Abstract
IS150, a member of the widespread IS3 family, contains two consecutive out-of-phase open reading frames, orfA and orfB, that partially overlap. These open reading frames encode three proteins, InsA, InsB, and the InsAB protein, which is jointly encoded by both open reading frames by means of programmed translational frameshifting. We demonstrate that the InsAB protein represents the IS150 element's transposase. In vivo, the wild-type IS150 element generates circular excision products and linear IS150 molecules. Circular and linear species have previously been detected with mutant derivatives of other members of the IS3 family. Our finding supports the assumption that these products represent true transposition intermediates of members of this family. Analysis of the molecular nature of these two species suggested that the circular forms are precursors of the linear molecules. Elimination of InsA synthesis within the otherwise intact element led to accumulation of large amounts of the linear species, indicating that the primary role of InsA may be to prevent abortive production of the linear species and to couple generation of these species to productive insertion events.
The 1,443-bp Escherichia coli transposable element IS150 (23) belongs to a large group of insertion sequences, the IS3 family, whose members have been found in more than 40 bacterial species (www-is.biotoul.fr/is/IS_infos/IS3_family.html). These elements are related at the level of their gene products and in addition share structural features, like the terminal dinucleotides TG…CA and the typical arrangement of their two long open reading frames, orfA and orfB, that utilize different translational reading frames and partially overlap (14). In addition to their products InsA and InsB, a third protein, InsAB, is jointly encoded by the two open reading frames of IS150 by means of remarkably efficient programmed translational frameshifting to the −1 phase that occurs within the region of the overlap (33) (Fig. 1A). If the classical concept of a gene is applied, IS150 thus possesses three genes, insA, insB, and the insAB gene. Synthesis of an equivalent InsAB transframe product has also been observed for other members of the IS3 family, and the sequence motif A6G/C could be defined as the frameshifting window (IS911 [17], IS3 [25], and IS2 [11]). This mode of frame splicing (namely, synthesis of one or two smaller proteins, each of which shares identity with the larger protein) is likely to be a common strategy in the IS3 family. The deduced InsA amino acid sequences exhibit a helix-turn-helix motif which is thought to recognize the terminal inverted repeats of the element (20) and at the C termini a leucine zipper motif which facilitates dimerization (9, 14). Within the InsB moiety there is a conserved amino acid motif, designated DDE or D,D35E, which has been shown to represent the catalytic site of the related integrases of retroviruses (1). Other common features of members of the IS3 family appear to be the potential to be excised as minicircles in which the terminal inverted repeats are separated by a characteristic number of base pairs of flanking sequences and the potential to generate linear IS molecules. Both types of molecules were, however, observed only in the absence of a functional InsA protein.
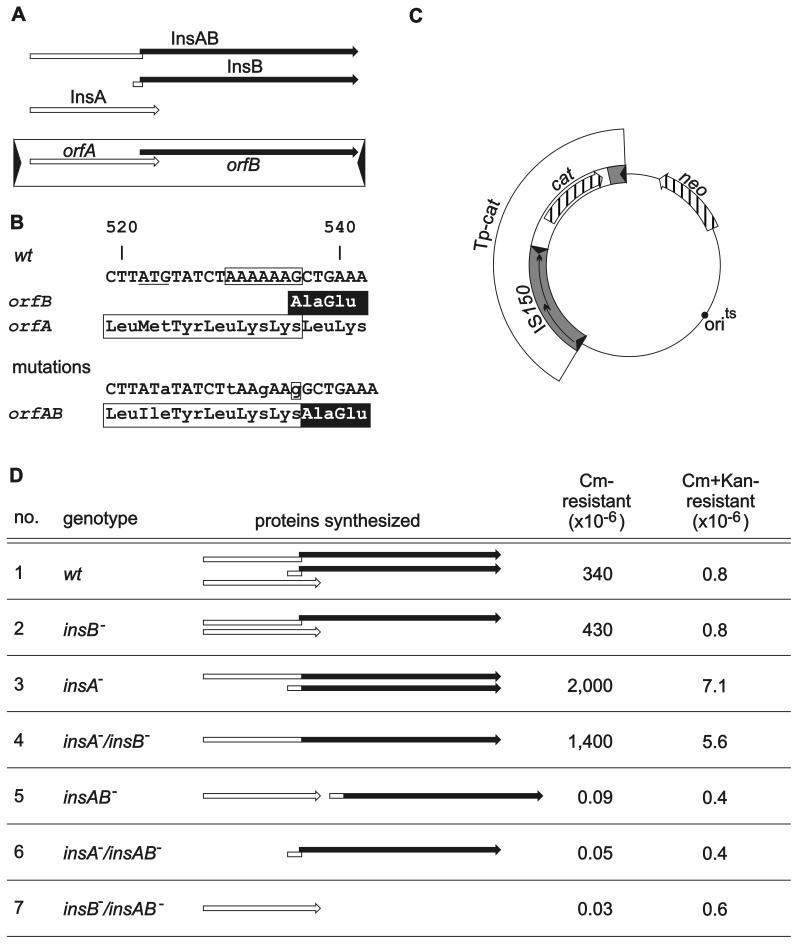
FIG. 1.
