Abstract
We have shown previously that a gene encoding a homologue to the eukaryotic dolichol-phosphate-d-mannose, protein O-d-mannosyltransferase, was required for φC31 infection of Streptomyces coelicolor. Here we show that a gene encoding the homologue to dolichol-phosphate-mannose synthase is also essential for phage sensitivity. These data confirm the role of glycosylation in the phage receptor for φC31 in S. coelicolor.
The temperate phage φC31 has been used to develop phage vectors for genetic manipulation of Streptomyces species, many of which are commercially useful producers of antibiotics and other bioactive secondary metabolites (4, 8). φC31 has a moderately broad host range and is able to infect around a third of the Streptomyces species tested (9, 18). The earliest event in phage infection is the recognition by phage tail proteins of some component on the host cell surface (7, 11, 15). While this initial step is often reversible, at some point a commitment to infection occurs in which a series of molecular changes in the phage results in the injection of DNA into the cell. Studies of the molecules on the cell surface recognized by the φC31 coat proteins will lead to a better understanding of the Streptomyces cell wall and to further development of phage vectors.
We have indicated previously that glycosylation of a Streptomyces coelicolor cell envelope protein is required for infection of the cell by φC31 (5). Mutants of S. coelicolor strain J1929 (ΔpglY and therefore sensitive to φC31 [1]) that are unable to support plaque formation by φC31cΔ25, a clear-plaque mutant of φC31, were isolated. The S. coelicolor mutants were unable to form infective centers after contact with phage but could release phage particles after being transfected with phage DNA. These observations indicated that the mutants are blocked early in phage infection, probably at the stage of receptor binding. The S. coelicolor mutants fell into three classes, designated I, II, and III. A gene isolated from the S. coelicolor ordered cosmid library, SCE87.05, complemented the class I mutants but not those of class II or III. SCE87.05 has significant similarity to the eukaryotic dolichol-phosphate-d-mannose (Dol-P-Man), protein O-d-mannosyltransferases (protein mannosyltransferase), suggesting that the receptor for φC31 infection is a glycoprotein generated through an O glycosylation pathway. This suggestion was supported by observations that the plant lectin concanavalin A could inhibit phage infection and that glycoproteins could not be detected by concanavalin A in Western blots of proteins from a mutant defective in SCE87.05. A mutant phage, φC31hc, could form plaques on class I and class II mutants of S. coelicolor but not on class III mutants. It was proposed that φC31 normally recognizes a cell wall glycoprotein but that φC31hc recognizes the unglycosylated protein. Thus, like the class I mutants, the class II mutants are predicted to be defective in the glycosylation pathway and the class III mutants are predicted to lack the protein target of glycosylation. Here we have tested part of this model with experiments to investigate the role of a gene, SC6D7.16, encoding a homologue of Dol-P-Man synthase, proposed to be required for the protein glycosylation pathway.
Dol-P-Man catalyzes the transfer of mannose from GDP-mannose to Dol-P in fungi and in mammals (10, 17). Dol-P-Man is then the substrate for the transfer of the mannose to a serine or threonine by the protein glycosyltransferase in the O- glycosylation pathway. The protein sequences of the fungal and mammalian Dol-P-Man synthases were used to search the S. coelicolor genome sequence (http://www.sanger.ac.uk/Projects/S_coelicolor/). The closest homologue was the predicted product of SC6D7.16 (Fig. 1). The cosmid SC6D7 was therefore introduced by protoplast transformation into two of the class II phage-resistant mutants, DT1029 and DT2021, and into the S. coelicolor class III mutant, DT2017, isolated previously (5). Seven individual transformants of each strain were tested for sensitivity to phage by using a plate assay. This was performed by preinoculating one-half of an R1M plate with φC31cΔ25 and then streaking spores from the transformants across the plate from the phage-free half to the phage-containing half. SC6D7 conferred sensitivity to phage φC31cΔ25 in approximately 50% of the transformants of DT1029 and DT2021 (the class II mutants). This incomplete complementation appears to be typical of complementation by cosmids, and it is believed that the mutant and wild-type alleles undergo homogenotization (5, 13). To confirm the complementation, we constructed a plasmid, pDT16, that carried only SC6D7.16. PCR was used to amplify the open reading frame and flanking DNA from cosmid SC6D7, and the PCR product was inserted into the integrating vector pSET152 (3). This plasmid was introduced by conjugation into a class I strain, DT1017; the class II strains DT1028, DT1029, DT1035, DT2008, and DT2021; and the class III mutant DT2017. Seven individual transconjugants from each mating were tested for sensitivity to phage by using the plate assay. Phage sensitivity was restored to all the transconjugants tested from the class II mutants DT1029, DT1035, and DT2021, and this was confirmed in a plaque assay (Fig. 2). The class II mutants DT1028 and DT2008 remained phage resistant (data not shown). As expected, pDT16 did not restore phage sensitivity to φC31cΔ25 in either the class I or class III mutant.
