Abstract
Ferric uptake repressor (Fur) proteins regulate the expression of iron homeostasis genes in response to intracellular iron levels. In general, Fur proteins bind with high affinity to a 19-bp inverted repeat sequence known as the Fur box. An alignment of 19 operator sites recognized by Bacillus subtilis Fur revealed a different conserved 15-bp (7-1-7) inverted repeat present twice within this 19-bp consensus sequence. We demonstrated using electrophoretic mobility shift assays that this 7-1-7 inverted repeat comprises a minimal recognition site for high-affinity binding by Fur. The resulting revised consensus sequence is remarkably similar to a related 7-1-7 inverted repeat sequence recognized by PerR, a Fur paralog. Our analysis of the affinity and stoichiometry of DNA binding by B. subtilis Fur, together with a reinterpretation of previously described studies of Escherichia coli Fur, supports a model in which the 19-bp Fur box represents overlapping recognition sites for two Fur dimers bound to opposite faces of the DNA helix. The resulting recognition complex is reminiscent of that observed for the functionally related protein DtxR. Like Fur, DtxR contains a helix-turn-helix DNA-binding motif, recognizes a 19-bp inverted repeat sequence, and has a typical DNase I footprint of ∼30 bp. By envisioning a similar mode of DNA recognition for Fur, we can account for the internal symmetries noted previously within the Fur box, the tendency of Fur to extend into adjacent regions of DNA in a sequence-selective manner, and the observed patterns of DNA protection against enzymatic and chemical probes.
Escherichia coli Fur (ferric uptake repressor) is the prototype for a large and growing family of metalloregulatory proteins (15). While these proteins were originally recognized for their role in coordinating the expression of iron uptake functions in response to iron availability, it is now appreciated that Fur homologs may also have other functions. For example, in Bacillus subtilis there are three Fur homologs, the ferric uptake repressor (Fur), a zinc uptake repressor (Zur), and the peroxide regulon repressor (PerR) (5, 18). All three proteins require a bound divalent metal ion in order to bind DNA; Fur responds in vivo to iron, Zur responds to zinc, and PerR responds to either iron or manganese. We have not yet discovered any examples of cross-recognition of DNA targets among these three paralogs, suggesting that they control mutually exclusive regulons (22).
The molecular basis for DNA recognition by Fur has been controversial. Studies of E. coli originally led to the proposal that Fur recognizes a 19-bp inverted repeat sequence designated the Fur box (GATAATGATAATCATTATC) (12). Studies in which synthetic oligonucleotides were used confirmed that this sequence is sufficient for Fur-mediated repression (6). Moreover, closely related sequences are found in Fur-regulated control regions in a variety of organisms. Indeed, a perfect Fur box consensus sequence is associated with the bacillibactin (dihydroxybenzoate) siderophore (dhb) operon in B. subtilis and is bound with high affinity by Fur (4, 31). A search of the B. subtilis genome with this sequence identified numerous operons associated with known or putative iron uptake functions (21), many of which are now known to be controlled by Fur (1). Similarly, genome searches by using a weight matrix based on this 19-bp consensus sequence have identified numerous likely Fur-regulated operons in several proteobacterial species (29).
While the evidence linking this 19-bp inverted repeat to recognition by Fur is quite compelling, it is difficult to explain how a small dimeric DNA-binding protein like Fur can interact with such an extended operator region. Most proteins that utilize a helix-turn-helix (HTH) DNA-binding motif interact with operators closer to 12 bp long than to 19 bp long (20). Similarly, the extent of the Fur interaction with DNA, as visualized by using DNase I footprinting, is typically 30 bp and corresponds to three turns of the helix rather than the expected two turns of the helix. Finally, Fur and Fur homologs often display extended regions of DNA binding, particularly at higher concentrations of protein (27). The relationship between the 19-bp Fur box consensus sequence and the tendency of Fur to polymerize on the DNA has not been resolved.
In an attempt to explain some of these unusual features, Escolar et al. proposed an alternative view of the Fur-DNA interaction (13). They noted that the 19-bp Fur box can also be viewed as a head-to-head-to-tail repeat of a simple hexamer, GATAAT. In studies performed with synthetic oligonucleotides, they demonstrated that Fur bound tightly to repeated arrays of hexamers as long as they contained a minimum of three GATAAT motifs. They therefore proposed a model in which GATAAT is the minimal recognition unit for Fur. According to this model, the symmetric AT-AT core within each hexamer could potentially interact with a dimeric Fur protein, and a minimum of three bound dimers bind cooperatively along the DNA to arrays of repeated sequences. Escolar et al. noted that this mode of DNA binding is virtually unknown among prokaryotic regulators and resembles instead the binding of Zn finger-containing proteins in eukaryotic systems (13, 15).
