Abstract
The complete genome sequences of two dairy phages, Streptococcus thermophilus phage 7201 and Lactobacillus casei phage A2, are reported. Comparative genomics reveals that both phages are members of the recently proposed Sfi21-like genus of Siphoviridae, a widely distributed phage type in low-GC-content gram-positive bacteria. Graded relatedness, the hallmark of evolving biological systems, was observed when different Sfi21-like phages were compared. Across the structural module, the graded relatedness was represented by a high level of DNA sequence similarity or protein sequence similarity, or a shared gene map in the absence of sequence relatedness. This varying range of relatedness was found within Sfi21-like phages from a single species as demonstrated by the different prophages harbored by Lactococcus lactis strain IL1403. A systematic dot plot analysis with 11 complete L. lactis phage genome sequences revealed a clear separation of all temperate phages from two classes of virulent phages. The temperate lactococcal phages share DNA sequence homology in a patchwise fashion over the nonstructural gene cluster. With respect to structural genes, four DNA homology groups could be defined within temperate L. lactis phages. Closely related structural modules for all four DNA homology groups were detected in phages from Streptococcus or Listeria, suggesting that they represent distinct evolutionary lineages that have not uniquely evolved in L. lactis. It seems reasonable to base phage taxonomy on data from comparative genomics. However, the peculiar modular nature of phage evolution creates ambiguities in the definition of phage taxa by comparative genomics. For example, depending on the module on which the classification is based, temperate lactococcal phages can be classified as a single phage species, as four distinct phage species, or as two if not three different phage genera. We propose to base phage taxonomy on comparative genomics of a single structural gene module (head or tail genes). This partially phylogeny-based taxonomical system still mirrors some aspects of the current International Committee on Taxonomy in Virology classification system. In this system the currently sequenced lactococcal phages would be grouped into five genera: c2-, sk1, Sfi11-, r1t-, and Sfi21-like phages.
Due to their economic impact on industrial milk fermentation, bacteriophages attacking dairy starter bacteria (Streptococcus thermophilus, Lactococcus lactis, and various species of Lactobacillus) became the focus of substantial research efforts which made them the best-investigated phage group with respect to complete genome sequences (10). Dairy phages are therefore a suitable test case for hypotheses on phage evolution and for a genomics-based phage taxonomy. The comparative analysis of S. thermophilus phage genomes has revealed a rather homogeneous group of phages with substantial DNA sequence sharing between all investigated temperate and virulent phage isolates (11). On the basis of the DNA packaging mechanisms two groups may be distinguished: cos site and pac site phages (35). These groups showed two clearly distinct structural gene clusters with phage isolates Sfi21 and Sfi11, respectively, as type strains (39). Comparative genomics revealed related phages in many other species and genera of low-GC-content gram-positive bacteria (Streptococcus pyogenes, Streptococcus pneumoniae, Lactococcus, Lactobacillus, Leuconostoc, Listeria, Staphylococcus, and Bacillus) (20). This observation led to the taxonomical proposal that Sfi21-like and Sfi11-like phages represent two new genera of phages in Siphoviridae. An intriguing observation was the fact that distinct Sfi21-like phage isolates were linked by a hierarchy of relatedness when S. thermophilus phages were used as a reference point (11). Sfi21-like phages from S. thermophilus were closely related at the DNA level essentially over the entire genome length (11); those from S. thermophilus and L. lactis shared a smaller degree of DNA identity limited to part of the structural genes (20); S. thermophilus and Lactobacillus gasseri Sfi21-like phages were related by a relatively high level of protein similarity over the structural genes, but significant DNA sequence homology was no longer detected (22). Notably, even Escherichia coli phage HK97 shared a functional gene map with Sfi21-like S. thermophilus phages over the structural genes in the absence of any sequence similarity (11). The observation of graded relatedness is a hallmark of evolving systems. Interestingly, the genetic diversity of Sfi21-like phages appears to increase between phages whose bacterial hosts are separated by greater phylogenetic distance. However, this observation might suffer from an observational bias by taking the very homogeneous S. thermophilus phage group as a reference point.
Comparative phage genomics opened the perspective of a natural taxonomy of Siphoviridae, i.e., implementation of a phage systematics that reflects (at least partially) the phylogenetic relationship between phages. To investigate this possibility in more detail for Sfi21-like Siphoviridae, we included further Sfi21-like phage genome sequences (S. thermophilus phage 7201 [51], Lactobacillus casei phage A2 [27, 28, 33, 46], and three L. lactis prophages [16]) into the comparative analysis. The analysis corroborated the proposal of a new Sfi21-like phage genus in lactic acid bacteria, yet the trend for a coevolution of phages and their host bacteria was not confirmed. Graded relatedness ranging from DNA, protein, and only functional gene map similarity for the structural genes was even observed for phages infecting a single host species, L. lactis. Comparative genomics of lactococcal phages also demonstrated a dilemma of phage taxonomy. On the basis of DNA homology, temperate lactococcal phages can be regarded as one or four phage species depending on whether the analysis is based on the nonstructural gene cluster or the structural gene cluster, respectively. On the basis of the mode of head morphogenesis, phage genomics can differentiate temperate lactococcal phages into at least two distinct phage genera widely distributed in low-GC-content gram-positive bacteria (Sfi21- and Sfi11-like phages). The reasons for this taxonomic dilemma and the prospect of a phage genomics-based taxonomy are discussed.
MATERIALS AND METHODS
All phage DNA sequences analyzed in the present report were retrieved from the database except for the complete sequence of L. casei phage A2. Nucleotide and predicted amino acid sequences were compared to those in the GenBank database. Additional database searches have been conducted using BLAST and PSI-BLAST (3) at the National Center for Biotechnology Information and FASTA (36). The dot plot matrix was calculated using DOTTER (50).
Phage A2 was propagated on L. casei ATCC 393 growing in MRS supplemented with 10 mM CaCl2 and MgSO4. Phage particles were purified by isopycnic centrifugation on a continuous CsCl gradient. DNase I-generated fragments were cloned in pUC18, and the DNA sequence was determined on both strands by the PCR cycle sequencing method (47). Gaps in the sequence were filled using PCR-amplified segments or direct sequencing using the whole phage DNA as a template.
Table 1 provides the pertinent characteristics of the phages investigated in this comparative study for an easier orientation.
TABLE 1.
