Abstract
The predicted amino acid sequence of Bacillus subtilis yfjS (renamed pdaA) exhibits high similarity to those of several polysaccharide deacetylases. β-Galactosidase fusion experiments and results of Northern hybridization with sporulation sigma mutants indicated that the pdaA gene is transcribed by EσG RNA polymerase. pdaA-deficient spores were bright by phase-contrast microscopy, and the spores were induced to germination on the addition of l-alanine. Germination-associated spore darkening, a slow and partial decrease in absorbance, and slightly lower dipicolinic acid release compared with that by the wild-type strain were observed. In particular, the release of hexosamine-containing materials was lacking in the pdaA mutant. Muropeptide analysis indicated that the pdaA-deficient spores completely lacked muramic δ-lactam. A pdaA-gfp fusion protein constructed in strain 168 and pdaA-deficient strains indicated that the protein is localized in B. subtilis spores. The biosynthetic pathway of muramic δ-lactam is discussed.
Bacillus subtilis produces spores under various nutrient limitations, and this provides a survival mechanism under adverse environmental conditions (25). On the addition of germinants, spores start to germinate and refractility quickly decreases (17). At the same time dipicolinic acid is released, and then cortex fragments containing hexosamine are released from the spores (17). The cortex is one of the most characteristic structures in spores and is degraded by germination-specific cortex lytic enzymes (GSLEs) (3, 17). SleB and CwlJ are candidate GSLEs (10, 15, 17), and it is believed that these proteins recognize the muramic δ-lactam structure, which is unique in spore cortex (7, 17, 24), because Bacillus cereus SleB, which is highly homologous to B. subtilis SleB and CwlJ, degrades normal cortex but not muramic δ-lactam-deficient cortex (13, 17; S. Makino and R. Moriyama, unpublished results).
Previously, two groups suggested that an N-acetylmuramoyl-l-alanine amidase homologue, CwlD, is associated with the biosynthesis of muramic δ-lactam (4, 18) because a cwlD-deficient mutant completely lacked muramic δ-lactam (4, 18) and expressed a germination-negative phenotype (21). The cortex of the mutant exhibited higher cross-linkage than the wild-type spores (4, 18). Under the B. subtilis functional analysis project, we analyzed the germination of spores from more than 500 disrupted strains (22). One of the mutations, yfjS, exhibited almost no germination (as measured by recovery of colonies from spores). The amino acid sequence similarity of YfjS with sequences in protein databases suggested it is a polysaccharide deacetylase homologue (BSORF database). In legumes, this enzyme is known as nodulation enzyme B (8), and two homologous gene products in yeast are chitin deacetylases (14).
In this paper we describe the germination profiles of the yfjS mutant and the yfjS gene, which is associated with muramic δ-lactam biosynthesis in the spore cortex.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains of B. subtilis and Escherichia coli used in this study are listed in Table 1. E. coli was grown on Luria-Bertani (LB) agar medium (19) at 37°C and then inoculated into LB medium. B. subtilis was grown on nutrient agar medium (8 g of Bacto nutrient broth/liter, 0.12 g of MgSO4 · 7H2O/liter, 1 g of KCl/liter, and 15 g of agar/liter; pH 7.0 to 7.2) at 30°C, inoculated into DSM (Schaeffer) medium (20), and then shaken at 37°C. If necessary, cells on the agar medium were precultured in NB medium (nutrient agar medium without agar) before cultivating in DSM medium. For muropeptide analysis, B. subtilis spores were produced in CCY medium (4, 26). If necessary, erythromycin, tetracycline, and ampicillin were added to the medium to final concentrations of 0.2, 5 (unless otherwise noted), and 50 μg/ml, respectively.
TABLE 1.
Bacterial strains used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| B. subtilis strains | ||
| 168 | trpC2 | D. Ehrlich |
| 168HR | trpC2 | Laboratory stock (S. J. Foster) |
| YFJSd | trpC2 yfjS::pM2fjS | pM2fjS→168a |
| HR(YFJS) | trpC2 yfjS::pM2fjS | pM2fjS→168HR |
| HR(YFJS CWLD) | trpC2 yfjS::pM2fjS cwlD::cat | AA107 (4)→HR(YFJS) |
| 1S86 | trpC2 spoIIA1 | BGSCb |
| 1S60 | leuB8 tal-1 spoIIG41 | BGSC |
| SpoIIIGΔ1 | trpC2 spoIIIGΔ1 | BGSC |
| 1S38 | trpC2 spoIIIC94 | BGSC |
| E. coli strains | ||
| JM109 | recA1 Δ(lac-proAB) endA1 gyrA96 thi-1 hsdR17 relA1 supE44 [F′ traD36 proAB+lacIqlacZ ΔM15] | Takara |
| C600 | supE44 hsdR17 thi-1 thr-1 lueB6 lacY1 tonA21 | Laboratory stock |
| Plasmids | ||
| pMUTIN2 | lacZ lacI bla erm | D. Ehrlich |
| pM2fjS | pMUTIN2::ΔyfjS (lacZ lacI bla erm ΔyfjS) | This study |
| pGEM3Zf(+) | lacZ bla | Promega |
| pGEMfjS | pGEM3Zf(+)::ΔyfjS (lacZ bla ΔyfjS) | This study |
| pUC119 | lacZ bla | Takara |
| pBluescriptII-SK(+) | lacZ bla | Stratagene |
| pHY300PLK | bla tet | Takara |
| pHYCM1 | pHY300PLK with a 292-bp citM promoter region (carrying a CG-to-AT dinucleotide change at +47 and +48 relative to the citM start point) | H. Yamamoto and M. Serizawa |
| pUCfjS | bla yfjS | This study |
| pBlfjS | bla yfjS | This study |
| pUCΔSfjS | bla ΔyfjS (lacking putative signal sequence) | This study |
| pBlΔSfjS | bla ΔyfjS (lacking putative signal sequence) | This study |
| pHYCMfjS | bla tet yfjS | This study |
| pHYCMΔSfjS | bla tet ΔyfjS (lacking putative signal sequence) | This study |
| pQBI63 | bla gfp | Takara |
| pHYGFP | bla tet gfp | This study |
| pHY300GFP14 | bla tet gfp | This study |
| pB1PRbaNG | bla ybaN-gfp | This study |
| pB1PRfjSG | bla yfjS-gfp | This study |
| pHYPRbaNG | bla tet ybaN-gfp | This study |
| pHYPRfjSG | bla tet yfjS-gfp | This study |
Sources shown before and after the arrows indicate donor DNA and recipient cells on transformation, respectively.