Genetic organization of IS150 (A), the mutations (B) introduced into an IS150 driven transposon (C), and the resulting transposition activities (D). (A) The element contains two overlapping open reading frames. The inverted repeats are represented by arrowheads. The three proteins synthesized are represented by the arrows above the element (open arrows, proteins encoded by orfA; solid arrows, proteins encoded by orfB), and the site of programmed translational frameshifting is represented by the offset. (B) At the top the DNA sequence of the region of frameshifting together with the amino acid sequence encoded by orfA (InsA protein) and the amino acid sequence jointly encoded by orfA and orfB after frameshifting (InsAB as well as InsB protein) is shown. The amino acid sequences of the transframe products are indicated by the box (sequence encoded by orfA) and by highlighting (sequence encoded by orfB). The A6G sequence essential for programmed frameshifting is enclosed in a box. The start codon of insB (ATG), arranged in the orfA phase, is underlined. At the bottom is a compilation of nucleotide exchanges and a 1-bp insertion introduced (i) to deactivate frameshifting, (ii) to fuse orfA and orfB, and (iii) to eliminate the insB start codon, together with the effect on the encoded amino acid sequence. Altered nucleotides are indicated by lowercase letters, and the G residue inserted is enclosed in a box. (C) Relevant structures of the IS150-driven transposon (TP-cat) together with the vector backbone encoding thermosensitive replication machinery and the neo gene as a selection marker. (D) Comparison of the transpositional activity of the TP-cat transposon carrying a wild-type IS150 element (wt) with the transpositional activities of transposons carrying various mutant elements. The IS150 genotypes are indicated on the left and are followed by schematic drawings of the different proteins synthesized by the various elements. The rates are the averages from four to six assays for at least two independent transformants. The deviations were less than 50% from the mean. Cm, chloramphenicol; Kan, kanamycin. The following plasmids were used: transposon 1, pFDX2339; transposon 2, pFDX4107; transposon 3, pFDX3949; transposon 4, pFDX3943; transposon 5, pFDX4104; transposon 6, pFDX4100; and transposon 7, pFDX4106.
In the case of IS150, synthesis of InsB is initiated at an AUG start codon within orfA and thus also requires translational frameshifting (33). While the InsAB protein turned out to represent the transposase in the cases that have been studied (IS2 [11], IS3 [25], and IS911 [17]), the in vivo role of the InsA and InsB proteins is less clear.
Here we report that circular and linear IS150 molecules are generated in vivo even from the wild-type element, further supporting the assumption that they represent natural intermediates in the transposition reaction. Molecular analysis of the linear and circular species indicates that the circular molecules likely represent the precursors of the linear forms in the transposition reaction. The lack of InsA led to a disproportionately large increase in formation of the linear species, while the rate of transposition increased only slightly. Elimination of synthesis of InsB resulted in no appreciable phenotype. We concluded that InsAB represents the transposase and that InsA may primarily function in coupling the formation of linear molecules to successive transposition.
MATERIALS AND METHODS
Plasmids and bacterial strains.
The relevant features of plasmids and the genotypes of strains are shown in Table 1. Plasmids were constructed by using standard recombinant techniques (21), and their complete annotated sequences, together with a short outline of their construction, can be retrieved from a website (http://www-rak.biologie.uni-freiburg.de/supplements/IS150-1.htm).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant structure and/or genotype | Reference or source |
|---|---|---|
| Strains | ||
| W3110 | F− λ− IN(rrnD-rrnE)I (= R1128) | 3 |
| R1607 | W3110 ΔlacI-lacZ lacY+ | K. Schnetz |
| R2124 | R1607 ΔIS150 | This study |
| R2129 | R2124 srl::Tn10 | This study |
| R2136 | R2124 recA | This study |
| Plasmids | ||
| pFDX2338 | neo, oriR pSC101-ts | This study |
| pFDX2339 | neo, oriR pSC101-ts, Tp-cat (IS150-cat-IS150 right end) | This study |
| pFDX3610 | tet, oriR pSC101-ts, tandem arrangement of adjacent sequences of the chromosomal IS150 copy | This study |
| pFDX3922 | neo, oriR pSC101-ts, IS150 | This study |
| pFDX3923 | pFDX3922 with an insA-insB-negative IS150 element | This study |
| pFDX3943 | pFDX2339 with an insA-insB-negative IS150 element | This study |
| pFDX3949 | pFDX2339 with an insA-negative IS150 element | This study |
| pFDX4100 | pFDX2339 with an insA-insAB-negative IS150 element | This study |
| pFDX4104 | pFDX2339 with an insAB-negative IS150 element | This study |
| pFDX4106 | pFDX2339 with an insB-insAB-negative IS150 element | This study |
| pFDX4107 | pFDX2339 with an insB-negative IS150 element | This study |
All of the bacterial strains used are derivatives of E. coli K-12. Strain W3110 carries a single chromosomal copy of IS150 (23). This copy was deleted in the Δ(lacI-lacZ) derivative R1607 according to Hamilton et al. (7) as follows. Strain R1607 was transformed with the temperature-sensitive plasmid pFDX3610, which carries 1,327 and 1,014 bp of the left and right sequences that immediately flank IS150 in the chromosome. The transformants were plated at a nonpermissive temperature (42°C) on Luria-Bertani (LB) medium plates containing tetracycline to select for integration of the plasmid into the chromosome by a single crossover event between the cloned sequences present on pFDX3610 and their identical chromosomal counterparts. The resulting colonies were inoculated into LB broth and incubated at 28°C to allow resolution of the cointegrates by a second crossover event, which could result either in deletion of IS150 or in reversion. The cultures were subsequently plated onto LB medium plates without tetracycline, and the plates were incubated at 42°C to allow segregation of the plasmids. Deletion derivatives were finally obtained by screening individual colonies for tetracycline sensitivity and for the presence of the correct deletion by PCR. This resulted in strain R2124, which was subsequently made recA by a two-step transduction. First, an srl::Tn10 allele was inserted by selection for tetracycline resistance. Subsequently, the recA allele was introduced by cotransduction with the wild-type srl gene and selection for growth on sorbitol, which resulted in strain R2136.
Growth conditions.
LB medium (15) was used as the standard medium. When necessary, antibiotics were added at final concentrations of 10 μg/ml (tetracycline), 20 μg/ml (chloramphenicol), and 40 μg/ml (kanamycin), unless otherwise indicated.
DNA preparation.
Alkaline lysis and subsequent plasmid purification by equilibrium centrifugation in CsCl-ethidium bromide density gradients were carried out by standard procedures as described previously (21). Cleared lysates were prepared as described by Clewell and Helinski (5), with the following modifications. Cleared lysates (4 ml) were treated with RNase A (final concentration, 60 μg/ml) on ice for 90 min. Sodium dodecyl sulfate (SDS) and proteinase K were added to final concentrations of 0.5% and 50 μg/ml, respectively. After incubation at 50°C for 30 min, samples were extracted with phenol-chloroform, and DNA was concentrated by isopropanol precipitation.