Characterization of the bacteriophages discussed in the current study
| Bacterial host | Strain | Phage | Origin | Genome size (bp) | Old classification | New classification | Lytic or lysogenic | Reference |
|---|---|---|---|---|---|---|---|---|
| L. casei | ATCC 393 | A2 | Cheese | 43,411 | Unclassified | Sfi21 group | Lysogenic | This work |
| L. lactis subsp. cremoris | UC509 | Tuc2009 | Cheese | 38,347 | P335-type | Sfi11 group | Lysogenic | Accession no. AF109874a |
| H2L | BK5-T | Cheese | 40,003 | BK5-T | Sfi21 group | Lysogenic | 20 | |
| 901-1 | TP901-1 | Cheese | 37,667 | P335 type | Sfi11 group | Lysogenic | 9 | |
| R1 | r1t | Cheese | 33,350 | P335 type | r1t group | Lysogenic | 53 | |
| IL1403 | bIL67 | Cheese | 22,195 | c2 type | c2 group | Lytic | 49a | |
| L. lactis subsp. lactis | MG 1363 | c2 | Cheese | 22,163 | c2 type | c2 group | Lytic | 37 |
| L. lactis subsp. cremoris | HID113 | sk1 | Cheese | 28,451 | 936 type | sk1 group | Lytic | 15a |
| L. lactis subsp. lactis | IL1403 | bIL170 | Cheese | 31,754 | 936 type | sk1 group | Lytic | 17 |
| IL1403 | bIL309 | Cheese | 36,949 | P335 type | Sfi21 group | Lysogenic | 16 | |
| IL1403 | bIL285 | Cheese | 35,538 | P335 type | Sfi21 group | Lysogenic | 16 | |
| IL1403 | bIL286 | Cheese | 41,834 | P335 type | Sfi21 group | Lysogenic | 16 | |
| S. aureus | ATCC 49775 | PVL | PVL-producing S. aureus | 41,401 | Unclassified | Sfi21 group | Lysogenic | 32 |
| S. thermophilus | B106 | 7201 | Yogurt | 35,466 | Unclassified | Sfi21 group | Lytic | 35 |
| SFi19 | Sfi19 | Yogurt | 37,370 | Sfi21 group | Sfi21 group | Lytic | 41 | |
| SFi21 | Sfi21 | Yogurt | 40,739 | Sfi21 group | Sfi21 group | Lysogenic | 41 | |
| L. innocua | CLIP11262 | Prophage 5 | 36,779 | Unclassified | Sfi21 group | Lysogenic | 29 |
GenBank accession no.
Nucleotide sequence accession number
The complete sequence of L. casei phage A2 was submitted to GenBank under the accession number AJ251789.
RESULTS AND DISCUSSION
The wide range of Sfi21-like phages: the case of Lactobacillus phage A2.
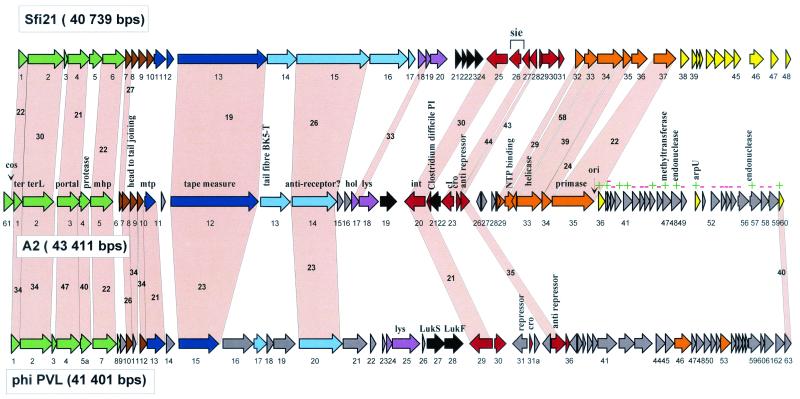
Until now only a single Sfi21-like phage was identified in the genus Lactobacillus. L. gasseri phage adh shared deduced amino acid sequence identity up to 61% with phage Sfi21 over the DNA packaging, head, tail, lysogeny, and DNA replication modules (22). The identical organization of the replication and lysogeny modules from L. casei phage A2 and phage Sfi21 (19, 28, 40, 46) suggested that phage A2 is a further member of this group. The sequencing of the A2 phage genome confirmed its attribution to the Sfi21-like phage group and demonstrated the wide distribution of this phage type in lactic acid bacteria. The predicted genetic map for the 43,411-bp-long phage genome and its sequence comparison to the genomes and encoded proteins of S. thermophilus phage Sfi21 (41) and the Staphylococcus aureus temperate phage PVL, another member of the Sfi21-like group (20, 32), are depicted in Fig. 1. In contrast to the phage-host coevolution hypothesis, phage A2 appears to be more closely related to Staphylococcus phage PVL than to Lactobacillus phage adh. With phage adh, A2 shared only three similar proteins across the complete, translated structural gene module (<30% identity at the deduced protein level).
FIG. 1.
Alignment of the genetic maps from the temperate cos site S. thermophilus phage Sfi21, L. casei phage A2, and S. aureus phage PVL, all members of the proposed Sfi21-like genus of Siphoviridae. The ORFs are color coded according to their predicted function deduced from a set of criteria consisting of either their database matches, the synteny argument with phage lambda (18), protein analysis (38), transcription analysis (55), or a mixture of these criteria. Green, predicted DNA packaging and head morphogenesis genes; brown, predicted head-to-tail joining genes; dark blue, tail morphogenesis genes; light blue, tail fiber genes; mauve, lysis genes; black, predicted lysogeny conversion genes; red, lysogeny genes; orange, DNA replication genes; yellow, transcription regulation genes (this region is relatively undefined in A2: ORFs with database match are denoted with a green +, and those lacking a match are denoted with a red −). Selected genes from the A2 phage genome are denoted. Genes encoding proteins that showed significant amino acid sequence similarity are linked by grey shading, and the percentage of amino acid identity is indicated. The phage genomes are depicted following the convention of placing the DNA packaging genes at the left end.
Below we provide a short comparative genome description for phage A2 (Fig. 1). The attribution of phage A2 to the Sfi21-like phage group is justified by sequence similarity of assumed DNA packaging, head morphogenesis, and two tail-associated proteins (tail tape measure and a putative antireceptor protein [23]) between A2 and Sfi21. Over the structural gene region, phage A2 is more closely related to Staphylococcus phage PVL than to Streptococcus phage Sfi21. This conclusion is based on a higher level of protein sequence similarity as well as a larger number of shared, homologous genes. Three observations are noteworthy: first, the putative joining gene (open reading frame 6 [ORF 6]) of A2 is shared with phage Sfi21 only, while homologues of the downstream located ORFs 7 to 9 and the major tail gene of A2 were only present in PVL. Second, at the corresponding position of the phage lambda tail genes G and T, only a single small ORF (ORF 11) without a database match is predicted for phage A2 as well as for the Sfi21 and PVL phages. Third, upstream of the genes predicted to represent the small and large terminase subunits (ORFs 1 and 2), a gene (ORF 61) was identified in phage A2 whose involvement in phage DNA packaging has been proven experimentally (27). No homologue of this gene is found at the equivalent position in other Sfi21-like phages (Fig. 1), while it was identified in L. casei phage PL-1.
Further deviations from the Sfi21 gene map were observed in A2. Of note are an insertion/deletion in the replication module (ORF 36 in Sfi21) and the insertion of the small ORF 21 upstream of the integrase gene in A2. The latter, ORF 21, is interesting in the controversy about the location of the excisionase gene in dairy phages, since in two lactococcal phages the excisionase gene was located one or two ORFs downstream of the cro-like gene (8; D. van Sinderen, unpublished data). However, no excisionase activity could be associated with this gene in vector integration experiments (4), and the location of the A2 excisionase is still unknown. Downstream of the replication module, two endonuclease genes (ORFs 48 et 57) are notable in A2. Related endonucleases are intron-associated in a number of phage systems (24). Also interesting is a methyltransferase gene in A2. Related methyltransferases were observed in Lactobacillus phage phig1e and Listeria phage A118, and they might be involved in the modification of phage and host DNA, but to our knowledge they are not involved in restriction-modification systems.