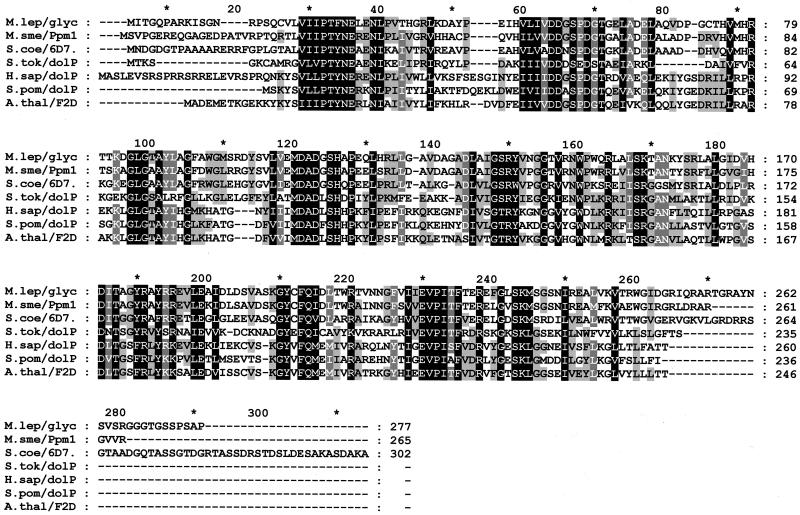
FIG. 1.
Alignment of fungal and mammalian Dol-P-Man synthases and SC6D7.16. The alignment was performed using CLUSTAL W (http://www.ebi.ac.uk/). Residues are shaded black, dark grey, and light grey to represent positions with 100, 80, and 60% similarity, respectively. Sequences are of M. smegmatis Ppm1 (M.sme/Ppm1, accession number CAC15463), a possible Mycobacterium leprae glycosyl transferase (M.lep/glyc, accession number CAC30390), a putative Sulfolobus tokodaii Dol-P-Man synthase (S.tok/dolP, accession number BAB67446), Homo sapiens Dol-P-Man synthase (H.sap/dolP, accession number CAB53749), Schizosaccharomyces pombe Dol-P-Man synthase (S.pom/dolP, accession number CAB11700), and the predicted products of S. coelicolor SC6D7.16 (S.coe/6D7, accession number CAB61668) and Arabidopsis thaliana F2D10.6 (A.thal/F2D, accession number AAF80640).
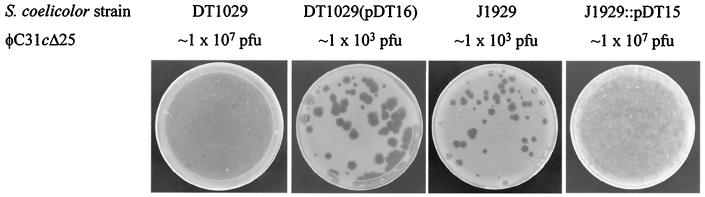
FIG. 2.