BGSC, Bacillus Genetic Stock Center.
Plasmids used for construction of a pdaA (yfjS) mutant.
To construct a pdaA disruption plasmid, an internal fragment of the pdaA gene was amplified by PCR using two primers, forward primer JS-HF (5′-GCCGAAGCTTGATGTGTTCAATATGCTGC; the HindIII site is underlined) and reverse primer JS-BR (5′-GCGCGGATCCTCATATCCGTTATCAAACGT; the BamHI site is underlined), with B. subtilis 168 DNA as a template. The PCR fragment was digested with HindIII and BamHI and then ligated to HindIII- and BamHI-digested pMUTIN2 followed by transformation of E. coli JM109. The resulting plasmid, pM2fjS, was used to construct a pdaA-deficient mutant. The digested PCR fragment was also ligated to HindIII- and BamHI-digested pGEM3Zf(+) to construct pGEMfjS. pGEMfjS was used to synthesize an RNA probe, and pM2fjS was also used for the transformation of E. coli C600 to produce concatemeric DNAs (6).
Construction of pdaA complementation plasmids.
pUCfjS was constructed by PCR with PQEJS (5′-GCCGGAGCTCATGAAGTGGATGTGTTCA; the SacI site is underlined) and JS-D (5′-GCGCGGATCCTTTACAAAGACGGCAGCC; the BamHI site is underlined) as primers and 168 DNA as a template. The amplified fragment containing the entire pdaA gene was digested with SacI and BamHI, followed by ligation to the corresponding sites of pUC119. The resultant pUCfjS was further digested with PstI and EcoRI, and the fragment containing the pdaA gene was ligated to the corresponding sites of pBluescript II SK(+), resulting in pBlfjS. The SalI fragment containing the yfjS gene from pBlfjS was ligated to the SalI site of pHYCM1 (described below), resulting in pHYCMfjS. pHYCM1 is a pHY300PLK derivative with a 292-bp citM promoter region (carrying a CG-to-AT dinucleotide change [mutated cre] at +47 and +48 relative to the citM start point) (30). Therefore, the citST-controlled gene in pHYCM1 is only expressed in the presence of citrate plus Mg2+ and is not repressed by glucose.
Plasmids containing a putative signal sequence-deleted pdaA gene were constructed basically as described above. A fragment containing the putative signal sequence-deleted pdaA gene (the start codon was also changed from GTG to ATG) was constructed using two primers, PQEspjS (5′-GCCGGAGCTCATGCCGAATGAGCCGATT; the SacI site is underlined) and JS-D. The amplified fragment was ligated to pUC119, resulting in pUCΔSfjS. Then, pBlΔSfjS and pHYCMΔSfjS were constructed as described for the construction of pBlfjS and pHYCMfjS.
Construction of GFP fusion plasmids.
A basic green fluorescent protein (GFP) fusion plasmid, pHYGFP, was constructed by insertion of the HindIII-BglII fragment from pQBI63 (rsGFP vector with red-shifted excitation wavelength; Takara) into the corresponding site of pHY300PLK. Then, a gfp-containing fragment was obtained by PCR amplification with two primers, GFP-HF (5′-GCCGAAGCTTTGTTTAAGAAGGAG; the HindIII site is underlined) and GFP-BgR (5′-GCGCGCAGCCAGATCTTCAGTTG; the BglII site is underlined), and pHYGFP as a template. After digestion with HindIII and BglII, the fragment was ligated to the corresponding site of pHY300PLK, resulting in pHY300GFP14. To construct an intermediate plasmid, pHYPRbaNG, a fragment containing the ybaN promoter, Shine-Dalgarno (SD) sequence, and the entire ybaN gene was obtained by PCR amplification with two primers, ybaN-SDB-U (5′-GCCGGGATCCGTGAAGAGACATTTTATCGG; the BamHI site is underlined) and ybaN-GFP2 (5′-GCGCTCTAGAACCTCCACCTCCGCTAGCCTTTACCTCTGCGGATTTG; the XbaI and NheI sites are underlined), and strain 168 chromosome DNA as a template. The amplified fragment was digested with BamHI and XbaI, followed by ligation to the corresponding site of pBluescript II SK(+), resulting in pBlPRbaNG. Then, a HindIII-XbaI fragment from pBlPRbaNG, containing the ybaN promoter, SD, and the ybaN gene, was ligated to HindIII- and NheI-digested pHY300GFP14, resulting in pHYPRbaNG. To finally construct a pdaA-GFP plasmid, a fragment containing the pdaA region was amplified by PCR using two primers, forward primer jT-HF (5′-GCCGAAGCTTGGGGAACGCCGTAAACAA; the HindIII site is underlined) and reverse primer jS-DG (5′-GCGCGAATTCGCTAGCCAAAGACGGCAGCCTCA; the EcoRI and NheI sites are underlined), with 168 DNA as a template. The PCR fragment was digested with HindIII and EcoRI and then ligated to the corresponding site of pBluescript II SK(+), followed by transformation of E. coli JM109. The resulting plasmid, pBlPRfjSG, was further digested with HindIII and NheI, and the digested fragment was ligated to the corresponding site of pHYPRbaNG, resulting in pHYPRfjSG. Therefore, pHYPRfjSG contains a region between yfjT and pdaA and the pdaA gene translationally fused with the gfp gene.
Mutant construction.
A pdaA-deficient mutant, YFJSd, was constructed by transformation of B. subtilis 168 with pM2fjS. Disruption of the pdaA gene by means of Campbell-type recombination was confirmed by PCR. Thus, the pdaA mutant was a pdaA-lacZ transcriptional fusion strain.