Labeling 5′ ends of linear IS150 molecules.
Labeling was accomplished by the exchange reaction (4) of T4 polynucleotide kinase. Three hundred femtomoles of DNA was mixed with 4 μl of [γ-32P]ATP (10 μCi/μl; Amersham Pharmacia Biotech) and 10 U of T4 polynucleotide kinase (New England Biolabs) in a final volume of 10 μl in the supplied buffer supplemented with 0.1 mM ADP. After incubation at 37°C for 45 min, samples were ethanol precipitated for further processing or mixed with 2 volumes of 1.5× denaturing buffer (1.5× Tris-borate-EDTA buffer, 40 mM EDTA, 10.5 M urea, 0.075% bromphenol blue). Four microliters was separated on a denaturing 16% polyacrylamide gel. Dried gels were exposed to X-ray film and to bioimaging plates and were analyzed by using a Molecular Imager FX (Bio-Rad) and the supplied software (Quantity one 4.1).
Southern hybridization.
Plasmid DNA was prepared by the cleared lysate method as described above. DNA samples that were to be hybridized were quantified on an analytical agarose gel by ethidium bromide staining of the samples and DNA standards of known concentrations. Subsequently, 100 fmol (325 ng) of plasmid DNA carrying the wild-type IS150 element and 1 fmol of plasmid DNA carrying the insA-negative element were separated on a 1% agarose gel. The upper part of the gel containing the plasmid DNA was removed before the DNA was transferred to a positively charged nylon membrane (Biodyne B; Life Technologies) as described previously (2). As a probe, a 238-bp PCR product (obtained with oligonucleotides 392 [5′-TCAATTGGAGTCAGACC-3′; IS150K:1365-1383] and 398 [5′-CGAATTCCCGGGATCAAAATATCCTTAAAGAAC-3′; IS150:1158-1178; heterologous nucleotides are underlined]) was labeled with [α-32P]dCTP (Amersham Pharmacia Biotech) by using the RadPrime DNA labeling system (Life Technologies). Hybridization was performed at 65°C overnight. The membranes were washed twice for 30 min at 65°C with 2× SSC-0.1% SDS and twice for 10 min at 65°C with 0.2× SSC-0.1% SDS, dried, autoradiographed, and quantified by bioimaging analysis as described above (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Transposition assay.
Transposition rates were determined essentially as described previously (33), except that cultures were grown at 28°C for 48 h before selective plating and chloramphenicol was added to LB medium plates to a final concentration of 30 μg/ml. In short, strain R2136 [ΔIS150 Δ(lacI-lacZ) recA] was transformed with plasmids containing the various IS150-driven cat transposons, thermosensitive replication machinery, and the neo gene as a selective marker. Transformants were selectively plated at 28°C to determine the titer and at 42°C to determine the numbers of singly and doubly resistant survivors. Chloramphenicol-resistant, kanamycin-sensitive colonies that appeared at 42°C represented simple transposition events.
RESULTS
Genetic analysis of IS150-encoded transposition functions.
In order to analyze the contributions of the different IS150-encoded gene products (Fig. 1A) to transpositional activity, we constructed mutant derivatives of IS150 which were defective in the individual insA, insB, and insAB genes. Expression of both insA and the transframe gene insAB at the same time was eliminated by a base substitution (G to A) within the shared ATG start codon. Synthesis of InsB was likewise eliminated by a substitution within the assigned start codon (Fig. 1B). The other mutations required manipulation of the frameshifting window within the region of overlap between orfA and orfB and are also shown in Fig. 1B. Synthesis of InsAB and InsB at the same time was eliminated by simultaneous replacement of the first and fourth A residues within the six-A sequence essential for translational frameshifting (34; unpublished data) by a T and a G, respectively. Additional insertion of a G residue downstream of this sequence led to an in-frame fusion of orfA and orfB which restored synthesis of InsAB and at the same time eliminated synthesis of InsA. Expression of insA and insB as an operon was achieved by cloning the two open reading frames of the in-frame fusion described above in tandem and providing the insB start codon with a ribosome binding site (AGGA). The various mutations were introduced into the IS150 moiety of an artificial transposon consisting of the cat gene (encoding chloramphenicol resistance) flanked by an entire IS150 on one side and by the right end of IS150, including its inverted repeat (IR-R), on the other side (Fig. 1C). These transposons were transferred onto a thermosensitive vector plasmid that is unable to replicate at 42°C and carries the neo gene conferring kanamycin resistance (Fig. 1C). The transpositional activity of the wild-type transposon was compared to that of mutant derivatives by determining the rate of chloramphenicol-resistant colony formers at 42°C. For this analysis we constructed a strain in which the copy of IS150 residing in the chromosome was deleted in order to eliminate any interference with the transposition assay by this copy.
The resulting transposition rates are shown in Fig. 1D. When the cat transposon flanked by the wild-type IS150 element was tested, the rate of singly resistant survivors on chloramphenicol plates exceeded that of doubly resistant colony formers by almost 500-fold, verifying the functionality of the transposon (Fig. 1D, lane 1). The absence of InsB caused no appreciable phenotype compared to the phenotype of the wild-type transposon. However, the absence of InsA led to ca. fivefold-enhanced transpositional activity (2 × 10−3) (Fig. 1D, lane 3). The rate did not change appreciably when in addition expression of insB was eliminated. The enhanced transpositional activities were paralleled by an increase in the formation of doubly resistant colonies. The possible causes of this increase are discussed below. However, the majority of the transposition events apparently were the result of cut-and-paste events.
All transposons that lacked InsAB (insAB negative, insA insAB negative, insB insAB negative) led to background rates that were less than 10−7. These events were not studied further. We concluded (i) that the InsAB protein is able to catalyze transposition without the aid of other element-encoded proteins and thus represents the transposase, (ii) that InsA may inhibit transposition (see below), and (iii) that InsB contributes no appreciable phenotype under our test conditions. Kanamycin-resistant, chloramphenicol-sensitive colonies were not observed in our assays (data not shown), verifying the transpositional functionality of the composite transposons.
In vivo generation of circular and linear IS150 molecules.
