Abstract
The FNR protein of Escherichia coli controls the transcription of target genes in response to anoxia via the assembly-disassembly of oxygen-labile iron-sulfur clusters. Previous work identified patches of surface-exposed amino acids (designated activating regions 1 and 3 [AR1 and AR3, respectively]) of FNR which allow it to communicate with RNA polymerase (RNAP) and thereby activate transcription. Previously it was thought that FNR lacks a functional activating region 2 (AR2), although selecting for mutations that compensate for defective AR1 or a miscoordinated iron-sulfur cluster can reactivate AR2. Here we show that the substitution of two surface-exposed lysine residues (Lys49 and Lys50) of FNR impaired transcription from class II (FNR box centered at −41.5) but not class I (FNR box centered at −71.5) FNR-dependent promoters. The degree of impairment was greater when a negatively charged residue (Glu) replaced either Lys49 or Lys50 than when uncharged amino acid Ala was substituted. Oriented heterodimers were used to show that only the downstream subunit of the FNR dimer was affected by the Lys→Ala substitutions at a class II promoter. Site-directed mutagenesis of a negatively charged patch (162EEDE165) within the N-terminal domain of the RNAP α subunit that interacts with the positively charged AR2 of the cyclic AMP receptor protein suggested that Lys49 and Lys50 of FNR interact with this region of the α subunit of RNAP. Thus, it was suggested that Lys49 and Lys50 form part of a functional AR2 in FNR.
The Escherichia coli FNR protein is a global transcription factor that controls the expression of target genes in response to oxygen starvation. It is structurally related to the cyclic AMP receptor protein (CRP), except for the presence of an N-terminal extension that contains three of the four cysteine residues that are essential for normal FNR function: C20, C23, C29, and C122 (17, 26). The essential cysteine residues are presumed to act as ligands for an oxygen-labile [4Fe-4S] cluster (6, 7, 10). Anaerobic acquisition of the iron-sulfur cluster converts monomeric FNR into a dimeric form containing two [4Fe-4S] clusters (10). This transition to the dimeric state enhances site-specific DNA binding and mediates transcription regulation by establishing direct FNR-RNA polymerase (RNAP) contacts (1, 2, 4, 5, 9, 13, 15, 29).
The activating contacts between FNR and RNAP involve two surface-exposed regions of FNR designated activating region 1 (AR1), which contacts the C-terminal domain of the α subunit of RNAP (αCTD) (13, 29), and activating region 3 (AR3), which contacts σ70 (9, 15) (Fig. 1). The contacts established between FNR and RNAP depend upon the architecture of particular promoters (Fig. 2). The AR1 contact is required for transcription activation at class I promoters (where the FNR box is located close to position −61 or further upstream), where contact is established between AR1 of the downstream subunit of the FNR dimer and αCTD (32). The AR1 contact is also used at class II promoters (where the FNR box is located at −41); however, in this situation it is the upstream subunit of the FNR dimer that makes contact with αCTD to promote transcription (8, 31). In addition to the AR1 contact, FNR has a second interaction with RNAP at class II promoters (32). In this situation, AR3 of the downstream subunit of the FNR dimer contacts a small region of the RNAP σ70 subunit (15). In vivo studies have indicated that the AR3-σ70 interaction makes the greatest contribution to transcription activation from class II promoters (1, 32).
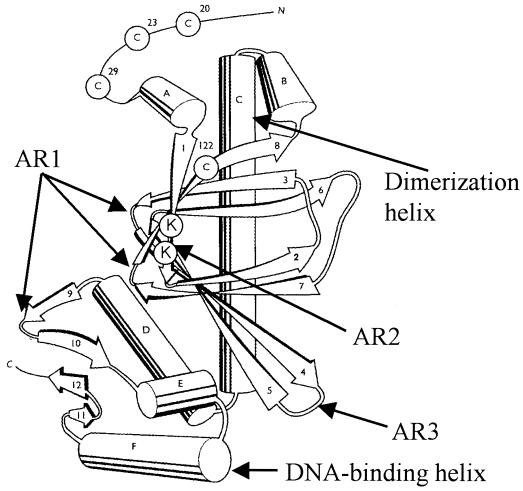
FIG. 1.
Locations of activating regions in FNR. The predicted structure of an FNR monomer based on CRP (25) shows the locations of the previously defined activating regions: AR1, responsible for contacting αCTD, and AR3, responsible for contacting σ70. The locations of lysine residues (K49 and K50) that form part of the newly identified AR2 are indicated. Also shown are the N and C termini, α helices (cylinders A to F), β strands (arrows 1 to 12), and essential cysteine residues (C20, C23, C29, and C122) that act as ligands for the oxygen-labile [4Fe-4S] cluster.