Transformation of E. coli and B. subtilis.
E. coli transformation was performed as described by Sambrook et al. (19), and B. subtilis transformation was performed by the competent cell method (1).
Spore germination.
B. subtilis 168 and YFJSd were cultured in DSM at 37°C until t48 (48 h after the onset of sporulation). Spores were washed until all cell debris and vegetative cells had been removed (10, 22). The spores were heat activated at 80°C for 20 min, unless otherwise noted, and then diluted with 10 mM Tris-HCl buffer (pH 8.4). Germination was initiated by the addition of l-alanine to 10 mM or AGFK (l-asparagine, d-glucose, d-fructose, and KCl) to final concentrations of 10, 1, 1, and 10 mM, respectively. At appropriate times, the A580 of the mixture was measured. If necessary, the amounts of dipicolinic acid and/or hexosamine released from spores during germination were determined (22). The supernatant (1 ml) was used for measurement of the released dipicolinic acid as described by Nicholson and Setlow (16). The rest of the supernatant was dried with a concentrator (model CC-180; TOMY), followed by measurement of the released reducing groups by the method of Elson and Morgan, with N-acetylglucosamine as a standard (16).
β-Galactosidase assay.
The β-galactosidase assay was performed basically as described by Shimotsu and Henner (23). One unit of β-galactosidase activity was defined as the amount of enzyme necessary to release 1 nmol of 2-nitrophenol from 2-nitrophenyl-β-d-galactopyranoside in 1 min.
Northern blot analysis.
B. subtilis cells (optical density at 600 nm of 15 to 20) cultured in DSM medium were harvested and then suspended in 1 ml of chilled killing buffer (20 mM Tris-HCl [pH 7.5] containing 5 mM MgCl2). After centrifugation at 11,000 × g for 2 min, the pellet was suspended in 1 ml of SET buffer (20% [wt/vol] sucrose, 50 mM EDTA, 20 mM Tris-HCl [pH 7.6]) containing lysozyme (final concentration, 6 mg/ml) (29). After incubation for 9 to 12 min at 0°C, the suspension was centrifuged at 11,000 × g for 2 min. The pellet was used for RNA preparation with Isogen (Nippon Gene) according to the manufacturer's instructions. Agarose-formaldehyde gel electrophoresis was performed as described by Sambrook et al. (19). The transfer of RNAs onto a nylon membrane (Magnagraph; Micron Separations) was performed with a vacuum blotter (model BE-600; BIOCRAFT). The DNA fragment used for preparing an RNA probe was amplified by PCR with M13 (−21) and M13RV as primers and with pGEMfjS DNA, containing the internal region of pdaA, as a template. The amplified fragment was digested with HindIII, and the resulting fragments were purified by phenol and chloroform treatment and precipitated with ethanol. The RNA probe was prepared with a DIG (digoxigenin) RNA labeling kit (Roche), and Northern (RNA) hybridization was performed according to the manufacturer's instructions.
RP-HPLC analysis of spore peptidoglycan.
Spores of B. subtilis strains 168HR (a laboratory strain of 168), HR(YFJS), and HR(YFJS CWLD) were produced in CCY medium, and cortex extraction from dormant spores, muropeptide separation by reverse-phase high-performance liquid chromatography (RP-HPLC), and amino acid and mass spectrometry (MS) analyses were performed as previously described (4, 5).
Phase and fluorescence microscopy.
Cells expressing green fluorescence were prepared as follows. After inoculation and incubation of YFJSd harboring pHYPRfjSG on LB agar plates containing erythromycin and tetracycline at 37°C for 10 h, each colony was suspended into 4 ml of NB medium containing tetracycline and then precultured at 37 or 30°C for 4 h. Then, the preculture was suspended in 50 ml of DSM medium containing tetracycline (final concentration, 10 μg/ml) to obtain a cell density (A600) of 0.01, followed by culturing at 37 or 30°C until t20. After centrifugation of the culture, cells were washed and stained with 4,6-diamidino-2-phenylindole (DAPI; final concentration, 1.0 μg/ml; Wako). For fluorescence microscopy, an Olympus BX61 microscope was used with a BX-UCB control unit and an UPPlan Apo Fluorite phase-contrast objective (magnification, ×100; numerical aperture, 1.3). The dichroic mirror cube units for GFP and DAPI contained a wide-band-pass (330 to 385 nm) excitation filter with a long-pass (420 nm) barrier filter (U-MWU2; Olympus) and a band-pass (470 to 490 nm) excitation filter with a narrow-band-pass (510 to 550 nm) barrier filter (U-MNIBA2; Olympus). The exposure times for phase-contrast, DAPI, and GFP detection were 0.1, 0.003, and 1 s, respectively. Cells were photographed by both fluorescence and phase-contrast microscopy by using a charge-coupled device camera (CoolSNAP HQ; Nippon Roper), and photo images were analyzed with MetaMorph software (Nippon Roper). Image overlays and micrograph figures were prepared with Adobe Photoshop software.
Complementation analysis.
Cultures of strains 168(pHYCM1), YFJSd(pHYCMfjS), YFJSd(pHYCMΔSfjS), and YFJSd(pHYCM1) were treated by heating (80°C, 20 min) and then plated on LB agar plates. After the LB plates had been kept at 37°C for 10 h, the colonies were counted. Each sporulation ratio was calculated on the basis of the number of spores and vegetative cells counted under the microscope. Each germination ratio was calculated on the basis of the number of colonies on an LB plate and the number of spores counted under a microscope.