Here we provide evidence for a revised view of Fur-DNA interactions in which each 19-bp Fur box corresponds to two overlapping inverted repeats that each bind a Fur dimer. This model was derived from an alignment of recently characterized DNA-binding sites for B. subtilis Fur (1) and the remarkable similarity between the deduced consensus sequence and the related sequence recognized by PerR, a Fur paralog. According to this model, each dimeric HTH-containing protein recognizes sequences in adjacent major grooves. The two dimers recognize sequences offset by 6 bp and are therefore envisioned to bind the DNA from opposing faces. A very similar arrangement has been observed in X-ray crystal structures determined for complexes of DtxR bound to its operator sites (30, 35).
MATERIALS AND METHODS
Purification of B. subtilis Fur protein
Fur protein was purified after overexpression in E. coli as previously described (4). As isolated, Fur protein binds DNA target sites with high affinity (1) and contains both zinc and iron (approximately 1 atom per monomer).
EMSA
Electrophoretic mobility shift assays (EMSA) were performed as previously described (4). Briefly, labeled DNA (10 pM) was incubated with the desired concentration of purified Fur. Protein-bound DNA and free DNA were separated by native polyacrylamide gel electrophoresis (PAGE) performed on an 8% polyacrylamide (19:1, acrylamide-bisacrylamide) gel electrophoresed at 160 V for 3 h, followed by drying and exposure to a phosphor screen. DNA fragments were 33 bp long and were prepared by labeling one strand and incubating it with a twofold excess of its complement for 5 min at 95°C, followed by slow cooling to room temperature.
Determination of protein oligomerization by native PAGE
Molecular weights of Fur-DNA complexes were determined for the protein bound to 33-bp fragments containing the consensus 7-1-7 sequence or the classical 19-bp Fur box [(7-1-7)2]. This was done by performing native PAGE as described by Orchard and May (28) and using a native PAGE kit (Sigma MW ND 500). Briefly, binding reaction mixtures containing 500 nM Fur (monomer) with target DNA and molecular weight markers were analyzed on a series of polyacrylamide gels (5 to 10% polyacrylamide; 19:1) electrophoresed in Tris-acetate buffer until the bromophenol blue bands in flanking sample lanes reached the bottoms of the gels. The gels were stained with Coomassie blue and dried, and then they were exposed to a phosphor screen. The distances from the top of the gel to the complexes or protein standards were measured and divided by the distance migrated by bromophenol blue for each gel to determine the relative mobility (Rf). The logarithm of the Rf was plotted against the gel concentration for each complex and protein standard, and best-fit lines were obtained. The negative slopes of these lines were then plotted against the molecular weights of the protein standards on a double-logarithmic scale, and a line of best fit was obtained. Interpolation of the graph by using the slopes of the lines from the protein-DNA complexes was used to deduce the approximate molecular weights of the complexes.
RESULTS AND DISCUSSION
Alignment of B. subtilis Fur box sequences.
B. subtilis Fur, purified after overexpression in E. coli, contains both iron and zinc (unpublished data) and binds with high affinity to a Fur box sequence in the regulatory region for the dihydroxybenzoate-derived siderophore (dhb) biosynthetic operon (4). This site is a perfect match (19 of 19 residues) with the classic Fur box, strongly suggesting that B. subtilis Fur has DNA selectivity similar to that observed for E. coli Fur. Indeed, sequence searches in which the classic 19-bp Fur box was used identified numerous candidate Fur-regulated operons, many of which have now been experimentally confirmed (1). However, other genes annotated as likely to encode functions related to iron transport are not associated with sites closely matching the classic Fur box.
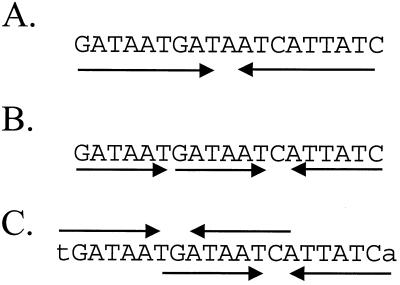
To further define the DNA sequences required for recognition by Fur, we performed a multiple-sequence alignment of 19 Fur-binding sites identified by DNA microarray-based mRNA profiling and confirmed by DNase I footprinting (1). As noted previously, the aligned sequences revealed conservation of a 15-bp core region (7-1-7 repeat) instead of the classic 19-bp inverted repeat (Table 1). Escolar et al. (13) presented a model in which the 19-bp inverted repeat is viewed as three GATAAT hexamers in a head-to-head-to-tail (6-6-1-6) orientation (Fig. 1). Clearly, the 7-1-7 heptamer motif which we propose is closely related to the hexamer motif described by these authors.
TABLE 1.