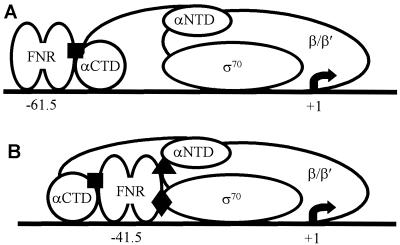
FIG. 2.
Organization of simple class I and class II FNR-dependent promoters. (A) At class I promoters, FNR binds to a site centered at −61.5 or further upstream. AR1 of the downstream subunit of the FNR dimer contacts αCTD (▪). (B) At class II promoters, FNR binds to a site centered at or near −41.5 and is thus embedded within RNAP; multiple interactions are possible. Two FNR-RNAP contacts were previously characterized at class II promoters. The AR1 surface of the upstream subunit of the FNR dimer contacts αCTD (▪), and the AR3 surface of the downstream subunit of FNR contacts σ70 (⧫). Also indicated is the contact between AR2 and αNTD (▴), which is used by CRP and now shown to be present in FNR.
CRP has evolved a different solution to the problem of activating transcription from class II promoters. In place of the AR3 contact, CRP has a distinct interaction between activating region 2 (AR2), a positively charged patch consisting of amino acids His19, His21, and Lys101, and a negatively charged region of the N-terminal domain of the RNAP α subunit (αNTD), consisting of Glu162, Glu163, Asp164, and Glu165 (3, 19, 22). In FNR, AR2 was considered to be inactive, but it has been shown that it can be reactivated by mutations that compensate for lesions in rpoA (14) and by mutations that restore activity to FNR variants with impaired AR1 (13) or a miscoordinated iron-sulfur cluster (21). Based upon the locations of the second-site substitutions identified in the latter study, it was suggested that two Lys residues (Lys49 and Lys50) may form AR2 of FNR (21). In this report, evidence is presented in support of this hypothesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, Western blotting, and β-galactosidase assays.
The bacterial strains and plasmids used in this study are listed in Table 1. Cultures of E. coli were grown in L broth (12) supplemented with appropriate antibiotics (ampicillin, 100 μg ml−1; chloramphenicol, 20 μg ml−1; tetracycline, 35 μg ml−1; and kanamycin, 25 μg ml−1). For estimating promoter activities in vivo, glucose (0.2% [wt/vol]) was included for anaerobic growth in sealed Bijou bottles. Cultures were grown at 37°C for 16 h before β-galactosidase activities were measured as described by Miller (18). To test whether the FNR variants used were present at similar levels, equivalent amounts of soluble protein were analyzed by Western blotting with anti-FNR serum, with detection by use of an anti-rabbit immunoglobulin G-alkaline phosphatase conjugate. Two independent cultures for each FNR variant were used, and the relative amount of FNR in each culture was determined by analysis of the blots with ImageMaster software (Amersham). Standard procedures were used for DNA isolation and manipulation (24). Site-directed mutagenesis was achieved by PCR with appropriate mutagenic primers. Automated DNA sequencing was used to confirm the authenticity of all the FNR and RpoA variants produced.
TABLE 1.
Bacterial strains, plasmids, and promoters
| Strain, plasmid or promoter | Relevant propertiesa | Reference or source |
|---|---|---|
| JRG1728 | Δfnr Δlac | 27 |
| pBR322 | General cloning vector; Apr Tetr | 24 |
| ptac85 | IPTG-inducible expression vector | 16 |
| pGS1121 | Like ptac85 but with a kan gene block cloned into the PstI site of the bla gene | N. Wyborn, University of Sheffield |
| pGS196 | pBR322 with a 1.6-kb fnr insert | 28 |
| pGS1566 | Like pGS196 but directs a K49E substitution | This work |
| pGS1567 | Like pGS196 but directs a K49A substitution | This work |
| pGS1568 | Like pGS196 but directs a K50E substitution | This work |
| pGS1569 | Like pGS196 but directs a K50A substitution | This work |
| pGS1578 | Like pGS196 but directs K49A/S73F substitutions | This work |
| pGS1579 | Like pGS196 but directs K50A/S73F substitutions | This work |
| pGS1580 | Like pGS196 but directs K49A/G85A substitutions | This work |
| pGS1581 | Like pGS196 but directs K50A/G85A substitutions | This work |
| pGS1425 (E165) | pGS1121-derived rpoA expression plasmid | This work |
| pGS1611 (A165) | Like pGS1425 but directs E162A/E163A/D164A/E165A substitutions | This work |
| pGS1612 (K165) | Like pGS1425 but directs E162A/E163A/D164A/E165K substitutions | This work |
| pGS1630 | Like pGS196 but directs K49A/K50A substitutions | This work |
| FF(−41.5) | FF(−41.5) melR::lac operon fusion in a low copy number; ColE1-compatible, broad-host-range vector pRW50 with a consensus FNR site centered at −41.5; Tetr | 32 |
| FF(−71.5) | Equivalent to FF(−41.5) except that the consensus FNR site is centered at −71.5 and the promoter has an improved −35 element; Tetr | 32 |
| YY(−41.5), YF(−41.5), FY(−41.5) | Equivalent to FF(−41.5) except that the consensus FNR site contains mutations such that one or both TTGAT (F) core motifs are altered to TTAAT (Y) to allow recognition by the FNR variant FNR-E209K | 1 |
| FF galΔ4 | Simple FNR-repressed lac reporter plasmid | 30 |
IPTG, isopropyl-β-d-thiogalactopyranoside.
