Abstract
Microbial virulence is known to emerge by horizontal gene transfer mechanisms. Here we describe the discovery of a novel filamentous prophage, designated CUS-1, which is integrated into the chromosomal dif homologue of the high-virulence clone Escherichia coli O18:K1:H7. An homologous chromosomal element (CUS-2) in Yersinia pestis biovar orientalis is integrated at the same relative location as CUS-1; both lysogenic E. coli and Y. pestis strains produce particles with properties expected of single-stranded DNA virions. CUSφ is epidemiologically correlated with the emergence of K1 strains with increased virulence and with the Y. pestis biovar responsible for the current (third) plague pandemic.
Most Escherichia coli strains residing in the large intestines of mammals are harmless commensals because they lack the virulence factors for causing disease in their hosts (5, 9). Another group of E. coli is responsible for intestinal infections, and diarrhea is the most common clinical presentation. Diarrheagenic strains synthesize toxins and specialized adhesins that distinguish them from the commensals, and the evolutionary origins of these virulence factors include plasmids, bacteriophages (phages), chromosomal accretions or islands, and transposons. Horizontal gene transfer mediated by mobile genetic elements thus is recognized as a primary mechanism for the emergence or reemergence of pathogenic microorganisms (15). A third group of E. coli strains does not cause localized intestinal disease but is responsible for the preponderance of extraintestinal (invasive) syndromes, including but not limited to bacteremia, sepsis (bacteremia with organ dysfunction), urinary tract infections, and meningitis (9). Sepsis is a significant reemerging disease in the rapidly aging United States population because of its high mortality and the associated economic losses, which exceed one billion dollars annually (12). This third group of extraintestinal pathogenic E. coli (ExPEC) is responsible for the majority of life-threatening E. coli infections (9-11, 19).
ExPEC (19), like other invasive pathogens that do not persist or multiply intracellularly, must have efficient mechanisms for escaping host innate immunity. Polysaccharide capsules are among the longest known and best understood virulence factors that allow such pathogens to resist antibody-independent (innate) destruction (21). In addition to capsules and other polysaccharides, ExPEC strains synthesize toxins (hemolysin, cytotoxic necrotizing factor), adhesins, and iron-scavenging systems found only sporadically in commensal E. coli strains (10). We hypothesized that in addition to these known pathogenicity factors there are unknown genes whose identification could help explain the strain-dependent virulence of certain ExPEC. For example, E. coli O18:K1:H7 is responsible for almost all cases of neonatal meningitis in the United States and the preponderance of uncomplicated cystitis in North American women (11). It is also the most virulent among all K1 strains in animal models of sepsis and meningitis (17). The molecular basis for this differential disease potential likely involves specialized virulence genes (10, 18) or some combination of such genes expressed in a particular genetic background (4).
To test for the existence of unknown systemic disease traits, we adapted the functional genomics method of signature-tagged mutagenesis (7) to the prototypic neonatal meningitis E. coli O18:K1:H7 strain RS218 (1) and the infant rat model of disease (6). In the process of validating this system, we detected a previously unidentified virulence gene, puvA, with 99% nucleotide identity to an uncharacterized open reading frame (ORF) in the Yersinia pestis strain CO92 genome (http://www.sanger.ac.uk/Projects/Y_Pestis). Here we show that puvA is shared by two novel genetic elements with similarity to filamentous phage. Molecular epidemiological analysis showed that among E. coli and Y. pestis strains these elements were located almost exclusively in E. coli strain O18:K1:H7 and Y. pestis biovar orientalis. We designated the E. coli and Y. pestis elements CUS-1 and CUS-2, respectively, to indicate that they were identified in the trimunicipality consisting of Champaign, Urbana, and Savoy, Ill.
ORFs flanking puvA.