Analysis of naturally occurring Fur boxes
| Operona | Left overlapb | 7-1-7 consensus sitesb TGATAAT-ATTATCA TGATAAT-ATTATCA(L) TGATAAT-ATTATCA(R) | Right overlapb |
|---|---|---|---|
| dhbABCKF | 6 | ATTGATAATGATAATCATTATCAATAGATTG | 2 |
| ydbN | 6 | ATTGATAATGATTATCAATATCGTTTGATTG | 2 |
| ykuN1 | 5 | GTTGACAATGAAAATCATTATCATTTAAAGT | 3 |
| ykuN2 | 5 | TATGATATTGAAAATCATTATCAACTAATGG | 0 |
| yuiI | 5 | GTTGATAGTGAAAATCATTATCATACATTGC | 1 |
| fhuB/D | 5 | GATGAAAAAGAGAATCATTATCATCTGTGAT | 1 |
| yoaJ | 5 | TCTTATAATGATAATGATTCTCATTTGAAGT | 3 |
| yclN | 4 | TATGTAAATGATAATGATAATCAATTACTAT | 2 |
| yxeB | 1 | TTATTAATTGATAATGATAATCATTACTAAT | 4 |
| yfmC | 1 | GTTACATGTGATAATGATTCTCATTACTAAA | 4 |
| yfiY | 3 | GATCTAAATGATAATGAATTTCAATATTGGG | 3 |
| ywbL | 3 | TTATACAATGATAATCATTTTCAATTATAGG | 2 |
| ybbB | 3 | ATTTTTATTGAAAATGATTATCAATTGAAAG | 2 |
| yfhC | 2 | TTATGAAATGATAATCATTTTCAATTGCATA | 3 |
| yusV1 | 1 | AAACTAATTGAAAATGATTTTCAAAGTCAGT | 3 |
| yfiZ | 3 | TTTGTTTTTGAGAATAATCCTCAATTAGGGA | 1 |
| feuABC ybbA | 2 | ATTCCAATTGATAATAGTTATCAATTGAACA | 2 |
| yhfQ | 1 | AAAATTGGTGATAATGATTCTCATTCCGTGT | 2 |
| ywjA | 2 | AGTATAATTGAGAAATATTATCAGTTATTTA | 1 |
Operons are sorted by the extent of match with one or more overlapping 7-1-7 motifs. Bases in bold are those that match the (extended) consensus of the central 7-1-7 dyad. Left indicates the position of a possible overlapping 7-1-7 binding site to the left of the aligned site; right indicates the position of a possible site to the right to the aligned site.
The operators were aligned by the best fit with the 7-1-7 consensus sequence. The extents to which the aligned operators also match an overlapping 7-1-7 consensus sequence were determined by noting the numbers of matches with the additional six bases (underlined) in the left (L) or right (R) shifted 7-1-7 consensus site. Operons with matches at four or more positions are indicated by boldface type.
FIG. 1.
Comparison of models to explain the Fur box consensus sequence. (A) The Fur box is classically defined as a 19-bp inverted repeat sequence, originally envisioned to bind a single Fur dimer. (B) An alternative view proposes that Fur binds to repeated arrays of three or more copies of the hexamer GATAAT (13, 15). According to this model, the classic Fur box is three GATAAT motifs in a head-to-head-to-tail (6-6-1-6) array. (C) We propose that the 19-bp Fur box results from two overlapping heptamer inverted repeats [(7-1-7)2] that together define a 21-bp sequence.
In most cases, the region of DNA protected against DNase I digestion by Fur extends beyond the core 7-1-7 heptamer repeat. The additional protected region can be as small as 3 bp or as large as 14 bp or more (1). Inspection of the aligned Fur box sequences revealed that many operators have at least one overlapping 7-1-7 motif (Table 1) with a six-base offset (either to the left or to the right of the sequence shown in the alignment). The presence of two overlapping 7-1-7 motifs [designated (7-1-7)2] generates the classic 19-bp Fur consensus sequence (Fig. 1C). For example, the first six operators in Table 1 all have matches at at least five of six residues with the additional bases in the left overlapping motif, and as a corollary, the central base of the 7-1-7 motif shown in the alignment is a C, corresponding to the conserved C in the left overlapping 7-1-7 motif. Similarly, the yxeB and yfmC operators appear to have right overlapping motifs and have the expected central G residue in the aligned 7-1-7 motif (Table 1). Finally, roughly one-half of the aligned operator sites do not have obvious overlapping heptamer repeats (less than four of six matches with the additional flanking bases) and may represent sites that have only one 7-1-7 motif. For these sites there is little apparent conservation of the central base in the 7-1-7 motif. Note that all of these sites are protected by as little as 10 nM Fur and therefore represent high-affinity binding sites (1).
The 7-1-7 heptamer motif represents the minimal recognition unit for Fur binding.
To determine the minimal sequences required for recognition by Fur, we synthesized a series of DNA oligonucleotides containing either a consensus Fur box [(7-1-7)2], a single heptamer repeat (7-1-7), or related hexamer sequences in either a direct (two 6-mers) or inverted (6-1-6) orientation (Table 2). As controls, we also tested a nonspecific DNA fragment and oligonucleotides containing a single heptamer or a single hexamer. Each of these sequences was incubated with purified B. subtilis Fur, and the bound complexes were separated by native PAGE in an EMSA. Fur failed to bind to the oligonucleotides containing single 6-mer or 7-mer repeats or to the nonspecific fragment and bound only weakly to the site with two 6-mers (Fig. 2 and data not shown). This is consistent with the results reported for E. coli Fur (13).