RESULTS
Substitution of Lys49 and Lys50 impairs transcription from a class II promoter.
The starting point for this work was the suggestion that Lys49 and Lys50 may constitute the AR2 surface of FNR (21). If this hypothesis is correct, then substitution of either Lys49 or Lys50 by neutral (Ala) or negatively charged (Glu) amino acids should impair transcription activation from a class II but not a class I promoter. This is because any AR2 contact made between FNR and RNAP would be formed only at a class II promoter (Fig. 2). Therefore, PCR-based site-directed mutagenesis was used to generate five FNR variants: FNR-K49A, FNR-K49E, FNR-K50A, FNR-K50E, and FNR-K49A/K50A. The ability of each of these variants to activate transcription from model class I, FF(−71.5), and class II, FF(−41.5), promoters was assessed by measuring the β-galactosidase activities of anaerobically grown cultures of JRG1728 (Δfnr Δlac) carrying plasmids expressing either wild-type fnr (pGS196) or genes for FNR-K49A (pGS1567), FNR-K49E (pGS1566), FNR-K50A (pGS1569), FNR-K50E (pGS1568), or FNR-K49A/K50A (pGS1630) and compatible low-copy-number plasmids carrying lacZ under the control of the FF(−71.5) or FF(−41.5) promoter (32).
The data obtained indicated that, as previously reported, both promoters were dependent on FNR for activity, with 37-fold enhancement at the class I promoter and 60-fold enhancement at the class II promoter when the activities obtained in the presence and absence of wild-type FNR were compared (Table 2). Replacing Lys49 of FNR with Ala reduced transcription by 32% at FF(−41.5) but only by 7% at FF(−71.5). Transcription from the class II promoter was further reduced to only 50% the wild-type level by the introduction of a Glu residue at position 49, while 96% of the wild-type activity was retained at the class I promoter (Table 2). Similarly, replacing Lys50 with Ala reduced transcription by 22% from the class II promoter but did not significantly change transcription from the class I promoter; the equivalent Glu-substituted protein retained only 69% of the wild-type FNR activity at the class II promoter, but activity at the class I promoter was relatively unaffected, at 84% of the wild-type FNR activity (Table 2). Replacing both Lys49 and Lys50 with Ala had a greater effect on transcription from the class II promoter (52% of the wild-type activity) than either of the individual Ala substitutions and caused a modest (10%) increase in expression from the class I promoter (Table 2).
TABLE 2.
Effect of predicted FNR AR2 variants on in vivo expression from simple model promotersa
| FNR variant | β-Galactosidase activity (Miller units) in the presence of:
|
Relative level of expression (%) | ||
|---|---|---|---|---|
| FF(−71.5) (class I) | FF(−41.5) (class II) | FF galΔ4 | ||
| None | 102 ± 8 | 88 ± 5 | 672 ± 14 | 0 |
| FNR | 3,809 ± 350 | 5,272 ± 209 | 38 ± 2 | 100 |
| FNR-K49A | 3,564 ± 113 | 3,599 ± 226 | 42 ± 5 | 93 |
| FNR-K49E | 3,640 ± 121 | 2,616 ± 116 | 11 ± 2 | 93 |
| FNR-K50A | 3,852 ± 203 | 4,122 ± 37 | 35 ± 1 | 108 |
| FNR-K50E | 3,195 ± 197 | 3,626 ± 437 | 47 ± 12 | 97 |
| FNR-K49A/K50A | 4,249 ± 99 | 2,770 ± 249 | 17 ± 1 | 122 |
β-Galactosidase activity was measured in at least three independent cultures of strains carrying plasmid pBR322-encoded FNR or the indicated FNR variants. All cultures were grown under anaerobic conditions in L broth supplemented with glucose (0.2%) at 37°C for 16 h. The reporter plasmids used were the simple FNR-activated class I, FF(−71.5), and class II, FF(−41.5), constructs (32) and the simple FNR-repressed FF galΔ4 construct (30). Values are means ± standard deviations. The relative levels of expression of FNR and FNR variants were determined by Western blotting. Measurements were obtained from two blots from independent anaerobic cultures. The values reported are the means of the two determinations, with the wild-type FNR value set at 100%.