RESULTS
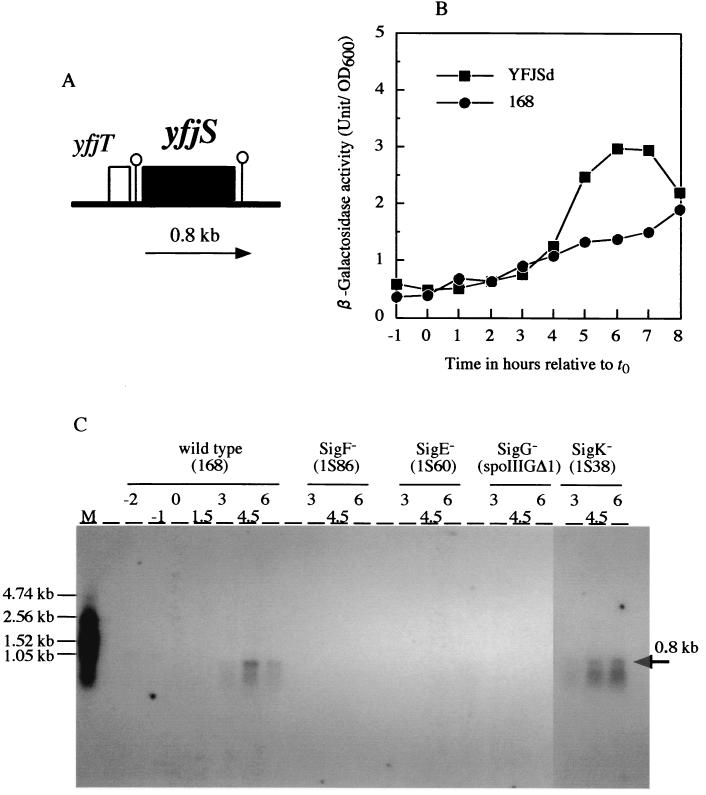
The B. subtilis genome contains five polysaccharide deacetylase homologues, one of which is the yfjS gene located at 74.2° on the B. subtilis chromosome (12, 31). YfjS (renamed PdaA) is a 263-amino-acid polypeptide with a predicted molecular mass of 30,069 Da. A consensus SD sequence (A-15AGGAG−10; the numbering is with respect to the translational start codon) is present. The N-terminal 21-amino-acid sequence seems to be a signal sequence, because a positively charged amino acid, lysine, at position 2, a hydrophobic core comprising positions 7 to 18, and a typical signal sequence cleavage site, A19XA21↓ (the arrow indicates the cleavage point), are present. The upstream gene is yfjT, but a good candidate for the rho-independent terminator (ΔG = −20.8 kcal/mol) is located between yfjT and yfjS. Another typical rho-independent terminator (ΔG = −18.0 kcal/mol) is present downstream of yfjS. Therefore, yfjS seems to be transcribed as a monocistronic mRNA (Fig. 1A).
FIG. 1.
(A) Physical map of the yfjT and yfjS (pdaA) genes of B. subtilis. Stem-loop structures indicate the putative terminators. The arrow indicates the position of yfjS mRNA. (B) β-Galactosidase activity of the pdaA-lacZ transcriptional fusion strain. Circles, wild-type strain 168; squares, pdaA-lacZ fusion strain YFJSd. (C) Northern blotting analysis with a pdaA RNA probe. Numbers indicate times after the onset of sporulation. Each RNA was isolated from strain 168, 1S86 (spoIIA), 1S60 (spoIIG), spoIIIGΔ1 (spoIIIG), or 1S38 (spoIIIC). Twenty micrograms of each RNA was separated on a 1% formaldehyde-agarose gel. The calculated size of the detected pdaA mRNA is indicated by an arrow. M, marker (RNA molecular weight marker 1, digoxigenin labeling [Boehringer Mannheim]).
Expression of the pdaA gene.
A pdaA-lacZ gene fusion strain, YFJSd, was constructed as described in Materials and Methods and cultured in DSM, and then β-galactosidase activity was determined. The activity was low but significant from t4 and was maximal at around t6 (Fig. 1B), which indicates that the pdaA gene is regulated by a sporulation-specific sigma factor. RNAs from four sigma factor-deficient (null) strains, 1S86 (SigF−), 1S60 (SigE−), spoIIIGΔ1 (SigG−), and 1S38 (SigK−), were analyzed by Northern blotting using an RNA probe containing the internal region of the pdaA gene. A weak hybridizing band was detected around 0.8 kb for the 168 strain and SigK mutant, but no such band was observed for the SigF-, SigE-, and SigG-deficient mutants (Fig. 1C). Since the pdaA gene consists of 789 nucleotide residues (excluding the stop codon) and the distance between the two rho-independent terminators is approximately 980 nucleotide residues, the size of the transcript (0.8 kb) reasonably matched the estimated size of monocistronic mRNA. These results indicate that pdaA was transcribed by σG RNA polymerase. Since PdaA seems to contain a signal sequence, it may be expected that the enzyme might be transported into the cortex layer and modify it during sporulation.
Characterization of a pdaA disruptant.