TABLE 2.
Model oligonucleotide substrates
| Oligonucleotide | Left overlapa | 7-1-7 Consensus sitesa TGATAAT-ATTATCA TGATAAT-ATTATCA(L) TGATAAT-ATTATCA(R) | Right overlapa | Low-mobility complexb |
|---|---|---|---|---|
| (7-1-7)2c | 1 | CGCAGTCGATAATGATAATCATTATCAGTCGCG | 6 | ++ |
| 9-1-9 | 4 | AGTCGACAATGATAATCATTATCATTCGCGCGC | 4 | ++ |
| 8-1-8 | 3 | AGTCGACGATGATAATCATTATCATCCGCGCGC | 3 | + |
| 8-1-7 | 3 | AGTCGACGATGATAATCATTATCACCCGCGCGC | 2 | +/− |
| 7-1-7 | 2 | CGCAGTCAGTGATAATTATTATCAGTAGTCGCG | 3 | − |
| 6-1-6 | 2 | CGCAGTCAGCGATAATTATTATCGGTAGTCGCG | 2 | − |
| Two 6-mers | 2 | CGCAGTCAGCGATAATGATAATTGGTAGTCGCG | 2 | NA |
| 6-mer | NA | CGCAGTCAGCGATAATCGCCGCTGGTAGTCGCG | NA | NA |
| nsd | NA | CGCAGTCAGCAGCGGCAGCCGCAGGTAGTCGCG | NA | NA |
As in Table 1, (L) indicates the position of a possible overlapping 7-1-7 binding site to the left of the central aligned site and (R) indicates the position of a possible site to the right to the aligned site. The extents to which the aligned 7-1-7 motifs (bases in bold) also have an overlapping site to either the right or the the left were determined by tabulating the matches with the six additional bases that would contribute to the overlapping site. Sites with matches at four or more positions are indicated by boldface type. NA, not applicable, since the oligonucleotide lacks a central 7-1-7 (or related) motif and therefore does not form specific complexes.
The presence of the lower-mobility complex is indicated for each of the model oligonucleotides as observed in EMSA experiments (Fig. 2).
Note that the (7-1-7)2 DNA contains a perfect match with the 19-bp Fur box consensus sequence but contains a C instead of a T for the first base of the left 7-1-7 inverted repeat.
ns, nonspecific DNA fragment.
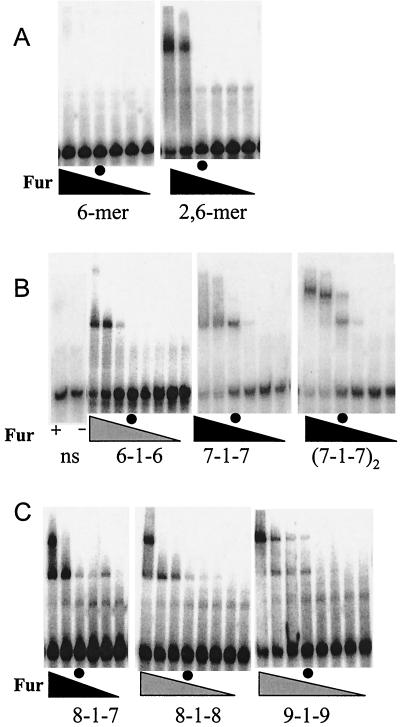
FIG. 2.
Binding of B. subtilis Fur to model oligonucleotide substrates. The final concentrations of Fur protein (monomer) in the reaction mixtures were 0, 1, 10, 100, 500, and 1,000 nM in the panels with six lanes (solid triangles) and 0, 10, 50, 75, 100, 200, 500, and 1,000 nM in the panels with eight lanes (shaded triangles). For ease of comparison between experiments, the lanes containing 100 nM Fur (monomer) are indicated by circles. (A) Binding of Fur protein to oligonucleotides containing either one or two copies of the GATAAT hexamer motif. (B) Comparison of Fur protein binding to the 6-1-6, 7-1-7, and (7-1-7)2 substrates. A nonspecific DNA fragment (ns) (see Table 2) was included as a control with either 1 μM Fur (lane +) or no added Fur (lane −). (C) Effect of additional flanking bases on formation of the lower-mobility complex (see Table 2 for a summary). Note that these studies were not done in parallel with those whose results are shown in panel B, so the absolute affinities cannot be compared.
The EMSA results indicate that Fur binds with similar affinity to either the 19-bp classic Fur box [(7-1-7)2] or to the single 7-1-7 heptamer repeat (Fig. 2B). In this experiment, approximately half-maximal binding was achieved with 100 nM Fur protein. In contrast, Fur binds with significantly reduced affinity to a 6-1-6 inverted repeat. These results suggest that the 7-1-7 motif is the minimal unit needed for high-affinity recognition by Fur. Comparable oligonucleotides were not included in the previous analysis of Escolar et al. (13).