To ensure that the effects on transcription activation described above were not due to defects in the ability to recognize and bind to FNR target DNA, an FF galΔ4::lacZ promoter fusion (30) was used to assess the ability of the FNR variants to repress transcription by simply occluding the promoter. Any DNA-binding defect in the variants would be exposed by increased transcription from the FF galΔ4 promoter compared to that in cultures containing unaltered FNR. The data showed that all the variants created were effective repressors of transcription from FF galΔ4 (Table 2). Thus, it was concluded that they were all unaffected in their ability to bind DNA. Western blotting was used to determine whether the amino acid substitutions affected the levels of FNR protein in the cultures. The blots revealed that all the variants of FNR were present at levels similar to that of the wild-type protein (Table 2). Therefore, the simplest explanation for the differences in the abilities of the Lys-substituted variants to activate transcription is that contact with RNAP is impaired. If this interpretation is correct, then like the situations for the previously characterized AR1 and AR3, more than one surface-exposed amino acid (at least Lys49 and Lys50) contribute to the AR2 contact of FNR.
Substitution of Lys49 or Lys50 affects the function of the downstream subunit of the FNR dimer.
Active, dimeric FNR binds DNA in a site-specific manner, targeting a 14-bp imperfect palindrome, TTGATNNNNATCAA (FF; F = TTGAT). Substitution of the key DNA-binding residue, Glu209, with Val, to generate FNR-E209V, permits the binding of FNR to TTAATNNNNATTAA (YY; Y = TTAAT) (1). This altered FNR-binding site was previously engineered into the class II FNR-dependent FF(−41.5) promoter, creating the FNR-E209V-activated YY(−41.5) promoter. The wild-type half-site (F) can also be placed alongside the mutant FNR half-site (Y) to create a promoter with either FY or YF regulatory sites. Such sites confer specificity for FNR-FNR-E209V heterodimers, and the orientation of the dimer relative to RNAP is determined by the orientation of the DNA-binding half-sites in the hybrid promoter. This system can be adapted to observe the consequences of combining one FNR-K49A or FNR-K50A subunit with one FNR-E209V subunit. This can be achieved by coexpressing FNR-E209V, which recognizes the Y half-site but retains both Lys49 and Lys50, with either FNR-K49A or FNR-K50A, which recognizes the F half-site. In this way, the effects of the Lys substitutions on the individual subunits of the FNR dimer can be determined. The plasmids expressing the FNR proteins are selected such that less FNR-E209V is present in the bacteria than FNR-K49A or FNR-K50A. Thus, most FNR-E209V in the bacteria should be present as heterodimers with the Ala-substituted variants. Therefore, JRG1728 (Δfnr Δlac) was transformed with a pLG339 derivative encoding FNR-E209V and with reporter plasmids containing the YY(−41.5), FY(−41.5), or YF(−41.5) promoter. The reporter strains were then transformed with pGS196, encoding FNR, pGS1567, encoding FNR-K49A, or pGS1569, encoding FNR-K50A. To test whether both FNR-K49A and FNR-K50A could form heterodimers with FNR-E209V as efficiently as wild-type FNR, anaerobic transcription from the YY promoter was measured in vivo.
The data showed that FNR-E209V alone yielded 4,050 ± 386 Miller units (mean ± standard deviation) from the YY promoter. The addition of wild-type FNR reduced this value to 446 ± 6 Miller units. The reduction is explained by the formation of heterodimers, which reduce the pool of FNR-E209V homodimers and thereby reduce occupancy of the YY promoter. The response of the YY promoter to the addition of FNR-K50A was similar to that obtained with FNR: a reduction in activity to 367 ± 23 Miller units, indicating that the introduction of the K50A substitution did not impair heterodimer formation. However, the data obtained upon the introduction of FNR-K49A indicated that heterodimers were formed, but not so readily (YY activity, 1,427 ± 15 Miller units). Thus, since both FNR variants formed heterodimers, if Lys49 and Lys50 are involved in an AR2-like contact, then the production of β-galactosidase should be greater when FNR-E209V (which recognizes the Y half-site and retains both Lys49 and Lys50) is the downstream subunit of the FNR dimer, because only this subunit can form the AR2-αNTD contact. Accordingly, the anaerobic activities of the YF and FY promoters revealed that when the downstream subunit contained either FNR-K49A or FNR-K50A, the relative level of transcription was reduced compared to the level obtained when this subunit was in the upstream position (Table 3). Moreover, FNR-K50A-containing heterodimers responded as expected in absolute terms (reduced activity at YF; the same activity at FY) compared to FNR-containing heterodimers. This result is consistent with the observation that FNR and FNR-K50A form heterodimers with FNR-E209V with similar efficiencies. For the apparently less readily formed FNR-E209V-FNR-K49A heterodimers, the relative activities of the YF and FY promoters were again consistent with AR2 functioning in the downstream subunit; however, in this instance, activities were manifested as similar activity at YF and enhanced activity at FY compared to the results obtained with FNR-containing heterodimers. Thus, these observations support the proposal that Lys49 and Lys50 of the downstream subunit of the FNR dimer form an AR2-like contact at class II promoters.