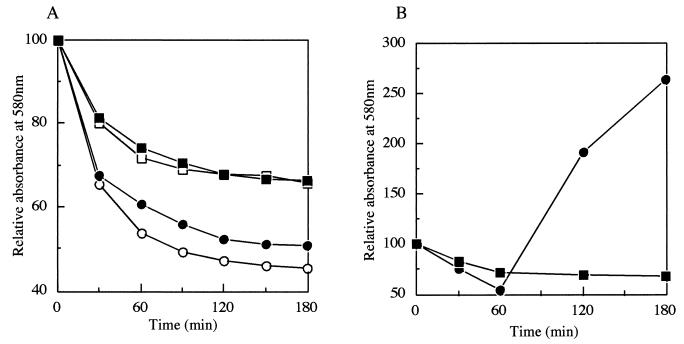
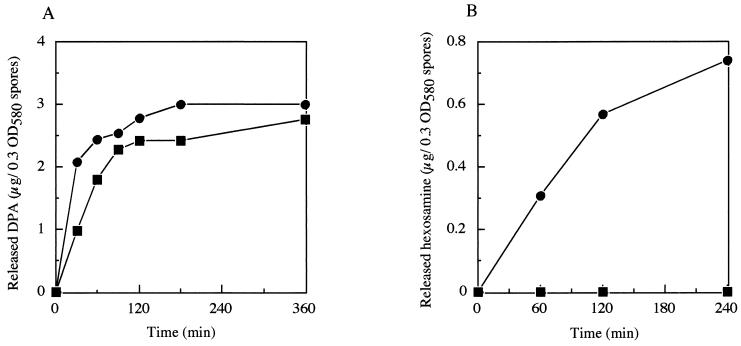
pdaA-deficient mutant YFJSd showed normal growth, cell separation, and motility and produced bright refractile spores. After heat activation at 80°C for 20 min, germination was measured by monitoring the decrease in the A580 of a spore suspension upon the addition of l-alanine or AGFK at 37°C (Fig. 2A). Spores of YFJSd responded to the germinants, and the A580 values of the spore suspensions slowly decreased by about 30%, in contrast with the decrease by about 50% for the wild-type strain (Fig. 2A). During the incubation, bright spores became phase-gray, and dipicolinic acid was released into the suspension. The amount released from YFJSd spores was not greatly different from that from the wild-type spores (Fig. 3A). However, there was a great difference in the release of hexosamine between the YFJSd and wild-type spores (Fig. 3B). A complete lack of hexosamine release has been reported for a mutant of cwlD, which is one of the homologous genes for cell wall-lytic N-acetylmuramoyl-l-alanine amidase. YFJSd did not show any outgrowth in the LB medium, like the cwlD mutant (ADD1) (Fig. 2B) (21).
FIG. 2.
Spore germination and outgrowth of B. subtilis YFJSd and 168. (A) Spore germination was monitored at 580 nm after the addition of l-alanine (closed symbols) or AGFK (open symbols) as a germinant. (B) Outgrowth of B. subtilis YFJSd and strain 168 spores in LB medium at 37°C was monitored at 580 nm. Circles, strain 168; squares, YFJSd.
FIG. 3.
Dipicolinic acid (DPA) (A) and hexosamine (B) released from B. subtilis YFJSd and 168 spores during germination. Released DPA and hexosamine were measured after the addition of l-alanine as a germinant. Circles, strain 168; squares, YFJSd.
Muropeptide analysis of pdaA-disrupted spores.
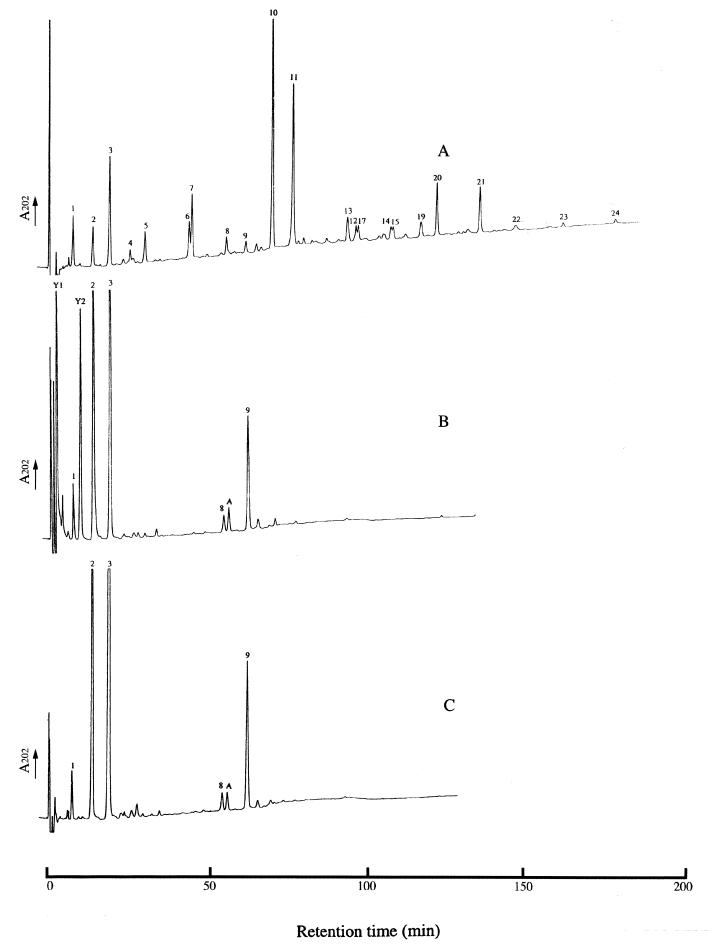
Since the pdaA- and cwlD-deficient spores have similar phenotypes and the cwlD-deficient spores completely lack muramic acid δ-lactam (4, 18), RP-HPLC of the spore cortex muropeptides from the B. subtilis wild-type strain, 168HR, and its congenic yfjS mutant, HR(YFJS), was performed (Fig. 4). There was a remarkable difference in peptidoglycan structure between the mutant and the wild-type strain. The HR(YFJS) peptidoglycan lacked all of the muropeptides containing δ-lactam (muropeptides 4 to 7 and 10 to 24), which represent about 70% of the total components in the wild-type strain (Fig. 4) (4). There are increases in muropeptides 2 and 3, which are, respectively, disaccharide alanine and disaccharide tetrapeptide (4). Some novel muropeptides (Y1, Y2, and A) were also detected in HR(YFJS). Amino acid analysis of muropeptides Y1 and Y2 revealed that they are peptidoglycan derived and contain only N-acetylglucosamine (GlcNAc) and N-acetyl muramic acid (MurNAc), the two amino sugars of glycan strands. MS analysis of muropeptide Y1 in the positive and negative modes gave, respectively, fragment ions at m/z 521.2 and 497.2. These values correspond, respectively, to [M+Na]+ and [M−H]− molecular ions. The Y2 MS value is nearly double that of Y1, and the fragment ions of positive [M+Na]+ and negative [M−H]− were observed, respectively, at m/z 999.4 and 975.2. Based on the amino acid composition and MS results, muropeptides Y1 and Y2 are, respectively, a disaccharide (GlcNAc-MurNAc) and a tetrasaccharide (GlcNAc-MurNAc-GlcNAc-MurNAc). The MurNAc residues at the reducing ends of the disaccharide (Y1) and the tetrasaccharide (Y2) and position 2 of the tetrasaccharide are unsubstituted. Digestion of the YFJSd cortex with a high concentration of lysozyme (2 mg/ml) resulted in a substantial decrease in muropeptide Y2 and an increase in muropeptide Y1 (data not shown). This demonstrates that the MurNAc-GlcNAc bond in muropeptide Y2 is susceptible to digestion, but only with a high concentration of hydrolase. Muropeptide A in Fig. 4 was coeluted with muropeptide A, previously detected for a cwlD mutant (4). Amino acid analysis and MS confirmed that this muropeptide is a disaccharide tripeptide disaccharide tetrapeptide and that it is identical to that found in the peptidoglycan of the cwlD mutant (4). This muropeptide is possibly amidated at the free carboxylic group of diaminopimelic acid.