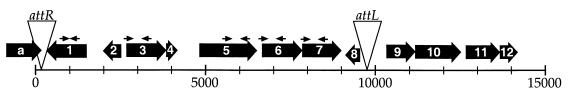
Following random transposon mutagenesis of an E. coli derivative (EV291) of strain RS218 (O18:K1:H7) and negative selection in the infant rat model of disease, we identified several genes not previously associated with systemic propagation or survival (6). Because the mutants were neither growth defective in vitro nor unable to colonize the large intestine, we concluded that the transposon insertions inactivated genes whose products may function during the extraintestinal phase of disease. One of these gene products, PuvA (accession no. AF345992), was nearly identical to a derived polypeptide of unknown function in Y. pestis strain CO92. Several ORFs surrounding puvA or its Y. pestis orthologues also were nearly identical to each other, suggesting that puvA is part of a larger, conserved chromosomal region in both E. coli and Y. pestis. Because the RS218 genomic sequence is incomplete (http://www.genome.wisc.edu), we constructed a cosmid library in pWEB (Epicentre, Madison, Wis.) that was subsequently screened by colony hybridization with a puvA-specific probe generated with the primers shown in Table 1. The resulting cosmid was sequenced with a puvA-specific primer (5′-ACCAACTAACTGGATGAACAG-3′), and the data were used to connect RS218 sequence contigs. The results indicated that the genetic unit that includes puvA is approximately 10 kb long and is inserted into the XerC recognition site of dif, a recombinational locus that functions in the resolution of chromosome dimers (Fig. 1).
TABLE 1.
PCR primers used in this study for analysis of CUSφ and control ORFs
| ORFa | PCR primers (5′ → 3′)b
|
Product size (bp)c | |
|---|---|---|---|
| Forward | Reverse | ||
| ORF1 | CGGGAAGTCTGATTTTGATG | AATGGTGGCGTTCTATCG | 308 |
| ORF3 | GGGCAACACATCTCTACATTGACG | TTTTGACCAGTCCAGTAAACCAGC | 433 |
| ORF5 | GAGATAGCCTAAGTTCCGACATCG | CTTCATTTCCCGCAGGTATTCC | 452 |
| ORF6 | CGGAGAGTTGTGACGAACATTTATGG | CTGGCGTTTATCTGTCTGCTTTTC | 539 |
| ORF7 | ATGTGCTTTCGCAGGGTCAGAG | CACTTCATCACTCATCGTATCCAGC | 516 |
| irp-2 | GTTGCTGTCCATCAAGCACG | GCCGGAAAGCCTGGCCTTTA | 1,243 |
| AltORF1 | CATGTAGAGAACGAATGTGATAACC | CCGCAAAGAATCTCAGATAACATT | 397 |
| pyk | GAAGTCACTGAACACACCGTTGTC | ACCGTCAAGGATGGCGTTTG | 509 |
ORF3, ORF5, ORF6, and ORF7 are present in both CUS-1 and CUS-2 as defined in the text, whereas AltORF1 of Y. pestis CUS-2 is located in the same relative genetic location as ORF1 but is not homologous. Note that the puvA (ORF5) primers do not flank the insertion in the previously described puvA::kan mutant (6).
Primers were purchased from IDT (Coralville, Iowa) and were used in PCRs at a final concentration of 8.75 pM 20-μl (total volume) mixtures containing PCR master mixture and MgCl2 purchased from Promega (Madison, Wis.). Amplification was carried out with a PTC-200 DNA Engine (MJ Research, Indian Valley, Nev.). The reaction conditions (35 cycles) for PCR amplification of ORF3, ORF5, ORF6, and ORF7 included denaturation at 94°C for 30 s, annealing at 61°C for 30 s, and extension at 72°C for 30 s. The annealing temperature was increased to 51°C for ORF1, pyk of Y. pestis, and irp-2.
Amplicons (products) were analyzed by electrophoresis in 0.7% agarose buffered with 40 mM Tris-acetate and 1 mM EDTA. The marker used was the 1-kb ladder purchased from Promega.
FIG. 1.
Genetic organization of CUS-1 in E. coli K1. CUS-1 is an approximately 10-kb genetic element integrated into the XerC recognition site (5′-ATTGGTGCGCATAATGTA-3′) of the strain RS218 dif locus (triangle labeled attR); there is a duplication (with one nucleotide difference) of this site at the other end of the phage (triangle labeled attL). The relative sizes (in kilobases) and the directions of transcription of the various ORFs are indicated by the large arrows. The small arrows over selected ORFs indicate PCR primers used to amplify the ORFs (Table 1). Each ORF is identified by a letter or number designation.