Also notable is an A2 gene (ORF 22) with links to the pathogenicity island of Clostridium difficile encoding toxin A. This gene is transcribed in the A2 prophage (33). Interestingly, the A2 gene is found at a position occupied by the superinfection exclusion gene (sie) in Sfi21. The sie gene is actually the most prominent transcript from the Sfi21 prophage (54). A sie function is not excluded for ORF 22 by the Clostridium link, since the predicted protein showed two membrane-spanning domains, which concurs with the data reported for the sie functions in lactococcal phages encoded at a corresponding genome position (44). ORF 19 without a database match is located between phage lysin and integrase genes. This region is occupied by toxin genes (ORF 27 and ORF 28) in phage PVL (32), and it is also one of the few transcribed regions in prophages from lactic streptococci (54). A2 ORFs 19 and 22 are, therefore, candidates for lysogenic conversion genes. While this position encodes superantigens, mitogenic factors, and toxins in pathogenic streptococci (S. pyogenes, Streptococcus equi) (21), no lysogenic phenotype has ever been demonstrated or searched for in dairy phages.
S. thermophilus phage 7201 shows graded relatedness with Sfi21-like streptococcal and lactococcal phages.
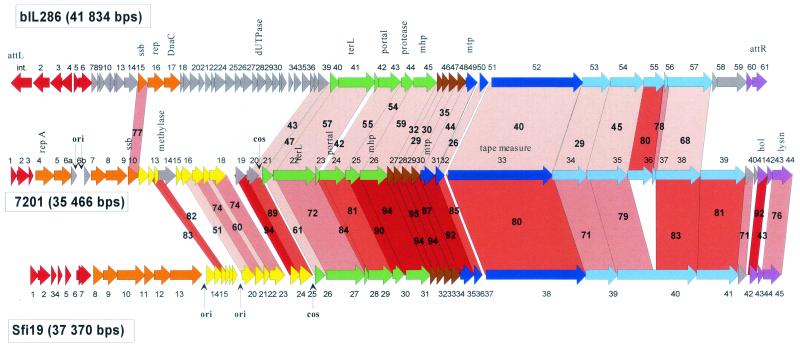
S. thermophilus phage 7201 is not inhibited by the Sfi21-type origin of phage replication (51). Since the DNA replication module is highly conserved in S. thermophilus phages (19), a distinct genome organization was suspected for phage 7201. The sequencing of the 35,466-bp-long phage 7201 genome revealed a distinct replication module in an otherwise standard Sfi21-like phage genome. The predicted genetic map is depicted in Fig. 2 and is compared to that of cos site virulent S. thermophilus phage Sfi19, an established member of the Sfi21 phage group (41). Note that the two virulent streptococcal phages are depicted in a hypothetical prophage configuration to facilitate the comparison with the L. lactis prophage bIL286 (top of Fig. 2). The database matches with phage DT1 and an S. thermophilus prophage remnant (54) identified the first three 7201 genes (ORFs 1, 2, and 3) as the lysogeny replacement module (Table 2). Over the next eight predicted genes (ORF 4 to 10), phage 7201 shared no similarity with any described S. thermophilus phages. However, with the exception of ORF 6a and 6b, the products of a DNA duplication event, all predicted proteins matched likely DNA replication proteins identified on plasmids or phages and prophages from pathogenic streptococci (51).
FIG. 2.
Alignment of the genetic maps from the L. lactis prophage bIL286 with the virulent S. thermophilus phages 7201 and SfiI9. The open reading frames are color coded according to their predicted function as in Fig. 1. Selected genes or genomic features are denoted. Genes encoding proteins that showed significant amino acid sequence similarity are linked by red shading, and the percentage of amino acid identity is indicated. The degree of amino acid identity (>90, >80, >70, and <70%) is reflected in the color intensity of the red shading. Note that the genomes of the virulent phages were rearranged to allow an easier comparison with the prophage bIL286. The natural ends of the streptococcal phage DNA flank the indicated cos sites.
TABLE 2.
Selected database matches for predicted genes from S. thermophilus phage 7201
| ORF | Match | Function | Best hit (% aa identity)a |
|---|---|---|---|
| 1 | S. thermophilus DT1 | cro-like | 94 |
| 2 | S. thermophilus Sfi16 remnant | Unknown | 90 |
| 3 | S. thermophilus DT1 | ORF 30 | 87 |
| 4 | S. aureus Mu50 | Replication initiator protein A (rep A) | 47 |
| 5 | L. lactis r1t | ORF 12 | 37 |
| 7 | S. pneumoniae MM1 | Erf like (ORF 9) | 34 |
| 8 | S. pneumoniae MM1 | ORF 10 | 35 |
| 9 | S. pyogenes SF370 | ssb (Spy 1830)b | 69 |
| 10 | S. pyogenes NIH1.1 | 45 | |
| 13 | L. lactis Tuc2009 | Putative methylase (ORF 18) | 63 |
| 14 | Mycobacteriophage L5 | Protein 64 | 49 |
aa, amino acid.
ssb, single-stranded-DNA binding protein.
For the remainder of the phage 7201 genome, covering ORF 11 to ORF 44, closely related genes were found at corresponding genome positions in the cos site S. thermophilus phages Sfi19. One exception is represented by ORFs 13 and 14, which showed a close match with a putative DNA methylase from L. lactis phage Tuc2009 and a putative transcriptional regulator from mycobacteriophage L5, respectively. The only other exception is provided by ORFs 36 and 37, which are closely related to corresponding genes in bIL286.
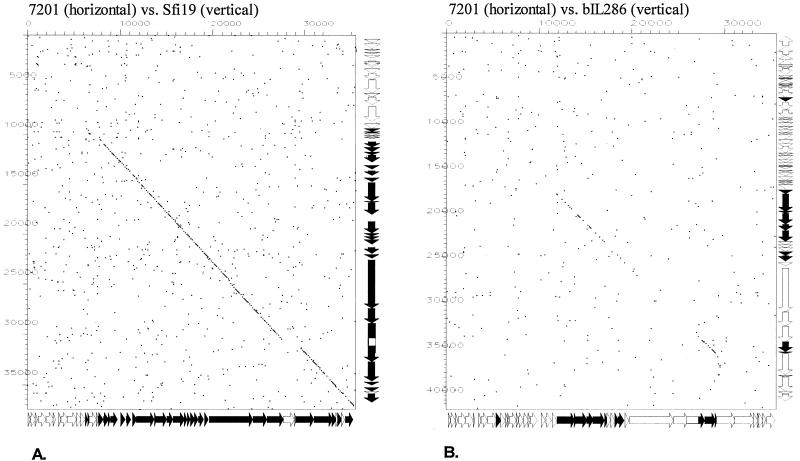
Sequence identity between S. thermophilus phages 7201 and Sfi19 was high and exceeded 80% at the amino acid level for proteins encoded by the head, tail, and part of the tail fiber genes (Fig. 2). At the DNA level, both phages could be aligned with few interruptions over three-quarters of the respective genomes (Fig. 3A). Proteins specified by the entire structural gene cluster of phage 7201 also displayed significant amino acid sequence similarity to the proteins encoded by the equivalent region of the Sfi21-like L. lactis prophage bIL286. However, the level of protein sequence identity between 7201 and bIL286 was lower than that between any two S. thermophilus phages belonging to the Sfi21 group. DNA sequence similarity between 7201 and bIL286 was limited to one DNA replication gene, several head genes, the genes involved in DNA packaging, the major tail gene, and two tail fiber genes (Fig. 3B). Due to the high level of sequence identity (85 and 80% base pair and amino acid identity, respectively, between 7201 ORF 36 and bIL286 ORF 55), this tail fiber gene is a candidate for lateral gene transfer between streptococcal and lactococcal phages.