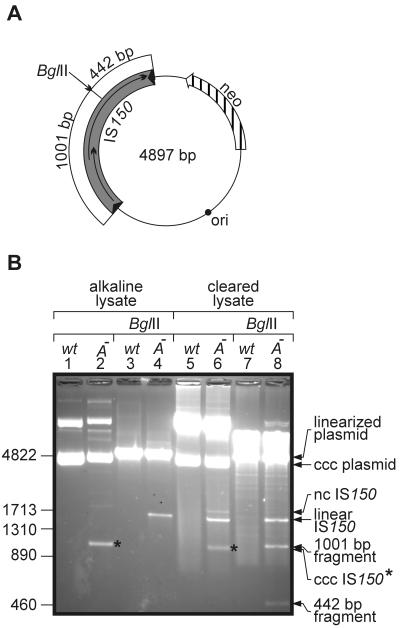
During our studies we detected DNA species in addition to the wild-type element when DNA preparations of plasmids carrying a copy of an insA-negative element were separated on agarose gels. Figure 2B, lanes 1 to 4, show examples of plasmid DNAs prepared by standard alkaline lysis and CsCl density gradient centrifugation, which should have yielded only closed circular DNA. Undigested DNA of the plasmid carrying the insA-negative element contained an additional species that migrated to a marker position at about 1,000 bp (Fig. 2B, compare lanes 1 and 2). This additional species shifted to about 1,450 bp when the DNA preparation was digested with BglII prior to gel electrophoresis (Fig. 2B, lane 4). BglII linearized the plasmid within the 1,443-bp IS150 (Fig. 2A). The decrease in migration velocity after digestion together with the apparent size suggested that the additional DNA species represented supercoiled minicircles consisting mainly of IS150. This suggestion was verified by additional restriction analyses (data not shown), as well as Southern hybridization and sequence analysis, as shown below.
FIG. 2.
Accumulation of low-molecular-weight DNA species generated in the presence of an insA-negative IS150 element. (A) Schematic drawing of plasmid used to transform strain R2124. Note that BglII cuts only once within the plasmid, at the site marked by an arrow, dividing IS150 into two arms consisting of 1,001 and 442 bp. (B) Agarose gel electrophoresis of plasmid DNA prepared from transformants harboring either plasmid pFDX3922 (wild-type IS150 element [wt]) or plasmid pFDX3923 (insA-insB-negative element [A−]). Plasmid DNAs were derived from alkaline lysates or cleared lysates, as indicated, and the DNAs, either undigested or after digestion with BglII, were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. The lengths of DNA standards (in base pairs) are indicated on the left. ccc, covalently closed circular; nc, nicked circle. The asterisks indicate the position of the covalently closed circular form of IS150 minicircles.
Faintly visible in the original of Fig. 2B, lane 2, was a DNA species that migrated at about 1,700 bp. From its position in the gel and because it disappeared upon digestion with BglII, we deduced that this DNA may represent nicked IS150 minicircles. The DNA samples used in the analysis were prepared by a standard method that involves alkaline denaturation and thus eliminates most of noncovalently closed DNA. In order to generally investigate the occurrence of DNA molecules other than covalently closed species, we repeated our analysis with DNA samples prepared by a cleared lysate method that does not include DNA denaturation steps. Separation of the samples is shown in Fig. 2B, lanes 5 to 8. Again, DNA carrying the insA-negative element contained the previously observed two forms of minicircles, the closed circular form and, this time more prominent, the nicked form, which were both absent in the wild-type sample (Fig. 2B, compare lanes 5 and 6). However, an additional species appeared with the mutant DNA, which was absent in the alkaline lysate (Fig. 2B, compare lanes 2 and 6) and had the same migration velocity as the linearized minicircle DNA (Fig. 2B, compare lanes 4 and 6), suggesting that it represented linear copies of IS150. This suggestion was supported by restriction analysis. When, for example, the DNA preparations were digested with BglII (Fig. 2B, lane 8), the two minicircle species (supercoiled and nicked circle) again migrated to the position expected for linear IS150 molecules. In addition, two bands, one at about 1,000 bp and, faintly visible, one at about 450 bp, appeared as expected after BglII digestion of a linear copy of IS150 (Fig. 2A). Identification of the ∼1,450-bp species present in the undigested preparation as linear IS150 molecules was confirmed by DNA sequencing (see below). In summary, we concluded that in the absence of the InsA protein large amounts of excised IS150 molecules that are linear and circular are generated in vivo.
Excision products are also generated by the wild-type element, but in different relative proportions.
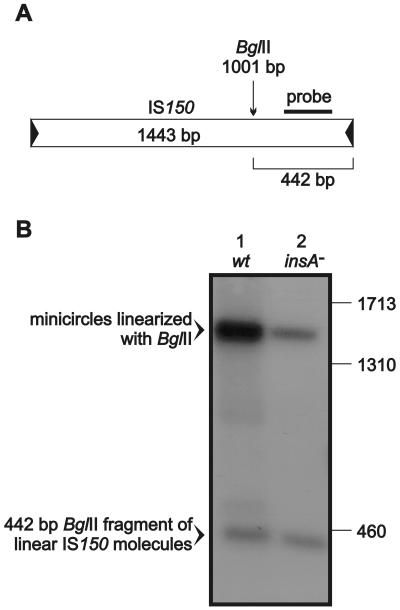
Next, we examined whether generation of the excision products is an artificial reaction that occurs only in the absence of InsA or whether it also takes place in the wild-type situation but at lower rates. As a first approach, we performed PCR using a pair of primers that were directed on both sides out of the element. Indeed, products could be generated in the presence of the wild-type element that corresponded in length to IS150 molecules with abutted left and right ends, which indicated that minicircles were present (data not shown). In order to quantitatively compare the generation of the different excision products from the wild-type element with the generation of the different excision products from the insA-negative derivative, we performed Southern hybridizations. Cleared lysate DNA preparations obtained from transformants harboring plasmids with the wild-type and insA-negative elements were digested with BglII (Fig. 3A), separated on an agarose gel, blotted, and hybridized. A 32P-labeled DNA fragment derived from the right arm of IS150 (Fig. 3A) served as a probe. The gel was loaded with a 100-fold excess of the wild-type DNA relative to the amount of the mutant DNA. The resulting autoradiograph (Fig. 3B) revealed that both linear and circular excision products were indeed generated from the wild-type element. However, phosphorimager analysis revealed that minicircles and linear molecules were generated from the insA-negative element in about equimolar amounts, while they were produced from the wild-type element at a 10:1 ratio. Moreover, the amount of linear molecules was ∼100 times larger (and the amount of minicircles was 10 times larger) in the mutant DNA preparation than in the wild-type preparation.