Complementation of a putative phage receptor mutant, S. coelicolor DT1029, by a plasmid, pDT16, containing SC6D7.16 and generation of a phage-resistant strain, J1929::pDT15, by insertional mutagenesis of SC6D7.16. DT1029 is a phage-resistant derivative of J1929 (ΔpglY [1]). Plasmid pDT16 was constructed by insertion of a fragment amplified from the cosmid SC6D7 with the primers DT25 (5′ GCGTCTAGAGGATCCCGAAGGGCGCCGGGCCCC 3′) and DT26 (5′ GCGGATATCGAATCCAGTGCCGCATAAGAGGGC 3′), cleaved with BamHI and EcoRV, and inserted into BamHI-EcoRV-cleaved pSET152 (3). pDT16 was introduced into strains of S. coelicolor by conjugation, and transconjugants were amplified on R2YE media containing apramycin (8). pDT15 was constructed with primers DT23 (5′ GCGAAGCTTGAATTCACGCGTGTGCGCGAGGGC 3′) and DT24 (5′ CGCTCTAGAGGATCCCTCCACCAGGATGTCGGG 3′) to amplify a 615-bp fragment internal to the SC6D7.16 open reading frame, and this fragment was inserted into pSET151 via HindIII and XbaI restriction sites introduced via primer composition (3). pDT15 was then introduced into J1929 by conjugation, and transconjugants were amplified by growth on R2YE media containing thiostrepton. Spores of DT1017(pDT16), DT1017, J1929, and J1929::pDT15 were plated in soft agar overlays containing yeast tryptone with the clear-plaque phage φC31cΔ25 (16) on Difco nutrient agar plates, and the plates were incubated overnight at 29°C. Phage sensitivity similar to that obtained with DT1029(pDT16) was also obtained with DT2021(pDT16) and DT1035(pDT16). Streptomyces strains and phage stocks were grown and maintained as described by Kieser et al. (8). Escherichia coli DH5α was used as the routine cloning host. Conjugations of plasmids into S. coelicolor were performed by using E. coli strain ET12567 (dam, dcm, and hsdM) (pUZ8002) containing either pDT16 or pDT15 as the donor (8). Cosmid SC6D7 from the M145 cosmid library of S. coelicolor A3 (2) was kindly provided by Helen Kieser (John Innes Centre, Norwich, United Kingdom).
To confirm the requirement for SC6D7.16 in phage infection, we constructed an S. coelicolor strain, J1929::pDT15, containing a targeted insertion in SC6D7.16. A DNA fragment internal to SC6D7.16 was amplified by using primers DT23 and DT24, inserted into the suicide vector pSET151 (3), and introduced into the phage-sensitive strain J1929 (1) by conjugation. Integration of this construct by insert-directed homologous recombination into S. coelicolor J1929 should give rise to a disrupted version of SC6D7.16. All seven transconjugants tested were resistant to φC31 by the plate assay. Spores were amplified and used for plaque assays with φC31cΔ25 and were resistant to infection, although a few very cloudy plaques could be observed at high phage titers (Fig. 2). As expected, introduction of pDT16 encoding the wild-type allele of SC6D7.16 into J1929::pDT15 resulted in phage sensitivity (data not shown). In addition, J1929::pDT15 spores could support plaque formation by φC31hc (data not shown). This phenotype is consistent with a class II phage resistance phenotype (5).
These observations indicate that the protein product of SC6D7.16 is essential for φC31 infection of S. coelicolor. SC6D7.16 is a homologue of the eukaryotic Dol-P-Man synthases (Fig. 1). In eukaryotes, Dol-P-Man provides the mannosyl residues in glycosylphosphatidylinositols and in N, O, and C glycosylation. A knockout of the single-copy gene DPM1, which codes for Dol-P-Man synthase, resulted in complete loss of protein mannosylation in Saccharomyces cerevisiae (12). Prokaryotes do not have dolichol. They contain instead other polyprenols, in particular undecaprenol phosphate (14). There is little information on the polyprenols in the Streptomyces cell envelope; however, the closely related bacteria Mycobacterium tuberculosis and Mycobacterium smegmatis contain a variety of polyprenol phosphates, which are covalently attached to mannose (Pol-P-Man) (6). The polyprenol phosphates in M. smegmatis have been shown to be used for several biosynthetic pathways for cell wall components, including mycolic acids, arabinogalactan, arabinomannan, and lipoarabinomannan (2, 6). Pol-P-Mans in mycobacteria are thought to be synthesized by Ppm1, a functional analogue to the eukaryotic Dol-P-Man synthase and the closest homologue to SC6D7.16 (Fig. 1).