TABLE 3.
Effect of the location of AR2 relative to RNAP on transcription activation by oriented FNR heterodimersa
| Heterodimer | Promoter activity (Miller units)
|
Relative activity (YF/FY) | |
|---|---|---|---|
| YF | FY | ||
| E209V (Y)-FNR (F) | 2,014 ± 85 | 2,281 ± 135 | 0.88 |
| E209V (Y)-K49A (F) | 1,867 ± 133 | 3,302 ± 250 | 0.57 |
| E209V (Y)-K50A (F) | 1,382 ± 101 | 2,271 ± 191 | 0.61 |
Promoter activities were measured by estimating β-galactosidase activities (Miller units) in at least three independent cultures of strains carrying plasmid pLG339-encoded FNR-E209V, which has native AR2 but altered DNA-binding specificity so that it recognizes the TTAAT (Y) half-site, and plasmid pBR322-encoded FNR, FNR-K49A, or FNR-K50A, which recognizes the TTGAT (F) half-site but carries the indicated amino acid substitution in the putative AR2. All cultures were grown under anaerobic conditions in L broth supplemented with glucose (0.2%) at 37°C for 16 h. The reporter plasmids used have been described previously (1). Values are means ± standard deviations.
Contribution of AR2 to transcription activation.
Having established that FNR possesses a functional AR2, the next step was to investigate its contribution to transcription activation at class II promoters. Therefore, FNR variants were created that combined amino acid substitutions that impair AR2 (Lys49→Ala or Lys50→Ala) with substitutions that impair AR1 (Ser73→Phe) or AR3 (Gly85→Ala). These variants with impaired AR1 and AR2 or AR3 and AR2 were tested for transcriptional activity at class I and II promoters, as were the corresponding variants with impaired AR1, AR2, or AR3 alone.
The data (Table 4) confirm the previous finding that the AR1 contact is operative at both classes of FNR-activated promoters (1), because the Ser73→Phe (AR1) substitution caused a 78% reduction in transcription from the class I promoter and a 61% reduction from the class II promoter. Combining an AR2-inactivating substitution (Lys49→Ala or Lys50→Ala) with the AR1-inactivating substitution caused a small reduction in transcription activation at the class I promoter. The data obtained for the AR3 (Gly85→Ala) variant also confirmed previous reports, showing a 1.6-fold increase in activity compared to that obtained with the unaltered protein at the class I promoter (20, 32). Combining the AR3- and AR2-inactivating substitutions reduced transcription from the class I promoter by 17.5% compared to that seen with the AR3 mutant, but the AR2-AR3 double mutant was still more effective than wild-type FNR (Table 4).
TABLE 4.
Effect of combinations of activating region mutations on in vivo expression from simple model promotersa
| FNR variant | β-Galactosidase activity (Miller units) in the presence of:
|
||
|---|---|---|---|
| FF(−71.5) (class I) | FF(−41.5) (class II) | FF galΔ4 | |
| FNR | 3,809 ± 350 | 5,272 ± 209 | 38 ± 2 |
| FNR-K49A | 3,564 ± 113 | 3,599 ± 226 | 42 ± 5 |
| FNR-K50A | 3,852 ± 203 | 4,122 ± 37 | 35 ± 1 |
| FNR-S73F | 838 ± 183 | 2,040 ± 140 | 17 ± 12 |
| FNR-S73F/K49A | 687 ± 6 | 1,031 ± 64 | 26 ± 4 |
| FNR-S73F/K50A | 626 ± 25 | 816 ± 59 | 87 ± 41 |
| FNR-G85A | 6,047 ± 115 | 1,081 ± 166 | 24 ± 4 |
| FNR-G85A/K49A | 4,952 ± 153 | 391 ± 29 | 24 ± 2 |
| FNR-G85A/K50A | 5,025 ± 585 | 360 ± 32 | 22 ± 6 |
β-Galactosidase activity was measured from at least three independent cultures of strains carrying plasmid pBR322-encoded FNR or the indicated FNR variants. All cultures were grown under anaerobic conditions in L broth supplemented with glucose (0.2%) at 37°C for 16 h. The reporter plasmids used were the simple FNR-activated class I, FF(−71.5), and class II, FF(−41.5), constructs (32) and the simple FNR-repressed FF galΔ4 construct (30). AR1 was impaired by the substitution Ser73→Phe, and AR3 was impaired by the substitution Gly85→Ala. Values are means ± standard deviations.