FIG. 3.
Southern hybridization demonstrating that IS150 minicircles and linear IS150 molecules are also generated from wild-type IS150 elements. (A) Design of the probe. Cleavage by BglII divides IS150 into two fragments. The 238-bp probe used should detect the 442-bp right fragment of linear IS150 molecules and a 1,443-bp fragment corresponding to linearized IS150 minicircles. (B) Plasmid DNA from cleared lysate preparations was digested with BglII prior to agarose gel electrophoresis, blotting, and hybridization. A 100-fold excess of plasmid pFDX3922 carrying the wild-type IS150 element (wt) relative to the amount of plasmid pFDX3923 carrying the insA-insB-negative element (insA−) was used (based on the amount of supercoiled and nicked circular plasmid DNA as described in Materials and Methods). The positions of marker bands (in base pairs) are indicated on the right. The strain used for transformation was R2136.
Structure of the termini of the linear IS150 molecules.
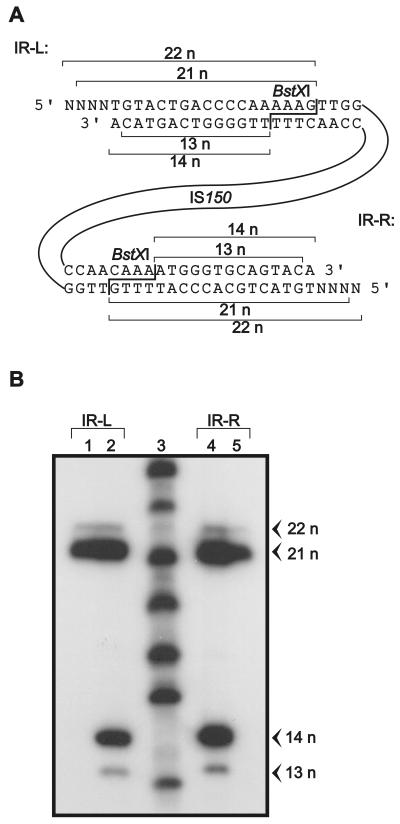
The presence of excision products in wild-type plasmid preparations suggested that they may represent intermediates in the transposition pathway. Knowledge of the exact molecular structure might therefore reveal details about the enzymatic reactions involved. For structural analysis of the termini of the linear excision products, we took advantage of the fact that the enzyme BstXI cleaves both inverted repeats of IS150 at identical positions. BstXI generates staggered cuts expected to release a 14-nucleotide fragment from each 3′ end and an 18-nucleotide fragment from each 5′ end when linear molecules are generated by blunt end cuts exactly at both termini (Fig. 4A). Determination of the lengths of these fragments should therefore reveal the exact positions of these cuts. To do this, we isolated the linear excision product from a gel, digested it with BglII, and separated the left arm of IS150 from the right arm on a second preparative gel. The arms were end labeled at their 5′ termini with 32P either before or after digestion with BstXI, and the resulting fragments were analyzed on a denaturing sequencing gel along with a ladder consisting of end-labeled oligonucleotides as markers. The resulting autoradiograph is shown in Fig. 4B. When labeling preceded digestion, only the terminal 5′ ends of the linear IS150 molecules were marked by labeling. This gave rise to a prominent band at 21 nucleotides and a minor band at 22 nucleotides with both arms (Fig. 4B, lanes 1 and 5), demonstrating that the two termini were processed identically. In the majority of cases the molecules possessed 3-nucleotide overhangs of flanking sequences at their 5′ ends, while a minor population (5%, as revealed by phosphorimager analysis) had 4-nucleotide protrusions (Fig. 4A shows the exact structure). When the arms were digested with BstXI prior to end labeling, the complementary strands were also labeled at the 5′ ends liberated by the enzyme. In this case two additional bands, a major band at 14 nucleotides and a minor band at 13 nucleotides, became apparent on the autoradiograph with both arms of IS150 (Fig. 4B, lanes 2 and 4). Again, the minor band accounted for 5% of the molecules. Thus, in 95% of all molecules the 3′ ends on both sides corresponded to the exact termini of the element, while in the other 5% they were recessed by one nucleotide (Fig. 4A). In summary, the data revealed that the majority of the linear IS150 molecules are molecules carrying three additional nucleotides at their 5′ termini. However, a minor fraction carry 4-base 5′ overhangs and/or are recessed by one nucleotide at their 3′ ends.
FIG. 4.
Molecular analysis of the lengths of the termini of the linear IS150 molecules. (A) Strategy and interpretation of the results. Restriction endonuclease BstXI cuts twice within IS150, once within each inverted repeat (IR-L and IR-R) and thus close to the termini, allowing precise determination of the lengths of the end fragments after digestion. (B) Determination of the lengths of BstXI-generated, 5′-end-labeled fragments derived from linear IS150 molecules. The linear species was isolated from a preparative agarose gel, and its left and right arms were separated by BglII digestion followed by another round of gel electrophoresis. Subsequently, the DNAs were end labeled with 32P either before (lanes 1 and 5) or after (lanes 2 and 4) digestion with BstXI, separated on a denaturing 16% acrylamide gel along with labeled oligonucleotides of known lengths, and autoradiographed. Labeling before digestion revealed the lengths of the 5′ end fragments of IS150, whereas labeling after digestion also revealed the lengths of the 3′ end fragments. The positions of the left inverted repeat (IR-L)-derived fragments (lanes 1 and 2) and the positions of the fragments derived from the right inverted repeat (IR-R) (lanes 4 and 5) are indicated in panel A. The source of the DNA was a cleared lysate of strain R2136 transformed with plasmid pFDX3923. n, nucleotides.