The data presented here, together with previous findings (5), indicate that SCE87.05 and SC6D7.16, which encode a putative glycosyltransferase and a putative Pol-P-Man synthase, respectively, are required for the synthesis of the φC31 receptor in S. coelicolor. Similar to the roles of equivalent proteins in eukaryotes, these enzymes probably catalyze two steps in a protein glycosylation pathway in S. coelicolor. The observation that not all of the class II mutants are complemented by SC6D7.16 suggests that DT2008 and DT1028 are defective in other genes, possibly those encoding other enzymes in the glycosylation pathway. It is clear from these studies that these proteins are not essential for the growth of S. coelicolor, implying that they are not exclusively required components of cell wall biosynthesis. The role of protein glycosylation is not known either in the mycobacteria or in Streptomyces, but it will hopefully become clearer once the targets for glycosylation have been characterized.
Acknowledgments
We are grateful to Helen Kieser for providing cosmid SC6D7.
We thank the BBSRC for funding (project grant 42/P09260).
REFERENCES
- 1.Bedford, D. J., C. Laity, and M. J. Buttner. 1995. Two genes involved in the phase-variable φC31 resistance mechanism of Streptomyces coelicolor A3(2). J. Bacteriol. 177:4681-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belanger, A. E., and J. M. Inamine. 2000. Genetics of cell wall biosynthesis, p. 191-202. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Chater, K. F. 1986. Streptomyces phages and their applications to Streptomyces genetics, p. 119-157. In S. W. Queener and L. E. Day (ed.), The bacteria. A treatise on structure and function, vol. 9. Antibiotic-producing Streptomyces. Academic Press, Inc., Orlando, Fla.
- 5.Cowlishaw, D. A., and M. C. M. Smith. 2001. Glycosylation of a Streptomyces coelicolor A3(2) cell envelope protein is required for infection by bacteriophage φC31. Mol. Microbiol. 41:601-610. [DOI] [PubMed] [Google Scholar]
- 6.Crick, D. C., M. C. Schulbach, E. E. Zink, M. Macchia, S. Barontini, G. S. Besra, and P. J. Brennan. 2000. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 182:5771-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haggård-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 9.Kobler, L., G. Schwertfirm, H. Schmieger, A. Bolotin, and I. Sladkova. 1991. Construction and transduction of a shuttle vector bearing the cos site of Streptomyces phage φC31 and determination of its cohesive ends. FEMS Microbiol. Lett. 62:347-353. [DOI] [PubMed] [Google Scholar]
- 10.Maeda, Y., S. Tanaka, J. Hino, K. Kangawa, and T. Kinoshita. 2000. Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J. 19:2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montag, D., H. Schwarz, and U. Henning. 1989. A component of the side tail fiber of Escherichia coli bacteriophage λ can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J. Bacteriol. 171:4378-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlean, P. 1990. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:5796-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piret, J. M., and K. F. Chater. 1985. Phage-mediated cloning of bldA, a region involved in Streptomyces coelicolor morphological development, and its analysis by genetic complementation. J. Bacteriol. 163:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigen and capsular polysaccharides, p. 104-122. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. I. ASM Press, Washington, D.C.
- 15.Sandmeier, H. 1994. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol. Microbiol. 12:343-350. [DOI] [PubMed] [Google Scholar]
- 16.Sinclair, R. B., and M. J. Bibb. 1988. The repressor gene (c) of the Streptomyces temperate phage φC31: nucleotide sequence, analysis and functional cloning. Mol. Gen. Genet. 213:269-277. [DOI] [PubMed] [Google Scholar]
- 17.Strahl-Bolsinger, S., M. Gentzsch, and W. Tanner. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426:297-307. [DOI] [PubMed] [Google Scholar]
- 18.Voeykova, T. A., E. V. Slavinskaya, A. V. Orekhov, and N. D. Lomovskaya. 1979. Identification of restriction and modification systems in Streptomyces strains. Genetika 15:1746-1756. [PubMed] [Google Scholar]