have been functionally characterized (VanSBRB and FsrCA) and four are most likely orthologues of functionally characterized systems (HK-RR02, HK-RR04, HK-RR07, and HK-RR08). For the remaining systems, notwithstanding the high level of similarity reported, no function assignment can be done at this time.
The two-component system HK-RR01 shares sequence similarity with YesM-YesN from B. subtilis (14) as well as a similar two-component pair from E. faecium (www.jgi.doe.gov). Apart from the unusual structural properties previously discussed, no biological function can be inferred for this system.
The HK-RR02 two-component system shares similarity with the LytSR system in Staphylococcus aureus and the LytST system in B. subtilis. The S. aureus LytSR system was first identified by Brunskill and Bayles (7) as a regulatory locus affecting the rate of autolysis and penicillin tolerance. Subsequent work has identified the genes regulated by the LytSR system as a bicistronic operon, lrgAB, whose gene products modulate murein hydrolase activity and penicillin tolerance (27). For B. subtilis, a comprehensive analysis of the two-component regulatory systems by use of DNA microarrays has revealed that LytST controls the expression of a bicistronic operon, designated ysbAB (37). The YsbB protein is highly related to the lrgB gene product. Downstream of the E. faecalis HK-RR02 system is an operon with genes that share strong similarity to those of the lrgAB and ysbAB operons, suggesting the possibility that this two-component system is the E. faecalis orthologue of the Lyt system of S. aureus and B. subtilis.
The HK-RR03 two-component system shares extensive sequence similarity with the B. subtilis YvqEC two-component system and a recently identified homologue in S. aureus, designated VraSR (39). In S. aureus, the overexpression of the response regulator VraR leads to an increased resistance to vancomycin. What this system senses for the cell and how its expression affects vancomycin resistance in S. aureus remain to be elucidated. Another interesting feature of this two-component system is that it is highly conserved in most gram-positive pathogenic bacteria, including S. aureus, S. pyogenes, and S. pneumoniae, with an amino acid identity of nearly 60 to 70% and a homology of 80 to 90% within the catalytic domain of the kinases and the response regulator sequences (17, 63, 64).
Of the 10 members of the group IIIA kinase-OmpR regulator family in E. faecalis, four could be assigned a putative functional role based on strong sequence similarity to proteins with known function.
HK-RR04 shares strong similarity with the PhoPR system of B. subtilis (54% amino acid identity with PhoP and 30% amino acid identity with PhoR) (32). The lower level of identity with the PhoR sensor kinase is due to the fact that no similarity exists between the amino-terminal ends of these two kinases. This may indicate that sensing modules for these proteins may have evolved independently.
The histidine kinase of the HK-RR07 system shares 45% amino acid identity and 70% similarity with the B. subtilis YycG kinase. This kinase, along with its cognate response regulator YycF, is highly conserved in gram-positive bacteria (4, 17, 25, 38, 40, 51, 63). It has been established that this two-component system is essential in B. subtilis (15), S. aureus (44), and S. pneumoniae (64). Consistent with these findings, the yycF gene has also been found to be essential in E. faecalis (L. Hancock and M. Perego, unpublished observation). A recent report has linked the YycFG system in B. subtilis to the regulation of the ftsAZ operon (23), which may, in part, explain the essential nature of this system. The nature of the signal to which YycG might respond remains a mystery, but some clues may be derived from the fact that it possesses a PAS domain (29). PAS domains are known to bind small regulatory molecules such as NAD, heme, flavin adenine dinucleotide, and, as reported more recently, ATP (3, 11, 54, 59, 61).
HK-RR08 is homologous to a recently described two-component system of S. pneumoniae, designated VncRS for its role in vancomycin tolerance (35% identity and 56% similarity between kinases; 62% identity and 79% similarity between response regulators) (53). Similarity to the VncRS system extends to the three open reading frames upstream of HK-RR08, which also seem to encode a putative ABC transport system (52). Between the two-component system and the ABC transporter, we identified a small open reading frame encoding a putative peptide of 43 amino acids. This is reminiscent of the pep27 gene upstream of VncRS, which encodes a 27-amino-acid peptide. This peptide has recently been shown to induce multiple cell death mechanisms, including antibiotic-induced autolysis, through modulation of the activity of the VncRS system (52).
HK-RR11 is 100% identical to both the histidine kinase and the response regulator of the VanB-type vancomycin resistance determinant. A comprehensive review of this system has recently been reported (8). This level of homology suggests that HK-RR11 was acquired by horizontal gene transfer (vancomycin resistance transposon).
For the remaining seven systems of group IIIA, two, HK-RR10 and HK-RR12, have similarities to known systems, but making a functional correlation is more hazardous.
The HK-RR10 system shows strong similarity to the LisRK system of Listeria monocytogenes (10) and the CsrRS system of S. pyogenes (16, 42). HK-RR10 has recently been implicated in the stress response and virulence of E. faecalis and was renamed EtaSR (62).