As previously stated, inactivating AR1 or AR3 reduced transcription from the class II promoter. Thus, it was predicted that FNR variants carrying an impaired AR1 or AR3 would be more reliant on the AR2 contact for transcription activation at a class II promoter. Consequently, impairing AR2 in these proteins should have a greater effect on transcription than the corresponding AR2 mutations when AR1 and AR3 remain fully active. When AR2 alone was impaired, transcription from the class II promoter was reduced by as much as 32% compared to that obtained with unaltered FNR (Table 4). When both AR1 and AR2 were impaired, transcription from the class II promoter was reduced by as much as 60% compared to that obtained when only AR1 was impaired (Table 4). When both AR3 and AR2 were impaired, transcription from the class II promoter was reduced by as much as 67% compared to that obtained when only AR3 was impaired (Table 4). Thus, when either AR1 or AR3 is impaired, FNR is more reliant on AR2 for transcription activation.
Once again, to ensure that the differences observed were not due to changes in the DNA-binding activities of the activating region variants, the repression of transcription from FF galΔ4 was tested. All the variants were as effective as the unaltered FNR protein at this promoter (Table 4). Therefore, it can be concluded that the effects on transcription activation were not due to changes in the affinity for target DNA.
Rescue of FNR variants with impaired AR2 by amino acid substitutions in αNTD.
AR2 of CRP is a positively charged patch that contacts a series of negatively charged amino acids referred to here as the 165 determinant (Glu162, Glu163, Asp164, and Glu165) in αNTD (3, 19). Because substituting either Lys49 or Lys50 with Glu has a greater effect on transcription from a class II promoter than the equivalent Ala substitution, it is likely that FNR AR2 establishes an interaction with RNAP that resembles that formed by CRP. Initially, a mutant rpoA gene was created by using PCR-based site-directed mutagenesis that directed the replacement of all four negatively charged residues (162EEDE165; designated E165) with the uncharged amino acid Ala (162AAAA165; designated A165). Combining the expression of this RpoA derivative (with an uncharged 165 determinant) with wild-type FNR reduced transcription from a class II promoter, suggesting that the 165 determinant contributes to transcription activation at FNR-dependent class II promoters (Fig. 3A). Introducing a positively charged Lys residue at position 165 (162AAAK165; designated K165) should create a clash with the positively charged AR2 of FNR; indeed, the expression of this RpoA variant further reduced FNR-mediated transcription from the class II promoter (Fig. 3A). Because the strategy used requires that three plasmids be maintained in these cultures, plasmid DNA was isolated from each culture and subjected to diagnostic restriction digestion. This process revealed that all three plasmids were present and that the relative level of each was unaffected by the rpoA mutations (data not shown). Thus, it was concluded that mutation of the 165 determinant reduces transcription from an FNR-dependent class II promoter by up to 28%, a value similar to the level of impairment caused by mutation of AR2 at this promoter. Transcription from a class I promoter was less affected by the substitutions in RpoA, with 3,067 ± 316 Miller units for wild-type RpoA, 2,702 ± 263 Miller units for RpoA-162AAAA165, and 2,819 ± 187 Miller units for RpoA-162AAAK165, suggesting that the 165 determinant is not required for FNR-mediated transcription activation at a class I promoter.
FIG. 3.
Partial rescue of impaired AR2 by amino acid substitutions in αNTD (RpoA). All strains carried a low-copy-number plasmid with the simple FNR-activated class II promoter, FF(−41.5), fused to lacZ (32) and plasmid pBR322-encoded wild-type FNR or FNR-K49E. In addition, RpoA or the indicated variants were supplied on plasmids (Table 1) encoding wild-type E165, A165, or K165. All cultures were grown under anaerobic conditions in L broth supplemented with glucose (0.2%) at 37°C for 16 h. β-Galactosidase activity was measured in at least three independent cultures of each strain.
