Abstract
Sulfate-reducing bacteria, like Desulfovibrio vulgaris Hildenborough, use the reduction of sulfate as a sink for electrons liberated in oxidation reactions of organic substrates. The rate of the latter exceeds that of sulfate reduction at the onset of growth, causing a temporary accumulation of hydrogen and other fermentation products (the hydrogen or fermentation burst). In addition to hydrogen, D. vulgaris was found to produce significant amounts of carbon monoxide during the fermentation burst. With excess sulfate, the hyd mutant (lacking periplasmic Fe-only hydrogenase) and hmc mutant (lacking the membrane-bound, electron-transporting Hmc complex) strains produced increased amounts of hydrogen from lactate and formate compared to wild-type D. vulgaris during the fermentation burst. Both hydrogen and CO were produced from pyruvate, with the hyd mutant producing the largest transient amounts of CO. When grown with lactate and excess sulfate, the hyd mutant also exhibited a temporary pause in sulfate reduction at the start of stationary phase, resulting in production of 600 ppm of headspace hydrogen and 6,000 ppm of CO, which disappeared when sulfate reduction resumed. Cultures with an excess of the organic electron donor showed production of large amounts of hydrogen, but no CO, from lactate. Pyruvate fermentation was diverse, with the hmc mutant producing 75,000 ppm of hydrogen, the hyd mutant producing 4,000 ppm of CO, and the wild-type strain producing no significant amount of either as a fermentation end product. The wild type was most active in transient production of an organic acid intermediate, tentatively identified as fumarate, indicating increased formation of organic fermentation end products in the wild-type strain. These results suggest that alternative routes for pyruvate fermentation resulting in production of hydrogen or CO exist in D. vulgaris. The CO produced can be reoxidized through a CO dehydrogenase, the presence of which is indicated in the genome sequence.
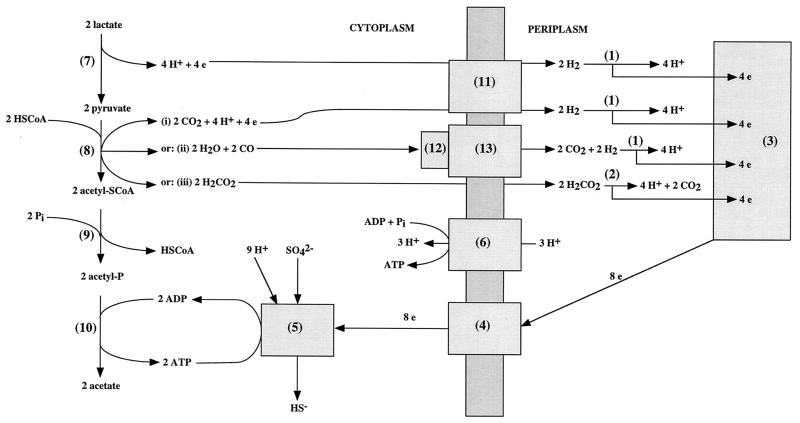
Desulfovibrio spp. derive energy for growth from redox reactions in which organic acids, alcohols, or hydrogen (the electron donors) are oxidized by sulfate, thiosulfate, or sulfite (the electron acceptors). A diagram indicating a possible mechanism for ATP synthesis in Desulfovibrio spp. growing on lactate, pyruvate, formate, or hydrogen and sulfate is shown in Fig. 1 (2, 14, 24). Growth on lactate- and sulfate-containing medium requires proton motive force-driven ATP synthesis (Fig. 1, reaction 6) because ATP formed by substrate level phosphorylation (Fig. 1, reactions 10) equals that required for activation of sulfate (Fig. 1, reaction 5). The hydrogen cycling hypothesis proposed by Odom and Peck (14) suggested that protons and electrons generated in reactions 7 and 8-i (Fig. 1) are converted to H2 by a cytoplasmic hydrogenase and that this H2 diffuses to the periplasm, where it is reoxidized. Cycling of 4 H2 molecules then yields eight periplasmic protons for every sulfate reduced, allowing a net synthesis of 2.7 ATP molecules per molecule of sulfate. When hydrogen is the exclusive electron donor, Fig. 1 indicates a net synthesis of 0.7 mol of ATP when 4 mol of hydrogen is used to reduce 1 mol of sulfate (1, 26). This is in reasonable agreement with the net production of 1 mol of ATP per mol of sulfate determined by growth yield studies (1).