FIG. 3.
Dot plot analysis for the S. thermophilus phage 7201 genome versus the S. thermophilus phage Sfi19 genome (A) and the S. thermophilus phage 7201 genome versus the L. lactis prophage bIL286 genome (B). The comparison window was 50 bp, and the stringency was 30 bp. Genes sharing nucleotide sequence similarity are highlighted in black on the gene maps (see Fig. 2 for an expanded view of the maps) of the phages depicted at the sides of the dot plot.
The 7201 sequence confirms the widespread distribution of Sfi21-like phages in S. thermophilus, and the sequence also confirmed the close link between streptococcal and lactococcal phages as already deduced from the Sfi21/L. lactis phage BK5-T phage comparison (20).
Dot plot matrix for lactococcal phages.
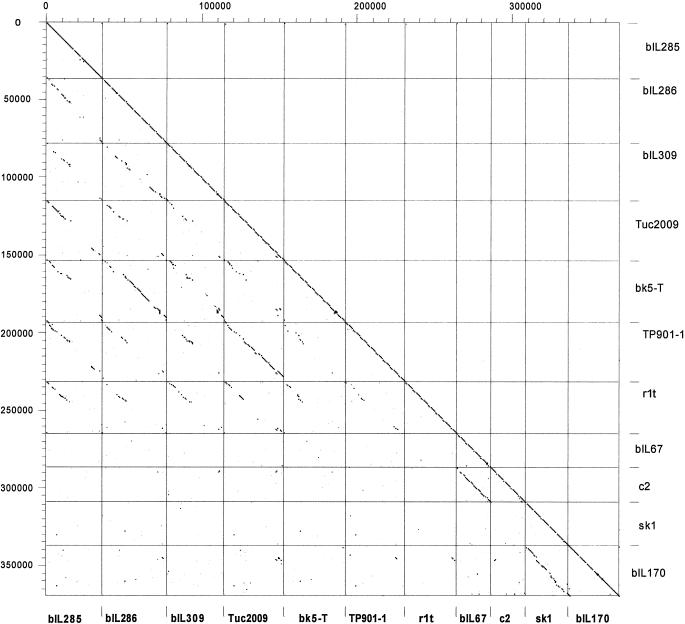
In apparent contradiction to the widespread distribution of Sfi21-like phages for S. thermophilus, Sfi21-/BK5-T-like L. lactis phages have only rarely been isolated from the cheese factory. However, Sfi21-like lactococcal phages were detected as prophages during the sequencing of the L. lactis strain IL1403 (16), increasing the number of complete lactococcal phage genomes to 11. It is, therefore, appropriate to study the DNA sequence relationships between all currently available L. lactis phages in the dot plot matrix displayed in Fig. 4. This analysis allows investigation into the genetic differences underlying the lactococcal phage species and genus classification established by phage taxonomists (7). Several observations are noteworthy. The virulent lactococcal phages represent two clearly separated groups with respect to DNA sequence relationships: c2-like phages (an International Committee on Taxonomy in Virology (ICTV)-accepted genus in Siphoviridae [43]) and sk1-like phages. Each group is represented by two sequenced members that share close sequence relationships with each other (c2 and bIL67 on one side and sk1 and bIL170 on the other side) but practically no DNA sequence relationships with any other group of lactococcal phages (Fig. 4). Both c2- and sk1-like phages are abundant for L. lactis, while phages with similar genome organization have, until now, not been isolated from any bacterial species other than L. lactis. Temperate lactococcal phages like r1t (53), Tuc2009 (J. F. M. L. Seegers, M. van de Guchte, M. Creaven, G. F. Fitzgerald, and D. van Sinderen, unpublished data), and TP901-1 (9) (currently classified as a P335 species of lactococcal phages) are also prominent factory isolates in contrast to temperate phages from the lactococcal BK5-T species (42). All temperate lactococcal phages are linked by DNA sequence relationships. This is most evident for the lysogeny, DNA replication, and putative transcriptional regulation genes, which are shared to a large extent by all seven temperate lactococcal phages (Fig. 4, diagonal line in top seven squares of leftmost column). However, with respect to DNA sequence homology over the structural genes, several groups could be differentiated within the temperate lactococcal phages. The first group is represented by BK5-T-like phages. This group contains three sequenced members: phages BK5-T, bIL286, and bIL309. They are clearly members of the proposed Sfi21-like genus of Siphoviridae. The second group is represented by phages Tuc2009 and TP901-1, while a third group is represented by a single phage isolate, r1t (Fig. 4). In the current taxonomic system of lactococcal phages, the latter two phage groups are classified together as P335 species. This classification leads to problems, however. Tuc 2009 and TP901-1 are pac site phages which are members of the recently proposed Sfi11-like genus of Siphoviridae (10, 11), which are as widely distributed in low-GC-content gram-positive bacteria as phages from the proposed Sfi21-like genus. In contrast, phage r1t is a cos site phage with a clearly distinct DNA sequence and genome organization of the structural gene module. Closely and distantly related phages were found in S. pyogenes and mycobacteria, respectively (20, 21), indicating that we might have here a further widely distributed structural gene module. Prophage bIL285 constitutes a fourth DNA homology group for the structural genes in temperate lactococcal phages. Therefore, at least six distinct structural modules can be defined for Siphoviridae from L. lactis phages, and more are likely to exist (e.g., in the 949 lactococcal phage species). What might be the reason for this much broader genetic diversity for phages from L. lactis than for those from S. thermophilus, which showed only two distinct structural modules for both virulent and temperate phages? Since L. lactis phages have been investigated for a longer time and by more laboratories, it might represent an observational bias. We judge this unlikely, since more than 200 S. thermophilus phage isolates have been investigated in several laboratories, revealing just two major phage types (12, 35). We prefer an alternative interpretation. Numerous plasmid-encoded phage inhibition systems have been described for L. lactis, while plasmids are rare for S. thermophilus, and even rarer are S. thermophilus plasmids that encode phage resistance mechanisms (26). Interestingly, one of the rare S. thermophilus plasmids (30) showed sequence similarity to the origin of replication of S. thermophilus phages (25). However, in contrast to the situation for E. coli, where phages can replicate as extrachromosomal elements (phage P1), no good evidence for prophage-like plasmids exists for lactic acid bacteria (15). L. lactis phages are therefore under a much greater selection pressure to diversify within the species or to accept “foreign” phage modules from the environment than S. thermophilus phages. We speculate that the latter process might be favored by the broader ecological range of L. lactis compared to S. thermophilus, which appears to be restricted to milk.