Similarity of ORFs flanking puvA to the genes of known filamentous phages.
Table 2 shows that the ORFs flanking puvA (ORF5) are predicted to encode homologues of filamentous phage polypeptides for replication, assembly, or secretion. Like puvA, ORF1 is predicted to encode a polypeptide with multiple membrane-spanning regions, but it has no putative function. With the exception of ORF1, the same genetic organization and insertion site (att) shown in Fig. 1 for CUS-1 were detected in Y. pestis biovar orientalis strain CO92 but not in the KIM5 (Y. pestis biovar medievalis) strain currently being sequenced (http://www.genome.wisc.edu). We designated this element CUS-2 because it is nearly identical to CUS-1 genetically. Y. pestis ORF1 (designated alternative ORF1 or AltORF1), like its topological equivalent in E. coli, is also predicted to encode a polypeptide of unknown function; however, as indicated above, this ORF lacks homology to E. coli ORF1. The results showed that the conserved E. coli and Y. pestis genetic elements are inserted into nearly identical att sites and differ by a single ORF. ORFa (hipA) defines one boundary of CUS-1 with E. coli K-12 DNA, whereas the other boundary is separated by a region that potentially encodes a type I pilus (indicated partially by ORF10 to ORF12 in Fig. 1). ORF9 (nucleoside diphospho sugar epimerase) also is present in the uropathogenic strain CFT073 genome but not in K-12 or enterohemorrhagic (O157:H7) strains. The absence of CUS-1 ORFs in the genomes of E. coli CFT073, O157:H7, and laboratory K-12 strains, Shigella strains, and all other genomes available in the BLAST server of the National Center for Biotechnological Information suggested that the CUS-1 element may be unique to K1 strains. The identification of ORFs that potentially encode polypeptides used for filamentous phage production was of further interest for at least two reasons. First, an insertion in E. coli puvA directly or indirectly affected the ability of EV291 to systemically propagate or survive in vivo, suggesting a possible role for puvA in pathogenesis (6). Second, the lysogenic filamentous phage CTXφ has been shown to be intimately involved with the pathogenesis of diarrheal disease caused by Vibrio cholerae because it encodes the cholera toxin (22). Therefore, if the genetic elements harboring puvA produce virions, they may encode transmissible virulence factors other than classical toxins, such as the lom and bor genes of phage lambda (3).
TABLE 2.
CUS-1 ORFs and proposed functions of their encoded products and selected flanking ORFs and proposed functions of their products
| ORF | Homologuea | Proposed function | % Identityb |
|---|---|---|---|
| ORFa | HipA (P23874) | High persistence to DNA or peptidoglycan inhibition | 95 (406) |
| ORF1 | None | Membrane protein | |
| ORF2 | TlcR (AAC38588) | Cryptic plasmid maintenance | 44 (75) |
| ORF3 | Gene II of I2-2 (NP039615) | Replication | 57 (344) |
| ORF4 | Gene V of I2-2 (NP039617) | Replication | 43 (100) |
| ORF5 | PuvA (AF345992) | Membrane protein | |
| ORF6 | Gene I of PhiLf (N003902) | Assembly | 27 (305) |
| ORF7 | SpsD (I39547) | Secretion | 24 (267) |
| ORF8 | Nucleocapsid (AF288649) | Assembly | 32 (61) |
| ORF9 | Epimerase (AP005277) | Nucleoside diphospho sugar epimerase | 34 (225) |
| ORF10 | Putative major fimbrial subunit (type I) (NP287648) | Pilus subunit | 84 (187) |
| ORF11 | Putative fimbrial chaperone (NP287649) | Pilus assembly | 97 (239) |
| ORF12 | Putative fimbrial usher (NP287650) | Pilus assembly | 93 (114) |
Only the most similar homologues are shown. The numbers in parentheses are accession numbers. Note that CUS-1 includes homologues of filamentous phage I2-2 and PhiLf gene products from enterobacteria and Xanthomonas, respectively. SpsD, TlcR, and the nucelocapsid homologues are from Aeromonas hydrophila, V. cholerae, and Hanta virus, respectively. The putative epimerase homologue is from Corynebacterium glutamicum.
The numbers in parentheses are the numbers of amino acid residues.