Stoichiometry of Fur-DNA complexes.
A second striking finding that emerged from the EMSA experiments is the difference in mobility between the complexes formed with the 7-1-7 motif and the 19-bp Fur box [(7-1-7)2] sites (Fig. 2B). While the 7-1-7 operator gives rise to a single lower-mobility band, which we interpret as binding of a single Fur oligomer to the 7-1-7 inverted repeat, the 19-bp Fur box gives rise to two bands. This is consistent with the suggestion that the classic 19-bp Fur box is actually two overlapping 7-1-7 heptamer repeats [(7-1-7)2] that bind Fur on opposing faces of the DNA helix.
To investigate the minimal sequence requirements for formation of the lower-mobility band, we tested a series of oligonucleotides containing additional consensus bases on either side of the 7-1-7 core element (Table 2). Increasing one repeat of the 7-1-7 site to produce an 8-1-7 sequence resulted in a lower-mobility band with 1 μM protein but not with 500 nM protein (Fig. 2C). This lower-mobility complex also formed with the 8-1-8 sequence, and the lower-mobility complex was the dominant species present with 1 μM Fur protein. If the repeat sequences were extended to form a 9-1-9 inverted repeat, the affinity was increased still further. In this case the lower-mobility complex was first apparent in the reactions with 100 nM Fur protein. This was presumably due to binding of one Fur oligomer to the central 7-1-7 motif and a second oligomer to either of the two imperfect motifs offset to either the left or the right. Note that in general there was a good correlation between the presence of a second, overlapping 7-1-7 repeat and the appearance of the lower-mobility complex (Table 2). These results are consistent with the idea that binding of an additional Fur oligomer requires overlapping sites with significant fit to the core consensus sequence.
To determine the stoichiometry of complexes formed between Fur and the 7-1-7 and (7-1-7)2 sites, we measured their apparent molecular weights by analyzing the mobilities of the complexes during native PAGE performed with gels containing different concentrations of polyacrylamide (28). When compared to the globular protein standards, the complex between Fur and the 7-1-7 fragment had an apparent molecular mass of 58 kDa, in close agreement with the expected mass of 54 kDa calculated for a dimer of Fur bound to a 33-bp DNA oligonucleotide (Fig. 3). In contrast, the complex between Fur and the (7-1-7)2 fragment had an apparent mass of 97 kDa, which is comparable to the expected mass of 89 kDa calculated for two dimers of Fur bound to DNA. In both cases the interpolated molecular weights of the Fur-DNA complexes were somewhat higher than the values calculated from the sums of the individual components, perhaps due to the fact that the protein-DNA complexes have a different shape than the globular protein standards employed. Similarly, when the stoichiometry of the Lrp-DNA complex was determined by this method, an 8-kDa overestimate was also obtained (11) These results support the idea that the 7-1-7 site binds one dimer of Fur, while the (7-1-7)2 site binds two dimers.
FIG. 3.
Determination of the stoichiometry of Fur-DNA complexes by native PAGE. (A) Logarithms of the relative mobilities of Fur-DNA and marker proteins (versus the mobility of bromophenol blue) as a function of polyacrylamide concentration. The complexes used were Fur-(7-1-7) (□) and Fur-[(7-1-7)2] (○). The marker proteins used were carbonic anhydrase (⧫), α-lactalbumin (▪), bovine serum albumin dimer (•), bovine serum albumin monomer (▾), and ovalbumin (▴). (B) Determination of the apparent molecular weights of the Fur-(7-1-7) and Fur-[(7-1-7)2] complexes. The negative slopes of the mobility lines in panel A were plotted against the molecular weights of the protein standards (solid symbols), and the apparent masses of the Fur-DNA complexes (open symbols) were determined by interpolation.
Binding of Fur to naturally occurring 7-1-7 and (7-1-7)2 sites.
Although we originally derived the 7-1-7 consensus sequence by alignment of naturally occurring operators for Fur (Table 1) (1), it is nevertheless clear than many of these operators also match the longer 19-bp Fur box consensus sequence and are therefore likely to be (7-1-7)2 sites. To determine if Fur interacts with different stoichiometries with naturally occurring representatives of the 7-1-7 and (7-1-7)2 classes, we synthesized oligonucleotides containing the sequences of the dhb and feu operators, respectively. The feu operator matches the 7-1-7 consensus sequence at 13 of 14 positions but matches the longer 19-bp Fur box sequence at only 13 of 19 positions. When assayed by EMSA, the preparations showed the same band patterns as the model 7-1-7 and (7-1-7)2 sequences derived from alignments (Fig. 4B). We concluded that two dimers of Fur recognize the dhb operator region, while a single dimer binds the feu operator. Inspection of the band pattern in the dhb experiment indicated that occupancy of the two sites results in little if any obvious cooperativity; complexes corresponding to both one and two bound dimers are readily observed. Note also that the feu site lacks the central G·C base pair usually found in overlapping arrays of 7-1-7 motifs. In general, conservation of this central position within the 7-1-7 motif correlates with the presence of an overlapping 7-1-7 motif (Table 1).