FIG. 4.
Dot plot matrix calculated for the complete genomes of the L. lactis prophages bIL285, bIL286, and bIL309, all from the genome sequence of L. lactis strain IL1403 (13), of the temperate L. lactis phages Tuc2009 (P335 lactococcal species, proposed Sfi11-like genus of pac site Siphoviridae), BK5-T (BK5-T lactococcal species, proposed Sfi21-like genus of cos site Siphoviridae), TP901-1 (P335 lactococcal species, proposed Sfi11-like genus of pac site Siphoviridae), r1t (P335 lactococcal species, new r1t/LC3-like genus (?) of cos site Siphoviridae) and of the virulent L. lactis phages bIL67 and c2 (both c2 lactococcal species, c2-like genus of Siphoviridae) and sk1 and bIL170 (936 species of lactococcal phages, new sk1-like genus (?) of Siphoviridae). The prophages and temperate phages are aligned so that the phage integrase is at the utmost left (top) of each indicated square, followed by the lysogeny and DNA replication modules; in the center are the structural genes (DNA packaging, head, and tail genes); at the right (bottom) of each indicated square are tail fiber and lysis genes. The virulent phages are given with the late structural genes at the right and the early nonstructural genes at the left. The BK5-T and c2 sequences were rearranged or inversed, respectively, with respect to the database entry. The left y axis provides a scale in kilobases; the right y axis and the bottom x axis identify the phage genomes that were compared in the corresponding square. The dot matrix was calculated using Dotter (50). The comparison window was 50 bp, and the stringency was 30 bp.
To explore the genetic diversity demonstrated by lactococcal phages, we performed a comparative genomics analysis for the L. lactis phages with an Sfi21-like gene map.
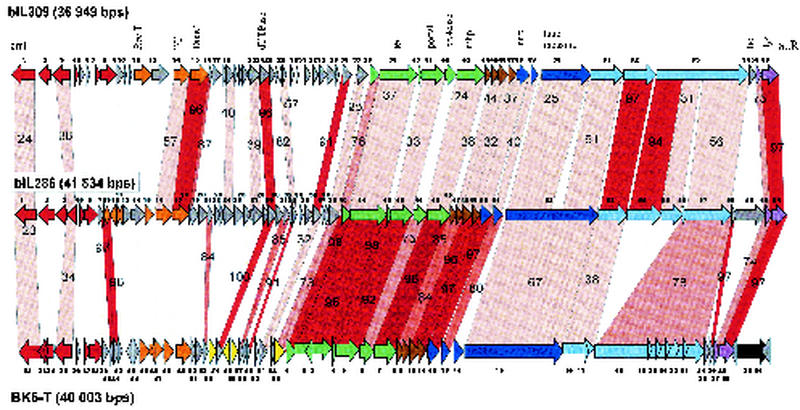
Prophages bIL286 and bIL309 are further examples of Sfi21-like L. lactis phages.
Over the entire late gene cluster, the L. lactis prophage bIL286 exhibited a high degree of protein sequence identity with phage BK5-T, an established member of the proposed Sfi21-like Siphoviridae genus (Fig. 5). In particular, the proteins encoded by the DNA packaging, major head and tail, and lysis genes were nearly identical between these two phages. Major differences between the late genes were limited to three regions. The first is the additional ORF 14 in BK5-T, located in a region where phage lambda shows frameshifting between the tail genes G and T. The putative tail fiber genes ORF 54 and 55 from bIL286 have no counterpart in BK5-T, while a gene specifying a closely related protein was detected in the L. lactis prophage bIL309. This is actually the second example where a single ORF in one phage has similarity to multiple ORFs in another phage (see also ORF 35 and 38 from 7201 compared to ORF 1291 in Sfi19) (Fig. 2). The large gene in phage Sfi19 encodes the likely receptor-recognizing tail fiber gene (39) and represents a multidomain protein, which is due to internal collagen-like repeats that are the target of recombination events (23, 39). It cannot be decided whether the fused or split genes represent the ancient gene constellation. A third difference was observed in the lysis module: bIL286 possesses a relatively large anonymous gene upstream of the holin gene (ORF 59), while a similarly sized gene was located downstream of the lysin gene in BK5-T (ORF 30). The latter gene is one of the few transcribed genes of the BK5-T prophage (6) and thus a candidate for a lysogenic conversion gene (54). Over the nonstructural gene region, only isolated genes shared sequence similarity between bIL286 and BK5-T.
FIG. 5.
Alignment of the genetic maps from the L. lactis prophage bIL309 with the L. lactis prophage bIL286 and temperate L. lactis phage BK5-T. The open reading frames are color coded according to their predicted function as in Fig. 1. Selected genes or genomic features are denoted. Genes encoding proteins that showed significant amino acid sequence are linked by red shading, and the percentage of amino acid identity is indicated. The degree of amino acid identity (>90, >80, >70, or <70%) is reflected in the color intensity of the red shading.
A qualitatively similar pattern of relatedness was observed when the L. lactis prophages bIL309 and bIL286 were compared (Fig. 5). However, important quantitative differences were observed. The degree of sequence similarity between bIL286 and bIL309 was, with only a few exceptions, lower than that between bIL286 and BK5-T. At the DNA level, sequence homologies were restricted to small genome segments, the longest covering two adjacent putative tail fiber genes (Fig. 4). In contrast, BK5-T and bIL286 phages shared DNA sequence similarity essentially over the entire structural gene cluster (Fig. 4).
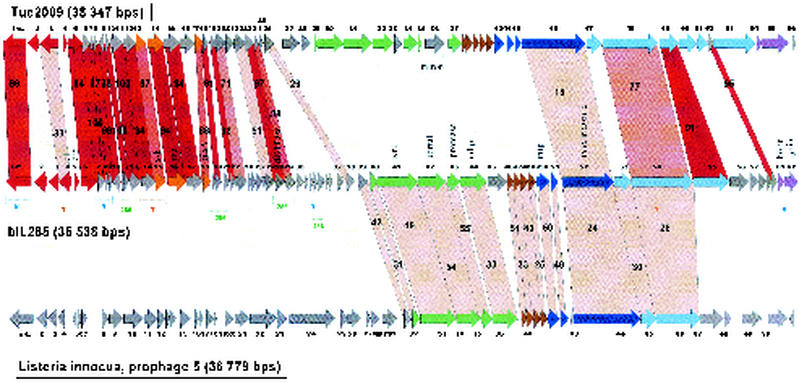
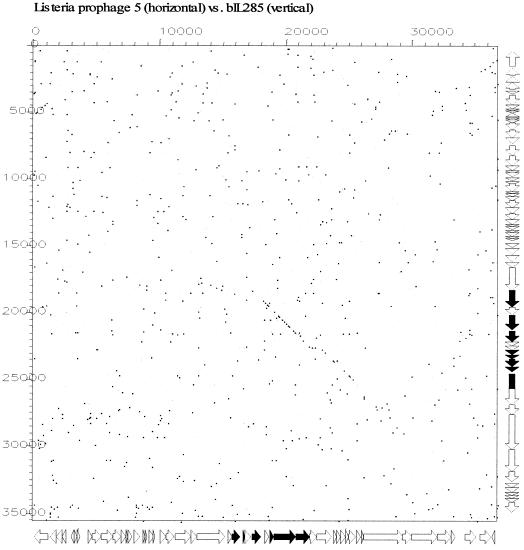
Prophages bIL285: a distant member of the Sfi21-like phage group.