Detection of CUSφ particles.
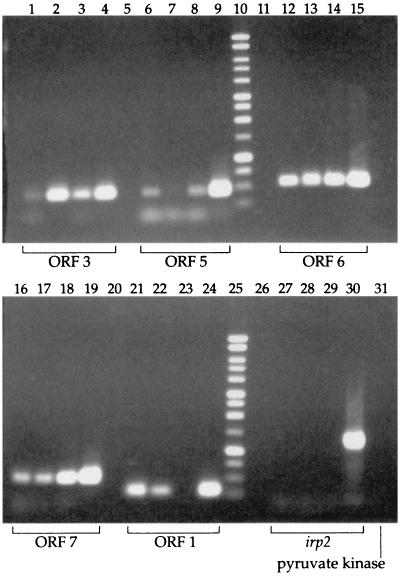
To determine if E. coli K1 and Y. pestis produce virions, filtered cell-free supernatants from RS218 or Y. pestis biovar orientalis spent culture media were treated with DNase I and RNase A and processed for phage banding in CsCl (19). By using the primer pairs (Table 1) indicated in Fig. 1, ORF1, ORF3, ORF5, ORF6, and ORF7 were amplified by PCR from the phage preparations. As controls for chromosomal contamination, the iron-regulated protein 2 (irp-2) and pyruvate kinase (pyk) genes from E. coli K1 and Y. pestis, respectively, were also tested. As shown in Fig. 2, each of the putative CUS-1 ORFs tested was amplified without evidence of chromosomal contamination. Although there was random variation in the relative amounts of some amplicons (Fig. 2, lane 7), the results represent one of six independent experiments in which puvA was specifically amplified without evidence of an irp-2 amplicon (Fig. 2, lanes 27 to 29), confirming that the results shown in Fig. 2 were not caused by chromosomal carryover. In contrast, compare the absence of an irp-2 signal in the phage preparations shown in Fig. 2, lanes 27 to 29, with the positive control results (lane 30). The results of this analysis also suggest that puvA is not required for the production of virions, because the same set of ORFs was successfully detected from puvA::kan mutant G-5 (Fig. 2, lanes 1, 6, 12, 16, and 21) as from wild-type strain RS218 (Fig. 2, lanes 2, 7, 13, 17, and 22). With the expected absence of an ORF1 amplicon from the Y. pestis preparation (Fig. 2, lane 23), all other CUS-2 ORFs tested were specifically amplified (Fig. 2, lanes 3, 8, 14, and 18) without evidence of chromosomal contamination by pyk (Fig. 2, lane 31). We concluded that both E. coli O18:K1:H7 and Y. pestis biovar orientalis produce particles with densities expected of intact phage.
FIG.2.
Identification of selected ORFs in phage preparations of E. coli and Y. pestis. Phage were purified (20) from E. coli strain RS218, the RS218 puvA::kan derivative G-5 (6), or Y. pestis biovar orientalis strain D30. Each group of four PCR lanes represents the results of PCR amplification performed with the appropriate primers (Fig. 1 and Table 1) and boiled phage preparations from G-5 (lanes 1, 6, 12, 16, and 21), RS218 (lanes 2, 7, 13, 17, and 22), Y. pestis (lanes 3, 8, 14, 18, and 23), or an RS218 genomic DNA control (lanes 4, 9, 15, 19, and 24). There was a blank lane between each group of lanes (lanes 5, 11, 20, and 26), and lanes 10 and 25 contained 1-kb ladders. Note the expected absence of ORF1 from the Y. pestis preparation (lane 23). In contrast, all other ORFs tested were successfully amplified, demonstrating homologies between the E. coli K1 and Y. pestis biovar orientalis phage particles; the signal for RS218 ORF5 (lane 7) was weak but clearly visible in the original. The control for contaminating genomic DNA was irp-2, which was negative (lanes 27 to 29). The positive control was chromosomal RS218 DNA (lane 30). Because the Y. pestis strain carried a deletion of irp-2, we used primers for pyk to control for chromosomal contamination (lane 31).