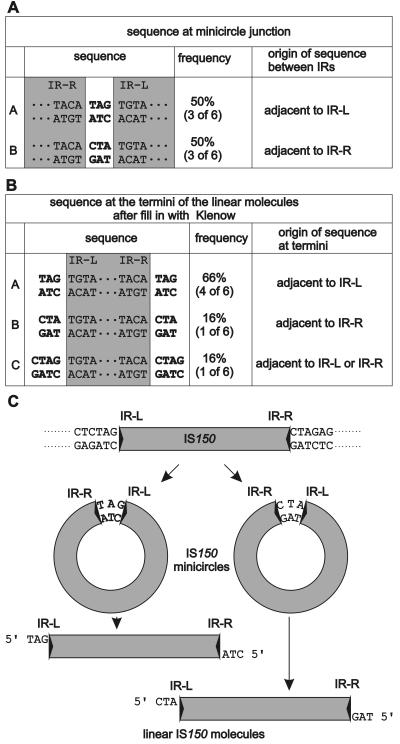
Minicircles likely represent the precursors of the linear excised IS150 molecules. In three cases the inverted repeats were separated by 3 bp originating from the vector sequences flanking the element's left end; in the other three cases they were separated by 3 bp originating from the vector sequences flanking the element's right end (Fig. 5C shows sequences of the flanking region). Essentially the same result was obtained when the junctions amplified by PCR from transformants carrying a plasmid with wild-type IS150 were analyzed. One of seven independent clones carried the 3 bp flanking the element's left border, while the other six clones contained the 3 bp flanking the element's right border (data not shown). Thus, no hybrid sequences consisting of nucleotides originating from the left and right flanking sequences were observed with both the wild-type element and the insA-negative derivative.
FIG. 5.
Sequences present between the abutted ends in minicircles (A) and at the termini of the linear IS150 molecules (B) and interpretation of the results (C). (A) Three base pairs (boldface type) bridge the inverted repeats (IRs) (shaded) in six cases analyzed. The data were derived from donor DNA immediately flanking the element on the right or left, as indicated. Minicircles generated from plasmid pFDX3923 were linearized with BglII, gel purified, cloned into the BamHI site of plasmid pUC19 (35), and sequenced across the circle junction with a primer located near the left inverted repeat (IR-L) and reading out of the element. IR-R, right inverted repeat. (B) Sequences found at the termini of linear IS150 molecules derived from the same preparation as in panel A after filling in of the recessed 3′ ends with Klenow polymerase and cloning into the SmaI site of plasmid pBluescript SK− (Stratagene). (C) Model explaining the precursor-product relationship of minicircles and linear molecules and summarizing the possible pathway of IS150 transposition. First, the element is removed from the donor DNA by an InsAB-catalyzed reaction. Evidence for the details of this reaction step came from analysis of figure-eight molecules that have been found in the case of IS2 (13) and IS911 (16) but were not detectable with IS150. In short, in these cases excision is initialized by a single-strand break catalyzed by the transposase that releases a 3′ hydroxyl end at one of the two inverted repeats, which subsequently attacks the donor DNA on the same strand three bases (and sometimes four bases) away from the other inverted repeat, resulting in circularization of this strand. An attack on the other strand, which may be catalyzed by host enzymes, leads to covalently closed double-stranded circles in which the inverted repeats are separated by a 3-bp segment that originates from either the left or the right of the donor DNA, as shown. These circles are then converted by two nicks to the linear forms that are integrated into the target DNA. At this stage the InsA protein may be important. It may partially inhibit this reaction until an interaction with target DNA ensures productive integration, and it may in addition take part in target acquisition, thus synchronizing these processes in order to prevent excess abortive formation of linear molecules.
To determine the terminal sequences of the linear excision products, the products were first cloned into a vector. Successful ligation required Klenow fill-in reactions but was not affected by dephosphorylation of the vector moiety, demonstrating that the 5′ ends of the linear IS150 molecules are phosphorylated. Six independent clones were used to determine the sequences that cover the junctions between the vector and the element (Fig. 5B). In all six cases the additional nucleotides attached to the 5′ end of the left terminus were complementary to those attached to the 5′ end of the right terminus. This demonstrated that linear molecules cannot be the products of an excision event due to staggered cuts at both ends within the donor DNA bearing the element but must be derived from the minicircles as a result of single-strand cuts occurring at both 3′ termini of IS150 (Fig. 5C). Thus, the linear molecules represent the products rather than the precursors of the minicircles. Interestingly, in one of the six cases the linear IS150 molecule was flanked by four nucleotides instead of three nucleotides on both sides. Due to the nature of this sequence we could not decide whether the nucleotides were derived from the left or right end of the flanking sequences. However, this variation points to some imprecision of the transposase, as indicated above. Whether this imprecision is due to the absence of the InsA protein in these experiments remains to be determined. It may be relevant, however, that upon transposition IS150 in most cases generates 3-bp direct repeats but occasionally generates 4-bp direct repeats (22; unpublished results).
DISCUSSION
Genetic analysis of the different gene products encoded by IS150 revealed that the transframe protein InsAB alone is necessary and sufficient to catalyze transposition, thus defining it as the element's transposase. This result is in accordance with data previously published for other members of the IS3 family of insertion sequences (see above). Also in agreement with these results is the finding that in the absence of InsA or its homologues large amounts of excision products accumulate in vivo. In the case of IS150 (Fig. 2), as well as IS2 (13), IS3 (25), and IS911 (18), the products were characterized as circular copies of the elements. In addition, linear IS molecules accumulated in the case of IS150 (Fig. 2), IS3 (24, 26), and IS911 (30).