FIG. 1.
Model for ATP synthesis in Desulfovibrio spp. growing on medium containing lactate and sulfate, pyruvate and sulfate, hydrogen and sulfate, or formate and sulfate. The reactions indicated are catalyzed by the following: 1, periplasmic (e.g., Fe-only) hydrogenase; 2, periplasmic formate dehydrogenase; 3, the cytochrome c3 network; 4, transmembrane electron transport (e.g., Hmc) complex; 5, enzymes that reduce sulfate to sulfide (ATP sulfurylase, adenosine phosphosulfate reductase, dissimilatory sulfite reductase, adenylate kinase, and pyrophosphatase); 6, ATP synthase; 7, lactate dehydrogenase; 8-i, pyruvate-ferredoxin oxidoreductase, 8-ii, CO formation from pyruvate (enzyme unknown), 8-iii, pyruvate-formate lyase; 9, phosphotransacetylase; 10, acetate kinase, 11, cytoplasmic, membrane-bound hydrogenase (e.g., Ech [16]); 12, CODH; 13, CO-dependent hydrogenase. CoA, coenzyme A.
One problem in acceptance of the hydrogen cycling hypothesis has been that a soluble cytoplasmic hydrogenase was never isolated from Desulfovibrio vulgaris. However, as discussed recently (16), the presence of a membrane-bound, nickel-containing hydrogenase with the active site facing the cytoplasm that could participate in hydrogen cycling (Fig. 1, reaction 11) is indicated by its genome sequence (http://www.tigr.org). A second problem is that hydrogen formation from lactate (Fig. 1, reactions 7 and 11) is energetically unfavorable (21). Also, although growth yield studies have shown a higher cell yield per mole of sulfate with lactate as the electron donor than with hydrogen, the difference is not compatible with a three- to fourfold increase in net ATP production. Thus, only a fraction of the protons and electrons generated in reactions 7 and 8-i of Fig. 1 may be used for generation of a proton motive force while the remainder is used directly for sulfate reduction.
Proton and electron movement leading to chemiosmotic ATP production during growth on lactate or pyruvate and sulfate is likely to be much more complex than indicated in Fig. 1. The genome sequence indicates that in excess of 100 gene products, including several membrane-associated redox protein complexes, could be involved. In the case of lactate dehydrogenase, experimental studies (19) have shown it to be an oxygen-labile, membrane-bound enzyme in Desulfovibrio that could directly contribute to chemiosmotic energy conservation. This topology is not indicated in Fig. 1. The hyd genes for Fe-only hydrogenase (Fig. 1, reaction 1) and the hmc genes for the high-molecular-mass cytochrome (Hmc) complex (Fig. 1, reaction 4), a large transmembrane electron transport complex, have been deleted (5, 16). These deletion mutations affected growth with hydrogen as the electron donor more strongly than growth with lactate and hydrogen. In the case of the hyd mutation, residual growth is caused by the presence of at least two other nickel-containing hydrogenases (the NiFe and NiFeSe hydrogenases) in the periplasm of D. vulgaris Hildenborough. In the case of the hmc mutation, the genome sequence indicates the presence of several other transmembrane electron transport complexes that may enable continued growth on hydrogen.
Organisms are frequently overdesigned and can compensate for the missing gene when a property such as growth rate is monitored (4). However, metabolite concentrations can often vary over orders of magnitude, even in mutations that are silent with respect to growth rate. A detailed analysis of metabolite concentrations in the hmc and hyd mutant and wild-type strains of D. vulgaris is therefore presented to gain more insight into the mechanisms that D. vulgaris employs for energy conservation.
MATERIALS AND METHODS
Growth of bacteria.