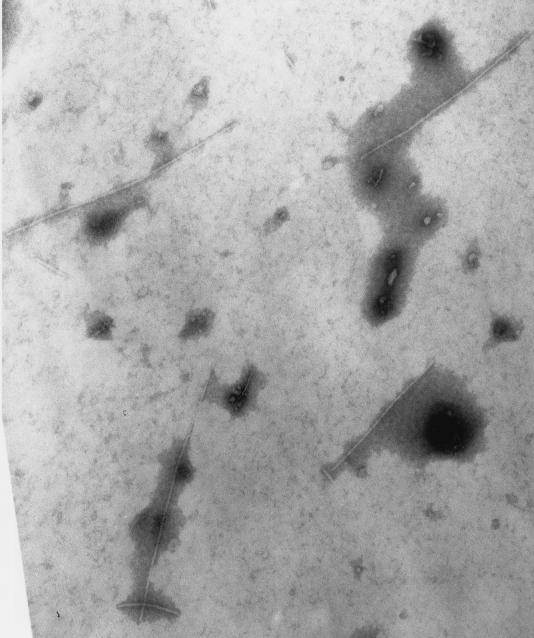
In an attempt to detect CUSφ morphology by transmission electron microscopy, droplets of phage samples were placed on Parafilm and transferred by capillary action to copper grids coated with formvar plastic and carbon. Excess sample was removed with filter paper, and the grids were placed on a 2% ammonium molybdate solution for 2 min. The grids were dried by removing the excess fluid with filter paper and placed into a box covered with Drierite crystals for 10 min. The grids were then examined at 75 kV with an Hitachi H600 electron microscope at a magnification of ×50,000. As shown in Fig. 3, filamentous particles which were consistent with the expected morphology of CUS-1 were detected in the RS218 sample. Similar particles were absent from control E. coli K-12 preparations. RS218 harbors prophage with lambdoid morphology (A. Nassar, C. Schouler, and M. Dho-Moulin, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. B-326, 2001), and some microscopic fields examined contained this type of particle. Therefore, while both E. coli K1 and Y. pestis biovar orientalis produced particles with densities expected of phage, additional evidence is needed to confirm the exact identity of the particles detected in Fig. 2 and 3. While the insertion mutation in puvA::kan mutant G-5 (6) did not block virion production (Fig. 2), we were unable to detect transmission of drug resistance to CUS-1-negative E. coli K1 strains by infection with phage preparations. Similarly, while we could detect an apparent replicative form of CUS-1 by PCR in standard plasmid preparations, we were unable to transfer kan by electroporation. Therefore, we are currently uncertain about the transmissibility of CUSφ or its potential receptor(s).
FIG. 3.
Transmission electron microscopy of CUS-1. E. coli K1 phage particles purified by CsCl (Fig. 2) appear as filamentous rods approximately 7 nm wide and 500 to 600 nm long. Magnification, ×100,000.
A Southern blot of DNA extracted from CUS-1 particles was positive when it was probed with the end-labeled antisense oligonucleotide (5′-CATGTCGGCAAGAAGTGCTGGCG-3′) but not when it was probed with the complement (5′-CGCCAGCACTTCTTGCCGACATG-3′), representing nucleotides 255 to 277 of puvA (accession no. AF345992). This result indicates that puvA is part of a genetic element that produces single-stranded DNA. Chromosomal DNA was positive when it was probed with either the sense or antisense oligonucleotide.
Molecular epidemiology of CUS-1.
The experimental and epidemiological evidence linking E. coli O18:K1:H7 with high virulence is unexplained at the molecular level. To determine if CUS-1 is correlated with this phenotype, we analyzed 149 different E. coli isolates for the presence of puvA by PCR. As shown in Table 3, almost all O18 strains tested were positive for ORF5 to ORF7 and thus CUS-1, whereas puvA was absent from all other K1 (non-O18) strains and most E. coli reference (ECOR) collection (14) strains. The invariant linkage of puvA with ORF6 and ORF7 (Table 2) is consistent with the unit inheritance of CUS-1. Two (#51 and #57) of the three CUS-1-positive ECOR collection strains were from the B2 group that, along with group D, includes serotype K1 and most other pathogenic E. coli strains. The third positive strain (#37) belongs to the nonaligned group. Several additional strains closely related to #51 or #57 were tested and were found to be negative for CUS-1 ORFs (Table 2), suggesting that CUS-1 is unlikely to have originated from any strain other than O18:K1 in the B2 group. On the basis of these results we think that it is more likely that #51 and #57 were lysogenized relatively recently, perhaps as a result of close association with CUS-1-positive K1 strains. Taken together, the prevalence data indicate the highly significant association of CUS-1 with E. coli O18:K1:H7 (Table 2), supporting our hypothesis that CUS-1 acquisition is correlated with virulence.