These results suggest that the positively charged AR2 of FNR (Lys49 and Lys50) contacts the negatively charged 165 determinant of αNTD and contributes to transcription activation at FNR-dependent class II promoters. To test this hypothesis further, the activities of the three RpoA 165 determinant variants (E165, A165, or K165) were estimated in the presence of FNR AR2 mutant FNR-K49E, in which one of the positively charged Lys residues is replaced by negatively charged Glu. Compared to the data obtained with wild-type FNR, the pattern of expression was reversed (Fig. 3B). Transcription was highest for K165 (RpoA-162AAAK165), intermediate for A165 (RpoA-162AAAA165), and lowest for E165 (RpoA-162EEDE165) (Fig. 3B). In contrast, in corresponding experiments with a class I promoter, transcription was essentially unaffected (2,708 ± 170 Miller units for E165, 2,403 ± 165 Miller units for A165, and 2,512 ± 134 Miller units for K165). These data are consistent with the positively charged AR2 of FNR, consisting of Lys49 and Lys50, interacting with a negatively charged region of RNAP αNTD that includes Glu165.
DISCUSSION
Many transcription factors activate transcription by making direct protein-protein contacts with RNAP. For the CRP family, three such contacts have been characterized and designated AR1, AR2, and AR3. Although FNR and CRP are members of the same family of transcription factors and the mechanisms that they use to activate transcription from class II promoters are similar, there are important differences in the balance of contacts made between the respective regulators and RNAP. For CRP, it has been shown that AR1 and AR2 contacts are the major contributors to transcription activation at class II promoters, because AR3-mediated activation is inhibited by an adjacent Lys residue (23). It has been argued that for CRP, the activating and inhibiting determinants of AR3 counteract each other such that the net contribution of AR3 to activation is so small that it is not evident unless the inhibitory determinant is removed (23). In contrast, FNR-dependent class II activation is predominantly governed by AR1 and AR3 contacts. However, it is now revealed that AR2 also makes a small but significant contribution to FNR-mediated transcription. Thus, whereas CRP relies entirely on AR1 and AR2 contacts, we suggest that FNR makes use of all three (AR1, AR2, and AR3) contacts in activating transcription from class II promoters. Recent work has revealed a further variation on this theme with the finding that for the CooA protein (a CO-sensing member of the CRP family of transcription factors from Rhodospirillium rubrum), AR2 and AR3 are both used and the disruption of either contact abolishes activation from a class II promoter when the heterologous E. coli RNAP is used (11).
The contacts made between a regulator and RNAP at class II-activated promoters play different roles in initiating transcription. The AR1 contact of CRP enhances the binding of RNAP to class II promoters, and the AR2 contact directly activates transcription by increasing the rate of open complex formation (19, 22). The AR1 contact of FNR accelerates the transition from the closed to the open complex (31), and AR3 is also thought to provide direct activation by interaction with σ70 (15). The evidence indicating that AR2 of FNR interacts with the same region of αNTD as CRP suggests that AR2 probably fulfills the same function in both regulators.
In conclusion, the experiments described here provide the first evidence for the existence of a functional AR2 in FNR. It would appear that two positively charged residues (Lys49 and Lys50) contribute to FNR AR2 and assist in activating transcription from class II promoters. The observation that the effects of amino acid substitutions in this region of FNR can be suppressed by substitutions in the 165 determinant of RpoA suggests that FNR can make an AR2-RpoA contact equivalent to that made by CRP (19). Thus, the evidence presented here adds further credence to the suggestions that all three activating regions were present in the ancestor of this family of transcription factors and that members of this family have since evolved an optimal balance of interactions that may be governed in part by the nature of the signal perceived and the way in which it is transduced.
Acknowledgments
We thank Neil Wyborn for constructing vector pGS1121.
This work was supported by the Biotechnology and Biological Sciences Research Council (United Kingdom).
REFERENCES
- 1.Bell, A., and S. J. W. Busby. 1994. Location and orientation of an activating region in the Escherichia coli transcription factor, FNR. Mol. Microbiol. 11:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Browning, D., D. Lee, J. Green, and S. Busby. 2002. Secrets of bacterial transcription initiation taught by the Escherichia coli FNR protein, p. 127-142. In D. A. Hodgson and C. M. Thomas (ed.), SGM symposium 61: signals, switches, regulons and cascades: control of bacterial gene expression. Cambridge University Press, Cambridge, United Kingdom.