HK-RR12 is homologous to the KdpDE two-component system from both gram-negative and gram-positive organisms (66). An unusual feature of this system is that, unlike other members of the class IIIA kinase-OmpR regulator systems, whose genes are arranged in the regulator-to-kinase order, it has the kinase-coding gene followed by the response regulator-coding gene. The kinase itself is unusual for the class IIIA family of sensor histidine kinases in that it possesses four predicted transmembrane domains; the other members of this family possess only two.
With regard to the four remaining members of the class IIIA kinase-OmpR regulator systems in E. faecalis (HK-RR05, HK-RR06, HK-RR09, and HK-RR13), HK-RR06 has homology to the B. subtilis YclKJ system, whose function is unknown; the other three members appear to be unique to this organism since no significant similarities were identified in other bacteria, with the exception that strong homologues to HK-RR06 and HK-RR13 were identified in the preliminary sequencing data from the E. faecium genome project. RR05 and RR13 actually show some similarities to known response regulators (Table 2), but the HK05 and HK13 proteins do not show sufficient similarities to the corresponding histidine kinase to justify any speculation on functional relationship.
According to Fabret et al. (14), HK-RR14 of E. faecalis strain V583 has a histidine kinase belonging to the class IV kinase family. Apart from the prediction that this kinase contains a single transmembrane domain, no other relevant structural features were evident and little sequence similarity with other two-component systems was found (Tables 1 and 2).
HK-RR15 corresponds to the recently described Fsr system of E. faecalis (48, 55, 56). This system has been shown to regulate the synthesis of two secreted proteins, gelatinase and a serine protease. Furthermore, in an intraperitoneal infection model, mutants with disruptions in the kinase gene fsrC or the response regulator gene fsrA are attenuated in the virulence of E. faecalis (55).
The kinase of the HK-RR16 system is predicted to contain seven transmembrane domains, and the region surrounding its phosphorylated histidine is similar to the one in FsrC. The RR16 response regulator is characterized by a DNA-binding domain that likely belongs to the LytTR (“litter”) family of transcriptional regulators (49). The E. faecalis RR02 and RR15 response regulators also belong to this family. Thus, this system may belong to the Agr kinase family (49). Apart from predicted structural findings, however, no functional data are available concerning the role of this system in the biology of E. faecalis.
The HK-RR17 system is the only one in E. faecalis with a cytoplasmic histidine kinase, but it does not have any significant similarity to known two-component systems.
Not surprisingly for a nonmotile organism, E. faecalis is devoid of a two-component system for chemotaxis and motility.
CONCLUSIONS
Using a genomics-based approach, we have identified the two-component signal transduction systems of E. faecalis V583 and have determined that widespread regulatory systems, found mainly among gram-positive organisms, are well represented in this strain. Enterococci are organisms capable of adapting to a number of host environments as either commensals or pathogens. Thus, it is not surprising that key regulators of physiological functions were identified together with putative pathogenic modulators. It is now abundantly clear from the analyses of several gram-positive bacterial genomes that most, but not all, two-component signal transduction systems are common to these related species. This suggests that the progenitor of the diverse present-day gram-positive genera had already amplified by gene duplication the two-component systems presently found in these bacteria before speciation. The conservation of catalytic domains of sensor kinases with the response regulators, their virtually invariant gene order, and the conserved interaction surfaces (31) provide strong evidence for such a conclusion. As the present-day genera evolved from the progenitor, the putative signaling domains became highly variable in sequence and structure, suggesting that the unique ecological niche that each species occupies contains specific signals that drive the diversity of the signal input domain. Furthermore, in any given species, one environment may activate a subset of two-component systems and, as a consequence, a certain pattern of genes, whereas a second environment may induce expression of another subset of genes. Thus, for opportunistic pathogens such as enterococci and its relatives, the capacity to survive in an environment depends on the signals and the genes that are connected to the two-component system that responds to the signals.
The two-component system conferring vancomycin resistance and three other systems, HK-RR05, HK-RR09, and HK-RR13, may be specific for the Enterococcus genus. Since it is likely that vancomycin resistance is horizontally transferred as a consequence of antibiotic therapy, the genus specificity of the three systems of unknown function also may have been acquired in a similar manner to allow the organism to cope with conditions within the gut and persist there. It will be of particular interest to determine whether the genes controlled by these systems have functions consistent with this speculation.
Acknowledgments
This work was supported, in part, by grants GM55594 and GM19416 from the National Institute of General Medical Sciences, National Institutes of Health, USPHS.
We thank Jim Hoch for helpful discussions.
Footnotes
Paper 14748-MEM from The Scripps Research Institute.