TABLE 3.
Prevalence of puvA (ORF5) in K1 and non-K1 E. coli
| Strains | Sourcea | % of isolates with ORF5 (no. tested)b | Pc |
|---|---|---|---|
| 018:K1:H7 | RS, DSMZ, or JJ | 85.7 (28) | |
| Other (non-O18) K1 | WV or JJ | 0.0 (38) | <0.0001 |
| ECORd | HO | 4.2 (72) | 0.0001 |
| ECOR-likee | JJ | 0.0 (11) | <0.0001 |
| ECORf | JJ | 11.8 (17) |
RS, Richard Silver, University of Rochester, Rochester, N.Y.; DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; JJ, James Johnson, University of Minnesota, Minneapolis,; WV, Willie Vann, Food and Drug Administration, Bethesda, Md.; HO, Howard Ochman, University of Arizona, Tucson.
Detected by PCR analysis with the puvA-specific primers given in Table 1. All O18:K1:H7 strains were also analyzed for ORF6 and ORF7 with the primers indicated in Table 1 and covaried with ORF5.
P values were determined by Fisher's exact test by comparison to the O18:K1:H7 strains; values of <0.01 were considered significant.
The ECOR collection (14) is considered representative of the worldwide diversity of E. coli strains.
#51 (O25:K+) and #57 (O2:K5/7:H1), two of the three ORF5-positive strains from the ECOR collection (both from the B2 group), are similar to strains BOS 031 and BOS 080 and to strains V27, H19, V6, V24, and H1, respectively, which unlike #51 and #57 were negative for puvA. Strains more distantly related to #51 (BOS 70, BOS 100, BOS 77, and BOS 56) were also tested and found to be negative.
ECOR strains #51 to #57, #59 to #66, #35, and #36. Strains #51 and #57 were kindly provided by an alternative source (James Johnson) and gave the same results as the equivalent strains obtained from Howard Ochman.
Molecular epidemiology of CUS-2.
Y. pestis is believed to have evolved from Yersinia pseudotuberculosis shortly before the first known human plague pandemic approximately 1,500 years ago (2). The three biovars (Y. pestis biovar antiqua, Y. pestis biovar medievalis, and Y. pestis biovar orientalis) associated with this and subsequent outbreaks of the plague are considered to be phylogenetically distinct on the basis of IS100 banding patterns and neighbor-joining analysis. However, DNA sequence analysis of six housekeeping loci from 36 different strains representing the global diversity of Y. pestis revealed no divergence (2), indicating the high degree of relatedness among all Y. pestis strains. In contrast, both puvA and AltORF1 were detected in all Y. pestis biovar orientalis isolates tested, which represented nine different geographic locations, but they were detected only sporadically in the other two biovars (Table 4). These data correlate the worldwide prevalence of CUS-2 with the emergence of Y. pestis biovar orientalis.
TABLE 4.
Prevalence of puvA (CUS-2) in Yersinia spp.