FIG. 4.
Analysis of the peptidoglycan composition by RP-HPLC. Samples of peptidoglycan from endospores of wild-type B. subtilis strain 168HR (A), a pdaA mutant HR(YFJS) (B), and a pdaA cwlD mutant HR(YFJS CWLD) (C) were digested with Cellosyl, and the resulting muropeptides were separated by RP-HPLC. The numbered muropeptides were previously identified (4).
The RP-HPLC spore cortex muropeptide profile of the pdaA cwlD mutant, HR(YFJS CWLD), is shown in Fig. 4C. The peaks of Y1 and Y2 are absent from the muropeptide profile of HR(YFJS CWLD), and the profile is identical to that of single mutant cwlD (4). CwlD was previously suggested to be the amidase that cleaves off muropeptide side chains and is involved in δ-lactam biosynthesis (2, 4, 18, 21).
Complementation with pdaA-containing plasmids.
pHYCMfjS and pHYCMΔSfjS, containing the entire pdaA gene and the signal sequence-deleted pdaA gene, respectively, were used to transform the pdaA-deficient mutant YFJSd. In the case of sporulation, the frequencies of YFJSd harboring pHYCMfjS and pHYCMΔSfjS were not significantly different from those of YFJSd and strain 168 harboring a control plasmid, pHYCM1 (Table 2). But in the case of germination, YFJSd(pHYCM1) almost completely lost the ability to germinate (germination ratio of 0.0008% on LB agar plates), whereas introduction of pHYCMfjS into the YFJSd strain complemented the germination-negative phenotype at nearly the wild-type level (Table 2). However, YFJSd(pHYCMΔSfjS) only partially complemented the germination-negative phenotype (0.019% germination). In this system, the pdaA gene was expressed with Mg-citrate probably both in the forespore and the mother cell. The results indicate that the signal sequence of pdaA is important for phenotype expression and they may suggest that the enzymatic reaction site is the cortex surrounded by two membranes.
TABLE 2.
Complementation of pdaA with a plasmid containing the entire pdaA gene or the putative signal peptide-deficient pdaA genea
| Strain | Plate assay | Microscopic observation
|
Germination ratio (%) | ||
|---|---|---|---|---|---|
| No. of outgrowing spores | Spores | Vegetative cells | Sporulation ratio (%) | ||
| YFJSd (pHYCMfjS) | 5.57 × 107 | 2.85 × 108 | 10.2 × 107 | 73.6 | 19.5 |
| YFJSd (pHYCMΔSfjS) | 9.8 × 104 | 5.15 × 108 | 6.72 × 107 | 88.5 | 0.019 |
| YFJSd (pHYCM1) | 2.0 × 103 | 2.53 × 108 | 11.5 × 107 | 68.8 | 0.0008 |
| 168 (pHYCM1) | 7.15 × 107 | 2.94 × 108 | 6.40 × 107 | 82.1 | 24.3 |
YFJSd is a pdaA mutant. pHYCMfjS and pHYCMΔSfjS contained the entire pdaA gene and the putative signal peptide-deficient pdaA gene, respectively, downstream of the citM promoter region. pHYCM1 is a control plasmid (lacking the pdaA gene). Expression of the genes was carried out by the addition of 2 mM Mg-citrate at t0.5, and spores were harvested at t48. The plate assay was performed as described in Materials and Methods. The representative results for two independent clones from each strain are shown.
Localization of PdaA-GFP.
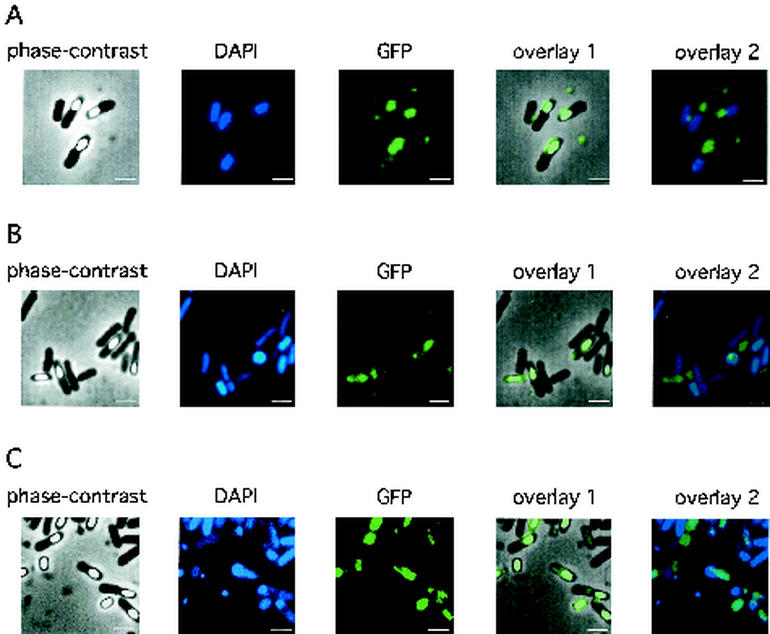
A GFP-encoding gene was fused to the 3′ terminal of pdaA, and the resultant plasmid (pHYPRfjSG) was introduced into the wild-type 168 strain. Then, 168(pHYPRfjSG) was cultured in DSM at 37°C and samples were taken at t20. Cells were stained with a DNA-staining fluorescent dye, DAPI, and then the fluorescence of GFP and DAPI was observed under a phase-contrast microscope equipped with a fluorescence apparatus. Strong fluorescence of GFP was found in sporangia at t7 (data not shown) and spores at t20 (Fig. 5A), but not in vegetative cells or mother cells (Fig. 5A; data not shown). pHYPRfjSG introduced into YFJSd also complemented the yfjS-deficient phenotype (plate assay was used; data not shown). Localization of GFP in YFJSd(pHYPRfjSG) was also found in sporangia and spores (Fig. 5B and C). These results indicate that the PdaA protein was localized in spores.