A detailed analysis revealed that in the IS150 minicircles the inverted repeats of the element are abutted and are separated by a short DNA segment, which in most cases is 3 bp long but sometimes is 4 bp long, that originates from the donor sequences flanking the element either on its left side or on its right side (Fig. 5A). The linear molecules carry single-stranded 5′ extensions at both inverted repeats that terminate with a phosphate group and are again mainly 3 bases long but are sometimes 4 bases long (Fig. 4). These single-stranded extensions in turn originate from the donor sequences that flank the element either on its left side or on its right side (Fig. 5B). This finding indicates that the linear molecules cannot be the products of staggered double-strand breaks in the flanking donor DNA but must be derived from the circular species by two single-strand cuts, each occurring at one of the two 3′ ends of IS150. Thus, the linear species must represent the products rather than the precursors of the minicircles. Similar results have been obtained in the absence of the InsA homologue with IS3 (24) and also with IS911. In the latter case linear species were detected in vivo and could also be generated in vitro (30). A third species, so-called figure-eight molecules that represent half-excision events in which only one strand of the element has become circularized, has been detected with IS2 (13) and IS911 (16), but this species was not observed in our preparations. These molecules were interpreted as precursors of the minicircles.
The finding that in the case of IS150 both species were also generated from the wild-type element (Fig. 3) supports the previous hypothesis that they may not represent artifacts that are generated only in the absence of InsA but rather are true transpositional intermediates. Interestingly, the absence of the InsA protein caused a disproportionate accumulation of these excision products. The 10-fold-higher rate of accumulation of minicircles observed in the absence of InsA (Fig. 3) roughly corresponds to the 5-fold-higher transposition rate observed with the insA-negative transposon (Fig. 1D). The amount of the linear species, however, increased 100-fold when InsA was absent.
The increase in circle formation and the parallel increase in transpositional activity were not necessarily caused by the absence of the InsA protein. These changes could at least in part be attributed to the higher rate of transposase synthesis as a result of the type of mutations that had to be introduced to make the element insA negative. These included in-frame fusion of orfA to orfB and simultaneous elimination of frameshifting, which together caused all ribosomes initiating at the orfA reading frame to synthesize the InsAB protein. Frameshifting within the wild-type IS150 element takes place at a rate of about 50% (32; the rate of 35 to 38% described by Vögele [33] was later corrected). Thus, the mutations that eliminate synthesis of InsA at the same time should lead to a doubling of transposase synthesis (as was indeed revealed by SDS-polyacrylamide gel electrophoresis of radiolabeled proteins [data not shown]). Recently, similar mutations were introduced into an IS911-flanked transposon with similar effects. Transpositional activity was stimulated about 80-fold (6). Since the frameshifting rate in IS911 is only 12.5% (17), the mutations in this case should lead to eightfold transposase overexpression, which could also in this case explain, at least in part, the higher transposition rates.
The large increase in formation of linear molecules observed in the absence of InsA may indicate that the InsA protein acts mainly after minicircle formation. Thus, the main activity of InsA could be to inhibit the abortive formation of linear molecules and to couple the generation of these molecules to productive insertion events. In vitro results obtained with IS911 minicircles as the substrate revealed that the InsA homologue is able to stimulate full transposition of the minicircles into a target plasmid, demonstrating at the molecular level that this protein is indeed active after minicircles are generated (29). The stimulatory effect was observed up to a certain ratio of the InsA homologues to the InsAB homologues, after which InsA was inhibitory. In preliminary studies we observed similar effects in vivo, where small amounts of InsA protein further stimulated transposition of an insA-negative IS150 transposon (data not shown).
No function could be attributed to the insB gene product. This was surprising, because the gene is regulated in a complex manner at the level of translation initiation (32, 33). However, identical negative results were obtained in vivo with mutations in the homologous gene of IS911 (17), while IS2 apparently does not even express the corresponding reading frame (10, 11). Only in the case of IS3 could a function be assigned to the gene product. In this case, the InsB homologue acted together with the InsA homologue as a coinhibitor of transposition (27). However, in this test all IS3 genes were highly overexpressed. Thus, the biological relevance of these results remains unclear. An in vitro activity could be detected in the case of IS911, however, in which the protein catalyzed part of the reversal of figure-eight formation (19).
In most cases in which transposition functions of members of the IS3 family of insertion sequences have been studied, heterologous expression systems have been used, and the different gene products have been delivered in trans to artificial transposons consisting of inverted repeats that flanked either a reporter gene or a selection marker. While the data obtained in this way provided important contributions to our understanding of the transpositional processes, they are difficult to compare. Indeed, quite different results were obtained for the relative transposition rates when the InsAB homologues were synthesized alone and when all IS genes were expressed; the values ranged from 105-fold stimulation in the case of IS2 (13) to 160-fold stimulation in the case of IS3 (27) to 60-fold inhibition in the case of IS911 (28). It remains to be determined whether these huge differences reflect true differences between the elements studied. In fact, 80-fold stimulation instead of 60-fold inhibition has been observed for IS911 in a more recent study, in which the mutation knocking out InsA synthesis was directly introduced into an IS911 transposon (6). We also have seen variable trans effects when studying IS150 (data not shown). It therefore appears to be important to avoid any unnecessary alterations when the in vivo functions of the IS150 genes are examined genetically. We took care to manipulate IS150 as little as possible and within the transposon itself. The results obtained support the pathway for transposition of elements of the IS3 family as it is now generally believed and shown in Fig. 5C.
Because of the elevated rate of doubly resistant colony formers observed in the absence of InsA (Fig. 1D), it could be argued that a small fraction of the reactions proceed via a replicative mode of transposition which cannot a priori be excluded. However, these additional events may represent simple transposition from dimeric plasmid structures that form at low rates even in recA-negative strains and may subsequently give rise to IS tandem structures that are highly active in transposition (31) and from more complex structures that may arise from reactions between the donor plasmid and the large amount of minicircles and linear molecules present under these conditions.
The linear molecules obviously are not degraded in vivo. The accumulation of these molecules in large excess over the minicircles, as observed in the absence of the InsA protein, indicates that they cannot be ligated efficiently by the host ligase. Moreover, unlike transposition intermediates of IS10 (8) and IS1 (12), they did not induce the SOS response (data not shown). Thus, an interesting question remains concerning the nature of the termini of the linear molecules, which are possibly protected as nucleoprotein complexes, in vivo.