| Strain(s)a | Geographic origin | Biovar | puvA |
|---|---|---|---|
| Y. pestis strains | |||
| CO92 and derivatives 1101, 1102, 9200, and CO92 (pgm), Alexander, Shasta, 76546NM (752), A12, 70-259-6, Yreka, NM610107 (684), Dodson, 1171 | United States | Y. pestis biovar orientalis | + |
| M-111 (74), EV76-51, EV76 (lot 4), H3, EV-76 (Paris), EV-76f | Madagascar | Y. pestis biovar orientalis | + |
| K2216-67VN, R1575-66VN (33), P1178VN (219), P824-67VN (31) | Vietnam | Y. pestis biovar orientalis | + |
| Stavropol, SEV, Russian Vaccine | Former USSR | Y. pestis biovar orientalis | + |
| Java2, Java9, CO3311 (770) | Indonesia | Y. pestis biovar orientalis | + |
| I-18 (71), I-254 (70), 195P-3 | India | Y. pestis biovar orientalis | + |
| PEXU | Peru | Y. pestis biovar orientalis | + |
| RFPBM-19 (350) | Burma | Y. pestis biovar orientalis | + |
| La Paz | Bolivia | Y. pestis biovar orientalis | + |
| PKR108, PKH10, KIM10 (pgm), KIM10var, KIM5 | Iran | Y. pestis biovar medievalis | − |
| Pestoides A, B, C, D, Aa, and Ba | Former USSR | Y. pestis biovar medievalis | − |
| Nicholisk 41 | Manchuria | Y. pestis biovar medievalis | − |
| PyHIRCabSal (366) | Yemen | Y. pestis biovar medievalis | − |
| A16, Antiqua | Congo | Y. pestis biovar antiqua | − |
| Pestoides E, F, and G | Former USSR | Y. pestis biovar antiqua | − |
| Nicholisk 51b | Manchuria | Y. pestis biovar antiqua | + |
| Angola | Angola | Y. pestis biovar antiqua | − |
| Yokohama | Japan | Y. pestis biovar antiqua | + |
| Nairobi | Kenya | Y. pestis biovar antiqua | + |
| Pestoides J | Former USSR | Atypical | − |
| Y. pseutotuberculosis (3 isolates) | United States | Unknown | − |
Due to the requirements of the Select Agents Act, all Y. pestis strains or DNA samples were analyzed on site by PCR at the United States Army Medical Research Institute of Infectious Diseases (Fort Detrick, Md.) or the University of Kentucky (Lexington). All Y. pestis samples also were analyzed for the presence of AltORF1 and gave results which were the same as the results for puvA.
The IS100 genotype is reported to be similar to Y. pestis biovar orientalis (13).
Y. pestis biovar medievalis is believed to have caused the second pandemic or Black Death in the 14th century (with sporadic outbreaks until the 19th century), while Y. pestis biovar antiqua strains constitute the oldest biovar, which was associated with the first or Justinian plague in the 6th century (2). Our data are consistent with the suggestion that Y. pestis biovar orientalis, which is responsible for the third pandemic, which began in the 19th century and continues today, did not arise from Y. pestis biovar medievalis but may instead have evolved from a CUS-2-lysogenized Y. pestis biovar antiqua strain (reference 13 and references therein). This suggestion is consistent with the idea that Y. pestis biovar antiqua is ancestral to both Y. pestis biovar medievalis and Y. pestis biovar orientalis but also indicates that Y. pestis biovar medievalis may have arisen from a prophage-negative Y. pestis biovar antiqua strain and that CUS-2 entered the lineage relatively recently. The absence of puvA and AltORF1, and thus the absence of CUS-2, from Y. pseudotuberculosis (Table 4) is consistent with recent lysogenization of Y. pestis biovar antiqua. Although it is claimed that there are no differences in virulence among the three biovars of Y. pestis (16), our data show that there is a perfect correlation between CUS-2 and strains of the third pandemic, which represents a major genetic difference between modern plague strains and most of their progenitors.
Conclusion.
Our data show that the limited distribution of CUS-1 in E. coli is paralleled by the distribution of CUS-2 in Y. pestis. Despite the dramatically different lifestyles of Y. pestis and E. coli K1, the virulence of both of these organisms is dependent on their ability to avoid innate host defense mechanisms. The selection for CUSφ thus may be causally related to the emergence of O18:K1:H7 and Y. pestis biovar orientalis if the decreased in vivo fitness of an E. coli K1 puvA mutant (6) is mimicked by a similar defect in Y. pestis, in which case the functions of CUSφ may be identical in the two pathogens. Our results provide the first demonstration of the presence of conserved lysogenic filamentous phage-like elements in separate bacterial species and identify the integration sites of these elements in homologues of dif (Fig. 1A), indicating that CUSφ may use the xer-encoded recombination machinery of their hosts. The results strongly indicate that filamentous prophage are more widely associated with the evolution of pathogenic microorganisms than could have been previously determined from the available evidence.