FIG. 5.
Localization of the PdaA-GFP fusion protein in the wild-type strain (A) or pdaA-deficient mutant YFJSd (B and C). The DAPI or GFP fluorescence of cells at t20 was detected as described in Materials and Methods. The pdaA-gfp-containing plasmid (pHYPRfjSG) was used for transformation of B. subtilis 168 or YFJSd. Spores and sporangia were collected at t20 at 37 and 30°C in DSM medium (B and C, respectively). The culture at 30°C exhibited stronger fluorescence in spores than that at 37°C. Overlay 1 is a phase-contrast image overlayed on a GFP image, and overlay 2 is a DAPI-stained fluorescence image overlayed on a GFP image. Bars, 2 μm.
DISCUSSION
The amino acid sequence of PdaA exhibits extensive similarity with those of polysaccharide deacetylases, including NodB chitooligosaccharide deacetylases, chitin deacetylases, and peptidoglycan N-acetylglucosamine deacetylase (8, 11, 14, 28). As to cellular function, these proteins act as bacterial nodulation signals in symbiosis (8), and they are involved in mature spore formation in yeast (14) and modification of peptidoglycan causing resistance to the hydrolytic action of lysozyme (28). Figure 6 shows the alignment of pdaA homologous genes. In Bacillus stearothermophilus, yfu2 is highly homologous with pdaA (60% identity over 256 amino acid residues). In B. subtilis, five paralogs (pdaA [yfjS], ybaN, yheN, yjeA, and ylxY) are present, three of which are highly homologous and also aligned in Fig. 6. In the genera Bacillus and Clostridium, none of the paralogs except pdaA dealt with in this paper were functionally characterized. The pdaA-deficient mutant showed the almost complete loss of germination, whereas the spores were fully refractive. This and other profiles, including dipicolinic acid release and the complete loss of hexosamine release from spores, led to the idea that the mutation may be associated with cortex maturation, especially muramic δ-lactam formation, as the previously characterized cwlD mutant completely lacks muramic δ-lactam (4, 18, 21).
FIG. 6.
Alignment of PdaA and homologous gene products. The polysaccharide deacetylase family members, including PdaA, are aligned with a protein families database of alignments and hidden Markov models (Pfam, Sanger Institute). A seed sequence is shown as the first line. The name and accession number in the SWISS-PROT 40 database are shown at the left and right, respectively, of the alignment. The positions of the first and last amino acid residues of the sequence with respect to the N-terminal amino acid residue are shown after the name. Shading indicates amino acid residues identical to the highly conserved amino acid residues (capital letters) of the seed sequence. NodB, nodulation protein B (chitooligosaccharide deacetylase); NODB_RHIME, NodB of Rhizobium meliloti; NODB_BRAEL, Bradyrhizobium elkanii; NODB_RHISN, Rhizobium sp. strain NGR234; NODB_RHITR, Rhizobium tropici; NODB_RHILV, Rhizobium leguminosarum bv. viciae; NODB_RHILT, R. leguminosarum bv. trifolii; NODB_RHIGA, Rhizobium galegae; NODB_RHILP, R. leguminosarum bv. phaseoli; NODB_AZOCA, Azorhizobium caulinodans; YFU2_BACST, B. stearothermophilus YFU2; YFJS_BACSU, B. subtilis PdaA; CDA1_YEAST, Saccharomyces cerevisiae chitin deacetylase 1 (CDA1); CDA2_YEAST, S. cerevisiae chitin deacetylase 2 (CDA2); CHDE_MUCRO, Mucor rouxii chitin deacetylase (CHDE); XYND_CELFI, Cellulomonas fimi xylanase D (XYND); AXEA_STRLI, Streptomyces lividans acetylxylan esterase (AXEA); Q59300_CELMI, Cellvibrio mixtus endo-β-1,4-xylanase; Q59674_PSUFL, Pseudomonas fluorescens endo-β-1,4-xylanase; O53444_MYCTU, Mycobacterium tuberculosis hypothetical protein; YJEA_BACSU, B. subtilis YjeA; XYNU_CLOTH, Clostridium thermocellum xylanase U (XynU); XYNC_CELFI, C. fimi endoxylanase (XynC); YHEN_BACSU, B. subtilis YheN; XYNA_RUMAL, Ruminococcus albus xylanase A (XynA). Two of the PdaA paralogs (YlxY and YbaN) are not aligned because of a lower sequence similarity.
A remarkable feature of the HR(YFJS) muropeptide profile is the total lack of products containing δ-lactam (Fig. 4). This peptidoglycan composition is similar to that of cwlD; however, two extra products are present in HR(YFJS). The new muropeptides have unsubstituted muramic acid residues that suggest that an amidase was cleaved off the side chains. YfjS is homologous to peptidoglycan de-N-acetylase, and thus it is likely to cleave the N-acetyl groups of muramic acid as an intermediate step of δ-lactam biosynthesis. Two mechanisms have been previously proposed for δ-lactam biosynthesis: an amidase action followed by transacetylation, and de-N-acetylation followed by transpeptidation (27). In light of the present results, the first step of δ-lactam formation appears to be the cleavage of muropeptide side chains, most likely that of tetrapeptides by CwlD, followed by de-N-acetylation of muramic acid residues by YfjS. The last step in δ-lactam formation would be via transpeptidase activity that gives the final structure. The fact that double mutant cwlD pdaA exhibits a muropeptide profile identical to that of the single mutant cwlD confirms that the first step of δ-lactam formation is the cleavage of muropeptide side chains by CwlD. The δ-lactam was recently shown to serve as a substrate recognition signal for the different GSLEs involved in cortex hydrolysis during germination (2, 5, 18). Spores lacking this cortex-specific moiety neither hydrolyze peptidoglycan nor outgrow (2, 5, 18). This correlates with the phenotype of the pdaA mutant spores.