Acknowledgments
Several of the site-specific mutations resulting in copies of IS150 defective in the insA, insB, and insAB genes were constructed by Caroline Welz in one of the precursor plasmids used in this study. Caroline Welz also took part in the initial characterization of excision products, and her contribution is gratefully acknowledged. We thank Karin Schnetz for construction of strain R1607 and Johannes Hölzle for construction of plasmids pFDX2338 and pFDX2339. Gabor Igloi provided the oligonucleotides and did the DNA sequencing. We are grateful to Christoph F. Beck for careful reading of the manuscript.
This work was supported by the DFG through the Graduiertenkolleg Biochemie der Enzyme and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Asante-Appiah, E., and A. M. Skalka. 1997. Molecular mechanisms in retrovirus DNA integration. Antiviral Res. 36:139-156. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bachmann, B. 1972. Pedigrees of some mutant strains of Escherichia coli K12. Bacteriol. Rev. 36:525-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkner, K. L., and W. R. Folk. 1977. Polynucleotide kinase exchange reaction: quantitative assay for restriction endonuclease-generated 5′-phosphoroyl termini in DNA. J. Biol. Chem. 252:3176-3184. [PubMed] [Google Scholar]
- 5.Clewell, D. B., and D. R. Helinski. 1969. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc. Natl. Acad. Sci. USA 62:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval-Valentin, G., C. Normand, V. Khemici, B. Marty, and M. Chandler. 2001. Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J. 20:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haniford, D. B., A. R. Chelouche, and N. Kleckner. 1989. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell 59:385-394. [DOI] [PubMed] [Google Scholar]
- 9.Haren, L., P. Polard, B. Ton-Hoang, and M. Chandler. 1998. Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. J. Mol. Biol. 283:29-41. [DOI] [PubMed] [Google Scholar]
- 10.Hu, S. T., J. H. Hwang, L. C. Lee, C. H. Lee, P. L. Li, and Y. C. Hsieh. 1994. Functional analysis of the 14 kDa protein of insertion sequence 2. J. Mol. Biol. 236:503-513. [DOI] [PubMed] [Google Scholar]
- 11.Hu, S. T., L. C. Lee, and G. S. Lei. 1996. Detection of an IS2-encoded 46-kilodalton protein capable of binding terminal repeats of IS2. J. Bacteriol. 178:5652-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lane, D., J. Cavaille, and M. Chandler. 1994. Induction of the SOS response by IS1 transposase. J. Mol. Biol. 242:339-350. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, L. A., and N. D. Grindley. 1997. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol. Microbiol. 25:517-529. [DOI] [PubMed] [Google Scholar]
- 14.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Polard, P., and M. Chandler. 1995. Bacterial transposases and retroviral integrases. Mol. Microbiol. 15:13-23. [DOI] [PubMed] [Google Scholar]
- 17.Polard, P., M. F. Prere, M. Chandler, and O. Fayet. 1991. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol. 222:465-477. [DOI] [PubMed] [Google Scholar]
- 18.Polard, P., M. F. Prere, O. Fayet, and M. Chandler. 1992. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 11:5079-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polard, P., B. Ton-Hoang, L. Haren, M. Betermier, R. Walczak, and M. Chandler. 1996. IS911-mediated transpositional recombination in vitro. J. Mol. Biol. 264:68-81. [DOI] [PubMed] [Google Scholar]
- 20.Prere, M. F., M. Chandler, and O. Fayet. 1990. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J. Bacteriol. 172:4090-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Schwartz, E. 1988. Gal+-Revertanten der polaren Insertionsmutation galOP-306::IS1: Molekulare Analyse der Mutationsereignisse und Charakterisierung eines neuen mobilen Promotorelementes. Ph.D. thesis. Universität Freiburg, Freiburg, Germany.
- 23.Schwartz, E., M. Kroeger, and B. Rak. 1988. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 16:6789-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekine, Y., K. Aihara, and E. Ohtsubo. 1999. Linearization and transposition of circular molecules of insertion sequence IS3. J. Mol. Biol. 294:21-34. [DOI] [PubMed] [Google Scholar]
- 25.Sekine, Y., N. Eisaki, and E. Ohtsubo. 1994. Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235:1406-1420. [DOI] [PubMed] [Google Scholar]
- 26.Sekine, Y., N. Eisaki, and E. Ohtsubo. 1996. Identification and characterization of the linear IS3 molecules generated by staggered breaks. J. Biol. Chem. 271:197-202. [DOI] [PubMed] [Google Scholar]
- 27.Sekine, Y., K. Izumi, T. Mizuno, and E. Ohtsubo. 1997. Inhibition of transpositional recombination by OrfA and OrfB proteins encoded by insertion sequence IS3. Genes Cells 2:547-557. [DOI] [PubMed] [Google Scholar]
- 28.Ton-Hoang, B., M. Betermier, P. Polard, and M. Chandler. 1997. Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J. 16:3357-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ton-Hoang, B., P. Polard, and M. Chandler. 1998. Efficient transposition of IS911 circles in vitro. EMBO J. 17:1169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ton-Hoang, B., P. Polard, L. Haren, C. Turlan, and M. Chandler. 1999. IS911 transposon circles give rise to linear forms that can undergo integration in vitro. Mol. Microbiol. 32:617-627. [DOI] [PubMed] [Google Scholar]
- 31.Turlan, C., B. Ton-Hoang, and M. Chandler. 2000. The role of tandem IS dimers in IS911 transposition. Mol. Microbiol. 35:1312-1325. [DOI] [PubMed] [Google Scholar]
- 32.Vögele, K. 1993. Molekulare Anaylse der Genexpression des bakteriellen Insertionselementes IS150. Ph.D. thesis. Universität Freiburg, Freiburg, Germany.
- 33.Vögele, K., E. Schwartz, C. Welz, E. Schiltz, and B. Rak. 1991. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 19:4377-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welz, C. 1993. Funktionelle Analyse des bakteriellen Insertionselementes IS150. Ph.D. thesis. Universität Freiburg, Freiburg, Germany.
- 35.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]