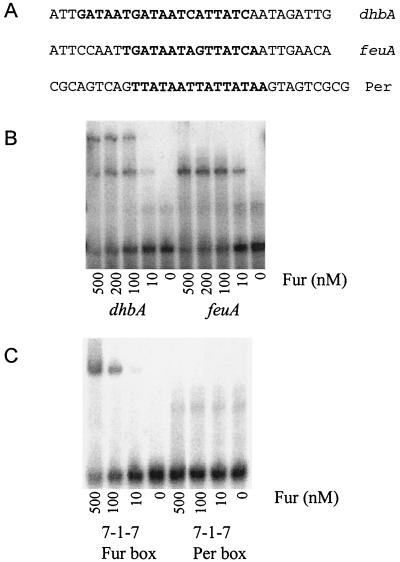
FIG. 4.
Binding of Fur to naturally occurring operator sites. (A) Sequences of the top strands of the DNA oligonucleotides representing the dhb, feuA, and Per box substrates. (B) Fur binds to the dhbA and feu operators with comparable affinities but forms the lower-mobility complex only with the dhb operator. (C) Fur binds to the 7-1-7 substrate but does not recognize the closely related Per box sequence.
Fur does not recognize a 7-1-7 Per box motif.
In light of the finding that Fur interacts with high affinity with a 7-1-7 sequence motif, it is interesting to consider the features that are likely to distinguish Per boxes from Fur boxes. PerR is 31% identical to Fur and is also a dimeric, metal ion-dependent, DNA-binding protein (5). In previous work we and other workers identified a total of nine Per boxes recognized by PerR (3, 8, 9, 16, 23). Alignment of these sequences supports our original suggestion that PerR recognizes a 7-1-7 inverted repeat motif, TTATAATnATTATAA (Fig. 4A). Like Fur, PerR often binds to extended regions flanking this core sequence. Binding appears to be sequence selective; at higher concentrations of PerR DNA binding may extend unidirectionally relative to this core (23).
A comparison of the consensus Fur and Per boxes reveals a remarkable similarity: the two heptamers are identical at six of seven positions. Indeed, the similarity between the Fur and Per boxes makes it difficult to assign genes to one regulon or the other based on genomic searches. For example, we originally suggested that ykvW might be a member of the Fur regulon based on the presence of a candidate Fur box in the regulatory region (22). It is now clear that ykvW (now called zosA) is actually a member of the PerR regulon (17). Here, we found that Fur does not recognize a Per box (Fig. 4C). Similarly, PerR binds tightly to a consensus Per box but does not recognize the related Fur box (data not shown). These results are consistent with in vivo analyses in which both lacZ reporter fusions and whole-genome transcriptional profiling were used. We have yet to find a gene that is regulated directly by both Fur and PerR in B. subtilis.
Comparison of the structure of the proposed Fur-DNA complex with the structure of DtxR-DNA complexes.
DtxR is the diphtheria toxin repressor of Corynebacterium diphtheriae and, like Fur, functions as an Fe(II)-dependent repressor of siderophore biosynthesis and transport functions (26, 33). The original identification of Fur as the ferric uptake repressor in E. coli and DtxR as the analogous protein in C. diphtheriae led to the suggestion that these two proteins may control iron uptake functions in gram-negative and gram-positive lineages, respectively. The actual situation is far more complicated: both the Fur and DtxR families of proteins control functions other than iron homeostasis, and both are widely distributed among bacteria (DtxR homologs are also found in some archaea [2]). For example, both B. subtilis and Staphylococcus aureus contain three distinct Fur homologs (Fur, PerR, and Zur), as well as an Mn(II)-sensing DtxR homolog (MntR) (22).
At the level of the primary amino acid sequence, the Fur and DtxR families are not very similar and are often assumed to have arisen independently. However, both proteins are dimeric, HTH-containing DNA-binding proteins thought to be related to the same superfamily of regulators (the CAP/LexA superfamily) (19, 25). While DtxR has been the subject of numerous structural studies performed with X-ray crystallography, a structure has yet to be reported for any member of the Fur family. Thus, the relationship between these families of proteins remains unclear. There are, however, obvious similarities. Both contain an amino-terminal DNA-binding domain linked to a metal-binding domain, and both bind two metal ions per monomer (15, 26).