An interesting case is presented by L. lactis prophage bIL285. Despite its lack of any DNA or protein sequence similarity with the structural gene cluster from any other sequenced lactococcal phage, the gene map suggested an Sfi21-like phage (Fig. 6). Diagnostic for this group is the protease/major head gene constellation presented by ORF 43 and ORF 44 in bIL285. Notably, the L. lactis prophage bIL285 and the Listeria innocua prophage 5 are closely related at the protein level for most of the predicted structural proteins. Exceptions were the bIL285 ORF 45 insertion/deletion and ORF 55, a probable tail fiber gene which we predict to be involved in host cell recognition since it is closely related to a corresponding gene in phage Tuc2009 (Fig. 6). The similarity between bIL285 and the Listeria prophage extended to the DNA sequence level (Fig. 7). This is unusual, since up to now DNA sequence similarity between phages from low-GC-content gram-positive bacteria have been limited to phages infecting the two closely related bacterial genera Streptococcus and Lactococcus (20, 21). Listeria, in contrast, is a more distant member of this branch of bacteria. Notably, lactococcal Sfi21-like phages cover genetic relationships ranging from close similarity at the DNA sequence level over relatedness at the protein sequence level to the sharing of a comparable gene map in the absence of any sequence similarity. This observation confirms the previously described graded relationships between phages in low-GC-content gram-positive bacteria and thus provides evidence for shared evolution of these phages that may be deciphered by comparative genomics. Apparently, at some point the Lactococcus, Listeria, and Streptococcus phages have been in the same host (which may be none of the above bacteria) and have shared their DNA. Since sequence relatives of the structural modules identified in all temperate lactococcal phages have also been found in Streptococcus and Listeria phages, it is unlikely that these structural modules have specifically evolved in the L. lactis species.
FIG. 6.
Alignment of the genetic maps from the temperate L. lactis phage Tuc2009 with L. lactis prophage bIL285 and L. innocua prophage 5. The open reading frames are color coded according to their predicted function as in Fig. 1. Selected genes from bIL285 are annotated. Genes encoding proteins that showed significant amino acid sequence similarity are linked by red shading, and the percentage of amino acid identity is indicated. The degree of amino acid identity (>90, >80, >70, or <70%) is reflected in the color intensity of the red shading. The brackets under the bIL285 gene map demarcate the patchwise sequence sharing of bIL285 genes with other lactococcal phages (B, BK5-T, T, TP901-1; 286, bIL286), possibly identifying genes whose proteins have to interact in a common function (see the text).
FIG. 7.
Dot plot analysis for the L. innocua prophage 5 (29) genome versus the L. lactis prophage bIL285 genome. The comparison window was 50 bp, and the stringency was 30 bp. Genes sharing nucleotide sequence similarity are highlighted in black on the gene maps (see Fig. 6 for an expanded view of the prophage maps) of the phages depicted at the sides of the dot plot.
The nonstructural gene module in temperate lactococcal phages: association of genes by comparative genomics—the case of bIL285.
All temperate lactococcal phages share substantial DNA sequence similarity over the nonstructural gene cluster. This sharing can be quite substantial as indicated in Fig. 6 for phages bIL285 and Tuc2009. However, the dot plot matrix demonstrated mainly a patchwise pattern of DNA sequence sharing for most phage comparisons. On the basis of the modular theory of phage evolution (5), one predicts involvement in related functions for those adjacent genes that share a high level of DNA sequence conservation in a context of sequence-unrelated genes. This rationale allows then a tentative subdivision of parts of the phage genome. The principle is illustrated for the lactococcal prophage bIL285 in Fig. 6. The comparison with L. lactis phage TP901-1 associated bIL285 ORFs 2, 3, 4, 5, and 6 (predicting lysogeny-related functions) and ORFs 14 and 15 (predicting related DNA replication functions), respectively, (Fig. 6, brackets T below bIL285 gene map) as partners in two distinct genetic functions. The comparison with BK5-T associated a different, but complementary, set of bIL285 ORFs (Fig. 6, brackets B below bIL285 gene map). From the latter comparison, ORFs 7, 8, and 9 are predicted to fulfill a common function (excisionase function to cooperate with the BK5-T like integrase?). Further groups of genes associated by prophage bIL286 comparisons are annotated in Fig. 6 (brackets 286 below bIL285 gene map in figure; unknown replication and transcription control functions?). While still conjectural, these associations provide a subdivision of the prophage genome organization that can serve as a basis for future genetic and biochemical experiments exploring the function of the predicted genes.
Outlook for phage taxonomy.
The current system from the ICTV classifies bacteriophages according to the type of genetic material (viruses with double- or single-stranded DNA or double- or single-stranded RNA) (43). This classification most likely represents phylogenetical distinct groups of phages. Within the group of bacteriophages with double-stranded DNA genomes a further subdivision is done according to morphology of the phage particle. A major group is the tailed phages of the order Caudovirales (1). This group represents the vast majority of all phages and is so characteristic that it is seen by many phage researchers as a monophyletic group. However, the contentious issue starts when the group of tailed phages is further subdivided into phage families. Currently, this division is mainly based on tail morphology (Myoviridae, Siphoviridae, and Podoviridae; phages with long contractile tails, with long noncontractile tails, and with short or no tails, respectively). Phage genomics has started to undermine this morphological subdivision, e.g., P22-like Podoviridae and lambda-like Siphoviridae share too similar a genome organization to justify their separation into distinct phage families (52).
The current taxonomic system has a hierarchical organization. Under the family classification come different phage genera (e.g., in Siphoviridae there are 6 genera accepted by ICTV: lambda-, T1-, T5-, L5-, c2-, and psiM1-like phages) and then phage species. It seems fair to say that the current phage classification system represents more our limited knowledge on the diversity of phages in nature than their true genetic diversity. Our limited knowledge is apparent at two levels. First, only a few detailed studies have been performed on the ecological complexity of phages in their natural environment (14), and second, we possess only a very small database of complete phage sequences. In addition, most sequences are from just three bacterial host systems: dairy phages, coliphages, and mycobacteriophages (13).