- 3.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 4.Green, J., and M. L. Baldwin. 1997. HlyX, the FNR homologue of Actinobacillus pleuropneumoniae, is a [4Fe 4S]-containing oxygen-responsive transcription regulator that anaerobically activates FNR-dependent class I promoters via an enhanced AR1-contact. Mol. Microbiol. 24:593-605. [DOI] [PubMed] [Google Scholar]
- 5.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44:1-34. [DOI] [PubMed] [Google Scholar]
- 6.Jordan, P. A., A. J. Thomson, E. T. Ralph, J. R. Guest, and J. Green. 1997. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 416:349-352. [DOI] [PubMed] [Google Scholar]
- 7.Kiley, P. J., and H. Beinert. 1999. Oxygen sensing by the global regulator FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 8.Lamberg, K. E., and P. J. Kiley. 2000. Fnr-dependent activation of the class II dmsA and narG promoters of Escherichia coli requires FNR-activating regions 1 and 3. Mol. Microbiol. 38:817-827. [DOI] [PubMed] [Google Scholar]
- 9.Lamberg, K. E., C. Luther, K. D. Weber, and P. J. Kiley. 2002. Characterization of activating region 3 from Escherichia coli FNR. J. Mol. Biol. 315:275-283. [DOI] [PubMed] [Google Scholar]
- 10.Lazazzera, B. A., H. Beinert, N. Khoroshilova, M. C. Kennedy, and P. J. Kiley. 1996. DNA-binding and dimerization of the Fe-S containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271:2762-2768. [DOI] [PubMed] [Google Scholar]
- 11.Leduc, J., M. V. Thorsteinsson, T. Gaal, and G. P. Roberts. 2001. Mapping CooA-RNA polymerase interactions: identification of activating regions 2 and 3 in CooA, the CO-sensing transcriptional activator. J. Biol. Chem. 276:39968-39973. [DOI] [PubMed] [Google Scholar]
- 12.Lennox, E. S. 1955. Transduction of linked genetic characters of host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 13.Li, B., H. Wing, D. Lee, H. Wu, and S. Busby. 1998. Transcription activation by Escherichia coli FNR protein: similarities to and differences from the CRP paradigm. Nucleic Acids Res. 26:2075-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lombardo, M. J. 1995. Genetic analysis of RNA polymerase/transcriptional activator interactions at the pepT promoter of Salmonella typhimurium. Ph.D. thesis. University of Illinois, Urbana-Champaign.
- 15.Lonetto, M. A., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J. Mol. Biol. 284:1353-1365. [DOI] [PubMed] [Google Scholar]
- 16.Marsh, P. 1986. ptac85, an Escherichia coli vector for expression of non-fusion proteins. Nucleic Acids Res. 14:3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melville, S. B., and R. B. Gunsalus. 1990. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J. Biol. Chem. 265:18733-18736. [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Niu, W., Y. Kim, G. Tau, T. Heyduk, and R. Ebright. 1996. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell 87:1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ralph, E. T., J. R. Guest, and J. Green. 1998. Altering the anaerobic transcription factor FNR confers a hemolytic phenotype on Escherichia coli K12. Proc. Natl. Acad. Sci. USA 95:10449-10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ralph, E. T., C. Scott, P. A. Jordan, A. J. Thomson, J. R. Guest, and J. Green. 2001. Anaerobic acquisition of [4Fe 4S] clusters by the inactive FNR(C20S) variant and restoration of activity by second-site amino acid substitutions. Mol. Microbiol. 39:1199-1211. [DOI] [PubMed] [Google Scholar]
- 22.Rhodius, V., D. West, C. Webster, S. Busby, and N. Savery. 1997. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 25:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodius, V. A., and S. J. W. Busby. 2000. Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J. Mol. Biol. 299:295-310. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science 253:1001-1007. [DOI] [PubMed] [Google Scholar]
- 26.Sharrocks, A. D., J. Green, and J. R. Guest. 1990. In vivo and in vitro mutants of FNR, the anaerobic transcriptional regulator of E. coli. FEBS Microbiol. Lett. 270:119-132. [DOI] [PubMed] [Google Scholar]
- 27.Spiro, S., and J. R. Guest. 1987. Activation of the lac operon of Escherichia coli by a mutant FNR protein. Mol. Microbiol. 1:53-58. [DOI] [PubMed] [Google Scholar]
- 28.Spiro, S., and J. R. Guest. 1988. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol. Microbiol. 2:701-707. [DOI] [PubMed] [Google Scholar]
- 29.Williams, S. M., N. J. Savery, S. J. W. Busby, and H. J. Wing. 1997. Transcription activation at class I FNR-dependent promoters: identification of the activating surface of FNR and the corresponding contact site in the C-terminal domain of the RNA polymerase α subunit. Nucleic Acids Res. 25:4028-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams, S. M., H. J. Wing, and S. J. W. Busby. 1998. Repression of transcription initiation by Escherichia coli FNR protein: repression by FNR can be simple. FEMS Microbiol. Lett. 163:203-208. [DOI] [PubMed] [Google Scholar]
- 31.Wing, H. J., J. Green, J. R. Guest, and S. J. W. Busby. 2000. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J. Biol. Chem. 275:29061-29065. [DOI] [PubMed] [Google Scholar]
- 32.Wing, H. J., S. M. Williams, and S. J. W. Busby. 1995. Spacing requirements for transcription activation by Escherichia coli FNR protein. J. Bacteriol. 177:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]