The similarities between Fur and DtxR are even more apparent when the structure of the operator sites is considered. Both proteins recognize operators initially defined as 19-bp inverted repeat sequences leading to a typical DNase I footprint of ∼30 bp. The DtxR operator is now viewed as two overlapping inverted repeats with 7-0-7 symmetry offset by 5 bp (30, 35). Thus, DNA recognition by DtxR is mediated by two proteins that bind to opposite faces of the DNA helix (Fig. 5). Recognition of DNA is mediated both by direct contacts between amino acids and the edges of the bases in the major groove (direct readout) and by contacts with the phosphate sugar backbone (indirect readout) (7).
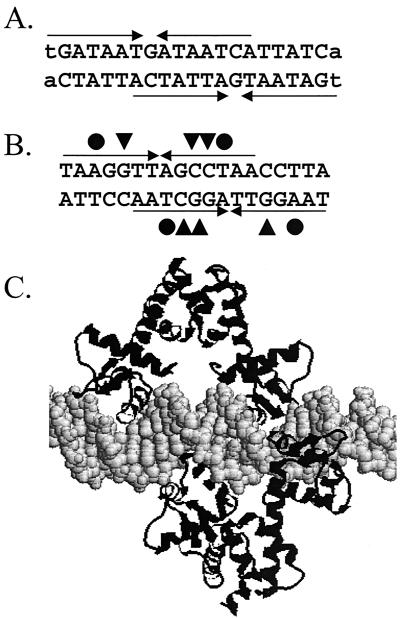
FIG. 5.
Comparison of the proposed Fur-DNA complex with the DtxR-DNA complex. (A) Two overlapping 7-1-7 heptamer motifs that generate the classic 19-bp Fur box binding site. (B) Two overlapping imperfect inverted repeats that generate the 19-bp binding site for DtxR (adapted from reference 7). The critical contacts for protein-DNA recognition include the interaction of DtxR Gln43 (triangles) with the G·C base pairs indicated and the interaction of a thymine methyl group with the Ser37-Pro39 pair (circles) (adapted from reference 7). (C) Model of the complex formed between DtxR and operator DNA, illustrating the role of two DtxR dimers in recognition.
The bases most important for protein-DNA interaction can be inferred in a variety of ways. For DtxR, a comparison of 21 in vitro-selected DtxR-binding sites identified a 19-bp T(A/T)AGGTTAG(G/C)CTAACCT(A/T)A consensus sequence (32). A similar pattern of conservation was noted when natural DtxR-binding sites were aligned (7). Analysis of aligned Fur-binding sites from E. coli also revealed a 19-bp inverted repeat consensus sequence. However, it has been argued that this region is actually an array of three hexamers and that each GATAAT hexamer represents an site of interaction for a Fur dimer (15).
Our results suggest that Fur-DNA complexes may be structurally similar to the complexes described for DtxR. Indeed, inspection of the detailed footprinting analyses reported for E. coli Fur (DNase I, hydroxyl radical, and missing thymine) supports a model in which Fur interacts with both faces of the DNA helix. For example, in studies in which synthetic hexamer repeats are used, Fur binding is severely reduced in templates lacking thymine residues at 6-bp intervals. Remarkably, DtxR has also been shown to contact thymine residues with similar periodicity by hydrophobic interactions among Ser37, Pro39, and the thymine methyl group (7). The model proposed here can also account for the documented propensity of Fur (and Fur homologs such as PerR) to bind to extended regions of DNA. Just as two dimers can bind to opposing faces of the helix to account for the 19-bp consensus sequence, three dimers can be envisioned to bind to DNA to generate an extended protected region containing an additional 6 bp. Indeed, just such an arrangement has been seen in several Fur-DNA complexes (10, 14).
Summary.
We propose that members of the Fur family of repressor proteins bind to a core sequence consisting of a 15-bp inverted heptamer repeat. This model represents a significant revision of previous models, including both the original suggestion that each 19-bp inverted repeat represents the binding site of a single dimer (12) and a revised model in which Fur recognizes repeated arrays of GATAAT (13, 15). Understanding the precise nature of the Fur-DNA complex should be useful for ongoing efforts to define Fur regulons by bioinformatic approaches. For example, analysis of the Fur regulon in Shewanella oneidensis has revealed that many apparent target genes do not contain an upstream sequence with a statistically significant match with the 19-bp Fur consensus sequence (e.g., a best fit of less than 10 of 19 residues) (34). Other investigators have attempted to identify Fur box sequences preceding iron-regulated genes by searching for repeated arrays of ATAAT (24). We suggest that more appropriate strategies may include searches for overlapping 7-1-7 inverted repeats. Since two such repeats regenerate the classic 19-bp consensus sequence and Fur often binds to multiple overlapping sites, searches with the classic 19-bp consensus sequence are likely to continue to be useful for analyzing bacterial genomes (1, 29).
Acknowledgments
We thank the members of the laboratory of J.D.H. for helpful discussions concerning this work.
This work was supported by grant MCB-9983656 from the National Science Foundation.