The present work suggests that a new phage genus should be added to the Siphoviridae family: the Sfi21-like phage genus. This group of phages is widely distributed in low-GC-content gram-positive bacteria, and it is characterized by a uniform genome organization (20). Sfi21-like phages infecting bacteria as different as lactic acid bacteria, staphylococci, listeria, bacilli, and clostridia are linked by numerous sequence similarities. The proposal of this new genus thus seems well founded.
However, the presented analysis goes beyond the proposal of a new phage genus—in fact, it questions some phage taxonomy principles. One of these principles is the species level of phage taxonomy. For example, the lactococcal phage species P335 (Table 1) and the BK5-T species share DNA sequence identity over the nonstructural genes (Fig. 4), which would justify their unification into one species. At the same time, similarities with streptococcal phages over the structural genes suggest a division of these temperate lactococcal phages into two distinct genera: according to their structural gene module phages, Tuc2009 and BK5-T should be classified into the proposed Sfi11-like and Sfi21-like genera, respectively, of Siphoviridae. We propose to replace the current lactococcal phage taxonomy (31) by one that is based on the genomic analysis of the structural gene cluster (Table 1).
This concurrent lumping and splitting of the same phage set is real and not compatible with a hierarchical taxonomical system. Phage species concepts should be the domain of geneticists and evolutionary biologists but not a prime concern of taxonomists (except for the notorious problem of weird naming of phage isolates). As already predicted by the modular theory of phage evolution (5) and vindicated by phage genomics, phages do not evolve along linear lines of descent (13). This is best illustrated by phages that are hybrids of currently distinguished phage families. For example, Shigella flexneri phage SfV is a lambda-like siphovirus with respect to its head genes but a Mu-like myovirus with respect to its tail genes (2). E. coli phage phi P27 is another example of such a lambda/Mu chimera (48). Comparative genomics has revealed prophages sharing head genes of a lambda-like siphovirus and tail genes of a P2-like myovirus (Xylella fastidiosa prophages XfP1 and XfP2). Comparative phage genomics has just started to delineate the evolutionary history of some individual phage modules, such as the structural gene module in Siphoviridae (11). The peculiar mode of phage evolution precludes depicting even in principle the genetic relationships between phages by a tree-like phylogeny (13). The development of web-like phylogenies will thus become necessary to illustrate evolutionary relationships between phages. However, the current phage taxonomy system is still based in its terminology and conception on Linnean thinking, and any hierarchical taxonomy will be unsuitable for biological systems demonstrating web-like phylogenies.
Some authors have argued that one should make a separation between taxonomy and phylogeny. The former revolves around the naming and does not necessarily have to deal with the phylogeny, although it traditionally does. One can therefore have taxonomy in the absence of phylogeny. Two solutions to phage taxonomy independent of phage phylogeny have been proposed. One is the phage proteomic tree, which classifies phage into taxa based on the overall similarity of about 100 completely sequenced phage genomes (49). The tree is the result of an objective calculation process conducted by a computer program and is applicable to any completely sequenced phage. Interestingly, the resulting taxa still matched relatively well the phage families and phage genera established by phage taxonomists. In another system, phages are classified into domains, divisions, families, and modi. The last are in some ways a shorthand description of the phage. For example, the above-mentioned SfV phage chimera is in this system characterized by the modi (i) HK97-like head genes, (ii) genes for Mu-like contractile tail, and (iii) integrase-mediated temperate phage (34). The usefulness of this approach will depend on the number of modi we will need to describe phages when we are confronting in the near future a thousand sequenced phage genomes. We propose a third approach to taxonomy based on the genomics analysis of a single, but prominent, phage module, which allows a rational hierarchical system (phages linked by different degrees of DNA sequence and then protein sequence identity and then sharing of a gene map in the absence of sequence similarity). There are some arguments in favor of basing a phage taxonomy system on the comparative genomics analysis of the DNA packaging-head gene cluster. The evolutionary analysis of the morphogenesis module is the most advanced of the different phage modules, and the structural gene order is apparently very conserved (11). Alternatively, one could base a phylogeny-inspired taxonomy system on phage tail genes (excluding tail fiber genes). This choice will assure a maximal overlap with the current phage taxonomy, which differentiates the majority of the isolated phages according to phage tail morphology. In fact, the hybrid phage SfV would have been classified as a myovirus by electron microscopy and a Mu-like myovirus by genomic analysis of the tail genes. The real taxonomic value of the different proposals will depend on the diversity of the bacteriophage genomes encountered in the environment. A systematic survey of prophages in genomes from low-GC-content gram-positive bacteria has up to now revealed only rather limited genetic diversity of the phage genomes (H. Brüssow, C. Canchaya, and C. Proux, submitted for publication). With respect to the structural genes, most could be classified as either Sfi21-like or Sfi11-like Siphoviridae, a few were r1t-like Siphoviridae, and further types were up to now limited to a single species (e.g., SPβ in Bacillus subtilis). In the sequenced genomes of gram-negative bacteria also, only a few prophage types could be differentiated with respect to the structural genes. Lambda-like, P2-like, P4-like, Mu-like, M13-like phages account for the majority of them (Brüssow et al., submitted). The taxonomy of temperate phages might thus not be a problem; however, only a minority of the phages have adopted the temperate lifestyle. In contrast, the sequencing of virulent phages still surprises us with genomes which have few if any database matches (45). Only the sequencing of more and ecologically representative virulent phages will show us if we reach some saturation of the sequence space or if we are confronted with a bewildering diversity of phage genomes which will complicate all approaches to phage taxonomy.
Acknowledgments
We are grateful for PB-EXP01-23 funding from FICYT (principado de Asturias) and the project BIO2201-3621 from CICYT (Spanish Ministry of Science and Technology).