While our manuscript was being revised, Huber and Waldor (8) reported that CTXφ uses the V. cholerae dif homologue for integration. This finding is in agreement with our data and suggests that a variety of chromosomal elements use the host's XerCD recombinational machinery for vertical transmission.
ADDENDUM IN PROOF
After acceptance of this article, a paper (T. Iida, K. Makino, H. Nasu, K. Yokoyama, K. Tagomori, A. Hattori, T. Okuno, H. Shinagawa, and T. Honda, J. Bacteriol. 184:4933-4935, 2002) was published which reports that the CTX-related phage f237 of Vibrio parahaemolyticus is integrated into the dif-like site of its host chromosome.
Acknowledgments
In addition to the individuals cited in Table 3, whom we thank for strains and information, we are indebted to Patricia Worsham (USAMRIID, Fort Detrick, Md.) and Susan Straley (University of Kentucky, Lexington) and their colleagues for sharing strains, DNA, laboratory space, or information. Due to the restrictions imposed by the Select Agents Act, we are especially grateful to members of the Straley lab for preparing the cell-free culture supernatant of strain D30 that was used for CUS-2 isolation. We thank Lou Ann Miller for the electron micrograph and Kerry Helms and Eric Deszo for assistance with graphics.
M.D.G. was supported in part by a Howard Hughes Undergraduate Research Fellowship, by an American Society for Microbiology Undergraduate Research Fellowship, and by a University of Illinois Minority Graduate Student Fellowship. This work was supported by NIH grant AI42015 to E.R.V.
REFERENCES
- 1.Achtman, M., A. Mercer, B. Kuscecek, A. Pohl, M. Heuzenroeder, W. Aaronson, and R. P. Silver. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barondess, J. J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346:871-874. [DOI] [PubMed] [Google Scholar]
- 4.Bonacorsi, S. P. P., O. Clermont, C. Tinsley, I. Le Gall, J.-C. Beaudoin, J. Elion, X. Nassif, and E. Bingen. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duriez, P., O. Clermont, S. Bonacorsi, D. Bingen, A. Chaventre, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671-1676. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez, M. D., C. A. Lichtensteiger, and E. R. Vimr. 2001. Adaptation of signature-tagged mutagenesis to Escherichia coli K1 and the infant-rat model of invasive disease. FEMS Microbiol. Lett. 198:125-128. [DOI] [PubMed] [Google Scholar]
- 7.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 8.Huber, K. E., and M. K. Waldor. 2002. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417:656-659. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, J. R., and T. A. Russo. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli.” J. Lab. Clin. Med. 139:155-162. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. R., T. T. O'Bryan, P. Delavari, M. Kuskowski, A. Stapelton, U. Carlino, and T. A. Russo. 2001. Clonal relationship and extended virulence genotypes among Escherichia coli isolates from women with a first or recurrent episode of cystitis. J. Infect. Dis. 183:1508-1517. [DOI] [PubMed] [Google Scholar]
- 12.McBean, M., and S. Rajamani. 2001. Increasing rates of hospitalization due to septicemia in the US elderly population. J. Infect. Dis. 183:596-603. [DOI] [PubMed] [Google Scholar]
- 13.Motin, V. L., A. M. Georgescu, J. M. Elliott, P. Hu, P. L. Worsham, L. L. Ott, T. R. Slezark, B. A. Sokhansanj, W. M. Regala, R. R. Brubaker, and E. Garcia. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 184:1019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 16.Perry, R. D., and J. D. Fetehrston,. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pluschke, G., A. Mercer, B. Kusecek, A. Pohl, and M. Achtman. 1983. Induction of bacteremia in newborn rats by Escherichia coli K1 is correlated with only certain O (lipopolysaccharide) antigen types. Infect. Immun. 39:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rode, C. K., L. Melkerson-Watson, A. T. Johnson, and C. A. Bloch. 1999. Type-specific contributions to chromosome size differences in Escherichia coli. Infect. Immun. 67:230-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Silver, R. P., and E. R. Vimr. 1990. Polysialic acid capsule of Escherichia coli K1, p. 39-60. In B. Iglewski and V. Miller (ed.), The bacteria, vol. 11. Molecular basis of bacterial pathogenesis. Academic Press, New York, N.Y.
- 22.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]