REFERENCES
- 1.Baikalov, I., I. Schroder, M. Kaczor-Grzeskowiak, K. Grzeskowiak, R. P. Gunsalus, and R. E. Dickerson. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053-11061. [DOI] [PubMed] [Google Scholar]
- 2.Bellomo, G., A. Mangiagle, L. Nicastro, and L. Frigerio. 1980. A controlled double-blind study of SF68 strain as a new biological preparation for the treatment of diarrhoea in pediatrics. Curr. Ther. Res. 28:927-934. [Google Scholar]
- 3.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutibonnes, P., J. C. Giard, A. Hartke, B. Thammavongs, and Y. Auffray. 1993. Characterization of the heat shock response in Enterococcus faecalis. Antonie Leeuwenhoek 64:47-55. [DOI] [PubMed] [Google Scholar]
- 6.Browse, J., and Z. Xin. 2001. Temperature sensing and cold acclimation. Curr. Opin. Plant Biol. 4:241-246. [DOI] [PubMed] [Google Scholar]
- 7.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coote, J. G. 2001. Environmental sensing mechanisms in Bordetella. Adv. Microb. Physiol. 44:141-181. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, P. D., N. Emerson, C. G. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado-Nixon, V. M., G. Gonzalez, and M. A. Gilles-Gonzalez. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39:2685-2691. [DOI] [PubMed] [Google Scholar]
- 12.Eaton, T. J., and M. J. Gasson. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evers, S., and P. Courvalin. 1996. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J. Bacteriol. 178:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A Streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flahaut, S., A. Hartke, J.-C. Giard, and Y. Auffray. 1997. Alkaline stress response in Enterococcus faecalis: adaptation, cross-protection, and changes in protein synthesis. Appl. Environ. Microbiol. 63:812-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flahaut, S., A. Hartke, J. C. Giard, P. Benachour, P. Boutibonnes, and Y. Auffray. 1996. Relationship between stress response toward bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol. Lett. 138:49-54. [DOI] [PubMed] [Google Scholar]
- 20.Franz, C. M. A. P., W. H. Holzapfel, and M. E. Stiles. 1999. Enterococci at the crossroads of food science? Int. J. Food Microbiol. 47:1-24. [DOI] [PubMed] [Google Scholar]
- 21.Franz, C. M. A. P., A. B. Muscholl-Silberhorn, N. M. K. Yousif, M. Vancanneyt, J. Swings, and W. H. Holzapfel. 2001. Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67:4385-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.French, G. L. 1998. Enterococci and vancomycin resistance. Clin. Infect. Dis. 27:75-83. [DOI] [PubMed] [Google Scholar]
- 23.Fukuchi, K., K. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 24.Giard, J. C., A. Hartke, S. Flahaut, P. Boutibonnes, and Y. Auffray. 1997. Glucose starvation response in Enterococcus faecalis. Res. Microbiol. 148:27-35. [DOI] [PubMed] [Google Scholar]
- 25.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vincente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez Diaz, R. Purcell, B. Remmel, M. Rose, T. Schleuter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 26.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 27.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu, Y., J. B. Hogenesch, and C. A. Bradfield. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40:519-561. [DOI] [PubMed] [Google Scholar]
- 30.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 31.Hoch, J. A., and K. I. Varughese. 2001. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 183:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 33.Itou, H., and I. Tanaka. 2001. The OmpR-family of proteins: insight into the tertiary structure and functions of two-component regulator proteins. J. Biochem. 129:343-350. [DOI] [PubMed] [Google Scholar]
- 34.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karchmer, A. W. 1999. Infective endocarditis, p. 621-635. In R. K. Root, F. Waldvogel, L. Corey, and W. Stamm (ed.), Clinical infectious disease. Oxford University Press, Inc., New York, N.Y.
- 36.Kim, D., and S. Forst. 2001. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147:1197-1212. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi, K., M. Ogura, K. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 39.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 40.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 41.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 42.Levin, J. C., and M. R. Wessels. 1998. Identification of esrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 43.Lewenstein, A., G. Frigerio, and M. Moroni. 1979. Biological properties of SF6, a new approach to the treatment of diarrhoeal diseases. Curr. Ther. Res. 26:967-981. [Google Scholar]
- 44.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 46.Moellering, R. C. 1992. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 14:1173-1176. [DOI] [PubMed] [Google Scholar]
- 47.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. L. Akkermans, W. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 49.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ninfa, A. J., and B. Magasanik. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 83:5909-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novak, R., E. Charpentier, J. S. Braun, and E. Tuomanen. 2000. Signal transduction by a death signal peptide: uncovering the mechanism of bacterial killing by penicillin. Mol. Cell 5:49-57. [DOI] [PubMed] [Google Scholar]
- 53.Novak, R., B. Henriques, E. Charpentier, S. Normark, and E. Tuomanen. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590-593. [DOI] [PubMed] [Google Scholar]
- 54.Pellequer, J.-L., K. A. Wagner-Smith, S. A. Kay, and E. D. Getzoff. 1995. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. USA 95:5884-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 58.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 59.Stephenson, K., and J. A. Hoch. 2001. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:15251-15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 70:1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 64.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, and M. Rosenberg. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 65.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treuner-Lange, A., A. Kuhn, and P. Dürre. 1997. The kdp system of Clostridium acetobutylicum: cloning, sequencing, and transcriptional regulation in response to potassium concentration. J. Bacteriol. 179:4501-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trovatell, L. D., and A. Schiesser. 1987. Identification and significance of enterococci in hard cheese made from raw cow and sheep milk. Milchwissenschaft 42:717-719. [Google Scholar]
- 68.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zientz, E., J. Bongaerts, and G. Unden. 1998. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 180:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]