REFERENCES
- 1.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol., in press. [DOI] [PubMed]
- 2.Bell, S. D., S. S. Cairns, R. L. Robson, and S. P. Jackson. 1999. Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol. Cell 4:971-982. [DOI] [PubMed] [Google Scholar]
- 3.Bsat, N., L. Chen, and J. D. Helmann. 1996. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J. Bacteriol. 178:6579-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bsat, N., and J. D. Helmann. 1999. Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the dhb operator in vitro and in vivo. J. Bacteriol. 181:4299-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Calderwood, S. B., and J. J. Mekalanos. 1988. Confirmation of the Fur operator site by insertion of a synthetic oligonucleotide into an operon fusion plasmid. J. Bacteriol. 170:1015-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. S., A. White, J. Love, J. R. Murphy, and D. Ringe. 2000. Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochemistry 39:10397-10407. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., and J. D. Helmann. 1995. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative stress gene. Mol. Microbiol. 18:295-300. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christoffersen, C. A., T. J. Brickman, I. Hook-Barnard, and M. A. McIntosh. 2001. Regulatory architecture of the iron-regulated fepD-ybdA bidirectional promoter region in Escherichia coli. J. Bacteriol. 183:2059-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, Y., M. A. Midkiff, Q. Wang, and J. M. Calvo. 1996. The leucine-responsive regulatory protein (Lrp) from Escherichia coli. Stoichiometry and minimal requirements for binding to DNA. J. Biol. Chem. 271:6611-6617. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537-547. [DOI] [PubMed] [Google Scholar]
- 14.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 2000. Evidence of an unusually long operator for the fur repressor in the aerobactin promoter of Escherichia coli. J. Biol. Chem. 275:24709-24714. [DOI] [PubMed] [Google Scholar]
- 15.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaballa, A., and J. D. Helmann. 2002. Identification of a peroxide-induced P-type ATPase reveals an important role of zinc as an antioxidant in Bacillus subtilis. Mol. Microbiol. 45:997-1005. [DOI] [PubMed] [Google Scholar]
- 18.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez de Peredo, A., C. Saint-Pierre, J. M. Latour, I. Michaud-Soret, and E. Forest. 2001. Conformational changes of the ferric uptake regulation protein upon metal activation and DNA binding; first evidence of structural homologies with the diphtheria toxin repressor. J. Mol. Biol. 310:83-91. [DOI] [PubMed] [Google Scholar]
- 20.Harrison, S. C., and A. K. Aggarwal. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59:933-969. [DOI] [PubMed] [Google Scholar]
- 21.Helmann, J. D. 1997. Metal cation regulation in Gram-positive bacteria, p. 45-76. In S. Silver and W. Walden (ed.), Metal ions in gene regulation. Chapman & Hall, New York, N.Y.
- 22.Herbig, A., and J. D. Helmann. 2002. Metal ion uptake and oxidative stress, p. 405-414. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives, 2nd ed. ASM Press, Washington, D.C.
- 23.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41:849-859. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, T., A. Schutz, M. Brosius, A. Volker, U. Volker, and E. Bremer. 2002. High-salinity-induced iron limitation in Bacillus subtilis. J. Bacteriol. 184:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm, L., C. Sander, H. Rüterjans, M. Schnarr, R. Fogh, R. Boelens, and R. Kaptein. 1994. LexA repressor and iron uptake regulator from Escherichia coli: new members of the CAP-like DNA binding domain superfamily. Protein Eng. 7:1449-1453. [DOI] [PubMed] [Google Scholar]
- 26.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect. Dis. 181(Suppl. 1):S156-S167. [DOI] [PubMed] [Google Scholar]
- 27.Le Cam, E., D. Frechon, M. Barray, A. Fourcade, and E. Delain. 1994. Observation of binding and polymerization of Fur repressor onto operator-containing DNA with electron and atomic force microscopes. Proc. Natl. Acad. Sci. USA 91:11816-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orchard, K., and G. E. May. 1993. An EMSA-based method for determining the molecular weight of a protein-DNA complex. Nucleic Acids Res. 21:3335-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohl, E., R. K. Holmes, and W. G. Hol. 1999. Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J. Mol. Biol. 292:653-667. [DOI] [PubMed] [Google Scholar]
- 31.Rowland, B. M., and H. W. Taber. 1996. Duplicate isochorismate synthase genes of Bacillus subtilis: regulation and involvement in the biosyntheses of menaquinone and 2,3-dihydroxybenzoate. J. Bacteriol. 178:854-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao, X., and J. R. Murphy. 1994. Determination of the minimal essential nucleotide sequence for diphtheria tox repressor binding by in vitro affinity selection. Proc. Natl. Acad. Sci. USA 91:9646-9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao, X., N. Schiering, H. Y. Zeng, D. Ringe, and J. R. Murphy. 1994. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, H. Lim, J. Yates III, C. C. Brandt, J. M. Tiedje, and J. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, A., X. Ding, J. C. vanderSpek, J. R. Murphy, and D. Ringe. 1998. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature 394:502-506. [DOI] [PubMed] [Google Scholar]