REFERENCES
- 1.Ackermann, H.-W. 1999. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez, M. A., M. Herrero, and J. E. Suarez. 1998. The site-specific recombination of the Lactobacillus species bacteriophage A2 integrates in Gram-positive and Gram-negative bacteria. Virology 250:185-193. [DOI] [PubMed] [Google Scholar]
- 5.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-491. [DOI] [PubMed] [Google Scholar]
- 6.Boyce, J. D., B. E. Davidson, and A. J. Hillier. 1995. Identification of prophage genes expressed in lysogens of the Lactococcus lactis bacteriophage BK5-T. Appl. Environ. Microbiol. 61:4099-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, V., S. Hertwig, H. Neve, A. Geis, and M. Teuber. 1989. Taxonomical differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization and protein profiles. J. Gen. Microbiol. 135:2551-2560. [Google Scholar]
- 8.Breüner, A., L. Bronsted, and K. Hammer. 1999. Novel organization of genes involved in prophage excision identified in the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7291-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronsted, L., S. Ostergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 10.Brüssow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 11.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 12.Brüssow, H., M. Frémont, A. Bruttin, J. Sidoti, A. Constable, and V. Fryder. 1994. Detection and classification of Streptococcus thermophilus bacteriophages isolated from industrial milk fermentation. Appl. Environ. Microbiol. 60:4537-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 14.Bruttin, A., F. Desiere, N. d'Amico, J.-P. Guérin, J. Sidoti, B. Huni, S. Lucchini, and H. Brüssow. 1997. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burrus, V., Y. Roussel, B. Decaris, and G. Guédon. 2000. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 66:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Chandry, P. S., S. D. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcs lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 16.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crutz-Le Coq, A.-M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 18.Desiere, F., S. Lucchini, and H. Brüssow. 1999. Comparative sequence analysis of the DNA packaging, head, and tail morphogenesis modules in the temperate cos-site Streptococcus thermophilus bacteriophage Sfi21. Virology 260:244-253. [DOI] [PubMed] [Google Scholar]
- 19.Desiere, F., S. Lucchini, A. Bruttin, M.-C. Zwahlen, and H. Brüssow. 1997. A highly conserved DNA replication module from Streptococcus thermophilus phages is similar in sequence and topology to a module from Lactococcus lactis phages. Virology 234:372-382. [DOI] [PubMed] [Google Scholar]
- 20.Desiere, F., C. Mahanivong, A. J. Hillier, P. S. Chandry, B. E. Davidson, and H. Brüssow. 2001. Comparative genomics of lactococcal phages: insight from the complete genome sequence of Lactococcus lactis phage BK5-T. Virology 283:240-252. [DOI] [PubMed] [Google Scholar]
- 21.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brüssow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 22.Desiere, F., D. Pridmore, and H. Brüssow. 2000. Comparative genomics of the late gene cluster from Lactobacillus phages. Virology 275:294-305. [DOI] [PubMed] [Google Scholar]
- 23.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 24.Foley, S., A. Bruttin, and H. Brüssow. 2000. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J. Virol. 74:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foley, S., S. Lucchini, M.-C. Zwahlen, and H. Brüssow. 1998. A short noncoding viral DNA element showing characteristics of a replication origin confers bacteriophage resistance to Streptococcus thermophilus. Virology 250:377-387. [DOI] [PubMed] [Google Scholar]
- 26.Forde, A., and G. F. Fitzgerald. 1999. Bacteriophage defence systems in lactic acid bacteria. Antonie Leeuwenhoek 76:89-113. [PubMed] [Google Scholar]
- 27.Garcia, P., J. C. Alonso, and J. E. Suarez. 1997. Molecular analysis of the cos region of the Lactobacillus casei bacteriophage A2. Gene product 3, gp3, specifically binds to its downstream cos region. Mol. Microbiol. 23:505-514. [DOI] [PubMed] [Google Scholar]
- 28.Garcia, P., V. Ladero, J. C. Alonso, and J. E. Suarez. 1999. Cooperative interaction of CI protein regulates lysogeny of Lactobacillus casei by bacteriophage A2. J. Virol. 73:3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 30.Janzen, T., J. Kleinschmidt, H. Neve, and A. Geis. 1992. Sequencing and characterisation of pST1, a cryptic plasmid from Streptococcus thermophilus. FEMS Microbiol. Lett. 95:175-180. [DOI] [PubMed] [Google Scholar]
- 31.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko, J., T. Kimura, S. Narita, T. Tomita, and Y. Kamio. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene 215:57-67. [DOI] [PubMed] [Google Scholar]
- 33.Ladero, V., P. Garcia, V. Bascaran, M. Herrero, M. A. Alvarez, and J. E. Suarez. 1998. Identification of the repressor-encoding gene of the Lactobacillus bacteriophage A2. J. Bacteriol. 180:3474-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. The imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Marrec, C., D. van Sinderen, L. Walsh, E. Stanley, E. Vlegels, S. Moineau, P. Heinze, G. Fitzgerald, and B. Fayard. 1997. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 63:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 37.Lubbers, M. W., N. R. Waterfield, T. P. J. Beresford, R. W. F. Le Page, and A. W. Jarvis. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lucchini, S., F. Desiere, and H. Brüssow. 1998. Evolution of Streptococcus thermophilus bacteriophage genomes by modular exchanges followed by point mutations and small deletions and insertions. Virology 241:345-356. [DOI] [PubMed] [Google Scholar]
- 39.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution model. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Similarly organized lysogeny modules in temperate Siphoviridae from low GC content Gram-positive bacteria. Virology 263:427-435. [DOI] [PubMed] [Google Scholar]
- 41.Lucchini, S., F. Desiere, and H. Brüssow. 1999. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260:232-243. [DOI] [PubMed] [Google Scholar]
- 42.Mahanivong, C., J. D. Boyce, B. E. Davidson, and A. J. Hillier. 2001. Sequence analysis and molecular characterization of the Lactococcus lactis temperate bacteriophage BK5-T. Appl. Environ. Microbiol. 67:3564-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maniloff, J., and H.-W. Ackermann. 1998. Taxonomy of bacterial viruses: establishment of tailed virus genera and the order Caudovirales. Arch. Virol. 143:2051-2063. [DOI] [PubMed] [Google Scholar]
- 44.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2002. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43:509-520. [DOI] [PubMed] [Google Scholar]
- 45.Mesyanzhinov, V. V., J. Robben, B. Grymonprez, V. A. Kostyuchenko, M. V. Bourkaltseva, N. N. Sykilinda, V. N. Krylov, and G. Volckaert. 2002. The genome of bacteriophage φKZ of Pseudomonas aeruginosa. J. Mol. Biol. 17:1-19. [DOI] [PubMed] [Google Scholar]
- 46.Moscoso, M., and J. E. Suarez. 2000. Characterization of the DNA replication module of bacteriophage A2 and use of its origin of replication as a defense against infection during milk fermentation by Lactobacillus casei. Virology 273:101-111. [DOI] [PubMed] [Google Scholar]
- 47.Murray, V. 1989. Improved double-stranded DNA sequencing using the linear polymerase chain reaction. Nucleic Acids Res. 17:8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage φP27 is not related to other STx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohwer, F., and R. Edwards. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed]
- 49a.Schouler, C., S. D. Ehrlich, and M.-C. Chopin. 1994. Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology 140:3061-3069. [DOI] [PubMed] [Google Scholar]
- 50.Sonnhammer, E. L., and R. Durbin. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1-10. [DOI] [PubMed] [Google Scholar]
- 51.Stanley, E., L. Walsh, A. van der Zwet, G. F. Fitzgerald, and D. van Sinderen. 2000. Identification of four loci isolated from two Streptococcus thermophilus phage genomes responsible for mediating bacteriophage resistance. FEMS Microbiol. Lett. 182:271-277. [DOI] [PubMed] [Google Scholar]
- 52.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella phage P22. J. Bacteriol. 182:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Sinderen, D., H. Karsens, J. Kok, P. Terpstra, M. H. J. Ruiters, G. Veneema, and A. Nauta. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343-1355. [DOI] [PubMed] [Google Scholar]
- 54.Ventura, M., A. Bruttin, C. Canchaya, and H. Brüssow. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology, in press. [DOI] [PubMed]
- 55.Ventura, M., S. Foley, A. Bruttin, S. Chibani Chennoufi, C. Canchaya, and H. Brüssow. 2002. Transcription mapping as a tool in phage genomics: the case of the temperate Streptococcus thermophilus phage Sfi21. Virology 296:62-76. [DOI] [PubMed] [Google Scholar]