The pdaA gene is transcribed by EσG RNA polymerase as a monocistronic mRNA (Fig. 1). Upstream of the pdaA gene are the sequences G−70CATA−66 and C−49AAAGTC−43 (the numbering is with respect to the translational start codon), with a spacing of 16, similar to the consensus sequences of −35 and −10 (GNATR and CATNNTA, respectively; R stands for A or G, and N stands for nonspecific nucleotide) of σG with a spacing of 15 (9). The SD sequence (AAGGAG) is located 14 to 9 bp upstream of the translational start codon. Coupled with the GFP fusion data, this shows that pdaA is a forespore-expressed gene. The product is exported into the developing cortex, where it acts with CwlD to produce muramic δ-lactam residues. How the final structure is attained is unknown and may require the action of an independent transpeptidase.
This is the first report that a polysaccharide deacetylase homologue is associated with Bacillus sporulation and germination. The other polysaccharide deacetylase homologues are now being investigated as to whether they are associated with differentiation or vegetative growth.
Acknowledgments
This research was supported by Grants-in-Aid for Scientific Research on Priority Areas Genome Biology (C) (12206005) and Scientific Research (B) (13460037) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (J.S.), the Biotechnology and Biological Sciences Research Council (A.A.), and the Royal Society (S.J.F.).
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atrih, A., and S. J. Foster. 1999. The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination. Antonie Leeuwenhoek 75:299-307. [DOI] [PubMed] [Google Scholar]
- 3.Atrih, A., and S. J. Foster. 2001. In vivo roles of germination-specific lytic enzymes of Bacillus subtilis 168. Microbiology 147:2925-2932. [DOI] [PubMed] [Google Scholar]
- 4.Atrih, A., P. Zöllner, G. Allmaier, and S. J. Foster. 1996. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J. Bacteriol. 178:6173-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atrih, A., P. Zöllner, G. Allmaier, M. P. Williamson, and S. J. Foster. 1998. Peptidoglycan structural dynamics during germination of Bacillus subtilis 168 endospores. J. Bacteriol. 180:4603-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canosi, U., G. Morelli, and T. A. Trautner. 1978. The relationship between molecular structure and transformation efficiency of some Streptococcus aureus plasmids isolated from Bacillus subtilis. Mol. Gen. Genet. 166:259-267. [DOI] [PubMed] [Google Scholar]
- 7.Foster, S. J., and D. L. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 8.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 9.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 10.Ishikawa, S., K. Yamane, and J. Sekiguchi. 1998. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J. Bacteriol. 180:1375-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kafetzopoulos, D., G. Thireos, J. N. Vournakis, and V. Bouriotis. 1993. The primary structure of a fungal chitin deacetylase reveals the function for two bacterial gene products. Proc. Natl. Acad. Sci. USA 90:8005-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 13.Makino, S., N. Ito, T. Inoue, S. Miyata, and R. Moriyama. 1994. A spore-lytic enzyme released from Bacillus cereus spores during germination. Microbiology 140:1403-1410. [DOI] [PubMed] [Google Scholar]
- 14.Mishra, C., C. E. Semino, K. J. McCreath, H. de la Vega, B. J. Jones, C. A. Specht, and P. W. Robbins. 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13:327-336. [DOI] [PubMed] [Google Scholar]
- 15.Moriyama, R., A. Hattori, S. Miyata, S. Kudoh, and S. Makino. 1996. A gene (sleB) encoding a spore cortex-lytic enzyme from Bacillus subtilis and response of the enzyme to l-alanine-mediated germination. J. Bacteriol. 178:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kindgom.
- 17.Paidhungat, M., and P. Setlow. 2002. Spore germination and outgrowth, p. 537-548. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 18.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc. Natl. Acad. Sci. USA 93:15405-15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekiguchi, J., K. Akeo, H. Yamamoto, F. K. Khasanov, J. C. Alonso, and A. Kuroda. 1995. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J. Bacteriol. 177:5582-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekiguchi, J., T. Fukushima, and S. Ishikawa. 2001. Selection and characterization of genes affecting germination, p. 211-213. In W. Shumann, S. D. Ehrlich, and N. Ogasawara (ed.), Functional analysis of bacterial genes: a practical manual. John Wiley & Sons, Chichester, United Kingdom.
- 23.Shimotsu, H., and D. J. Henner. 1986. Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes. J. Bacteriol. 168:380-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 25.Sonenshein, A. L. 2000. Endospore-forming bacteria: an overview, p. 133-150. In Y. V. Braun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 26.Stewart, G. S., K. Johnstone, E. Hagelberg, and D. J. Ellar. 1981. Commitment of bacterial spores to germinate. Biochem J. 198:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tipper, D. J., and J. J. Gauthier. 1972. Structure of the bacterial endospore, p. 3-12. In H. O. Halvorson, R. Hanson, and L. L. Campbell (ed.), Spores V. American Society for Microbiology, Washington, D.C.
- 28.Vollmer, W., and A. Tomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto, H., M. Mori, and J. Sekiguchi. 1999. Transcription of genes near the sspE locus of the Bacillus subtilis genome. Microbiology 145:2171-2180. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, H., M. Murata, and J. Sekiguchi. 2000. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 37:898-912. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto, H., S. Uchiyama, and J. Sekiguchi. 1996. Cloning and sequencing of a 40.6 kb segment in the 73°-76° region of the Bacillus subtilis chromosome containing genes for trehalose metabolism and acetoin utilization. Microbiology 142:3057-3065. [DOI] [PubMed] [Google Scholar]