D. vulgaris subsp. vulgaris Hildenborough (NCIMB 8303; isolated from clay soil near Hildenborough, United Kingdom) (17), D. vulgaris H801 (Δhmc Cmr) (5), and D. vulgaris Hyd100 (ΔhydAB Cmr) (16) were stored in Postgate's medium B (17) and grown in defined Widdel-Pfennig (WP) medium formulated as described by Widdel and Bak (25). Concentrations of electron donors (lactate, pyruvate, and formate) or the electron acceptor sulfate are described in Results. Data are reported for growth in 500-ml flat glass culture flasks containing 250 ml of WP medium and 250 ml of headspace flushed with 90% (vol/vol) N2 and 10% (vol/vol) CO2. These were closed with a soft rubber stopper, allowing repeated sampling of both the medium and the headspace. For growth, the flasks were placed flat on an orbital shaker and shaken at 60 rpm at an average temperature of 28°C. Cell densities were measured with a Shimadzu spectrophotometer as the optical density at 600 nm (OD600). An OD600 of 1 was determined to correspond to a cell dry-weight concentration of 0.309 g/liter. This value was higher than one used before (5, 16) that was based on data in the literature. In a typical experiment, single cultures of the wild-type strain and the two mutant strains (i.e., a total of three cultures) were compared. The experiment was repeated at least once to validate the significance of observed differences.
Analytical procedures.
For analysis of H2 and CO concentrations in the headspace of WP medium-containing flasks, 1-ml samples were withdrawn and appropriately diluted by injection in air-filled serum bottles. Aliquots (2.5 ml) of the diluted samples were then injected into the sample coil of a Shimadzu GC-8A gas chromatograph fitted with a Molecular Sieve 5A column and an RGD2 Reduction Gas Detector (Trace Analytical) as described previously (16). For analysis of samples that did not require dilution, 2.5 ml was withdrawn from the headspace and injected directly into the sample coil. H2 and CO concentrations were calculated by comparing the peak area of the diluted sample with that of an H2 or CO standard (20 ppm in helium). Headspace pressures were close to ambient (1 atm). Gas concentrations in parts per million can be converted to millimolar concentrations by multiplying by 4 × 10−5.
Duplicate 1-ml samples of the growing culture were taken and placed in microcentrifuge tubes every time a headspace analysis was done. One of these was used immediately for OD600 determination and then discarded. The others were stored frozen at −20°C. For sulfate concentration determinations, 150 μl of a thawed and homogenized sample was mixed with 350 μl of H2O and 500 μl of 20 mM Na2CO3 in a microcentrifuge tube. Following storage at 4°C for 1 h, the samples were centrifuged at 13,000 rpm for 4 min and filtered through a 0.45-μm-pore-size filter. Samples (∼0.5 ml) were then injected in a Sykam high-pressure liquid chromatograph equipped with an LCA A14 (3 by 250 mm) anion-separating column equilibrated with 10 mM Na2CO3, a suppression unit, and a conductivity detector (3). Concentrations were calculated from the peak areas of standard samples (150 μl of 28 mM sulfate in WP medium salts, or dilutions thereof, mixed with 350 μl of H2O and 500 μl of Na2CO3). The column was regenerated by elution with 0.2 M H2SO4 after every three samples.
To determine the concentrations of organic acids, 500 μl of thawed and homogenized sample was mixed with 500 μl of H2O in a microcentrifuge tube. Following centrifugation and filtration, samples were placed in 1-ml autosampler vials. The autosampler injected the samples (0.5 ml) into a Sykam high-pressure liquid chromatograph equipped with an ICSep WA-1 column (TransGenomic) equilibrated with 5 mM H2SO4 and a Linear TM UVIS204 UV-VIS detector set at 215 nm and/or a refractive-index detector. Concentrations were calculated from the peak areas of a mixture of standards (10 mM [each] succinate, lactate, formate, acetate, propionate, and isobutyrate) diluted similarly to the samples and run after every 10 samples. Pyruvate and fumarate standards were run separately at lower concentrations in view of their high UV absorbance. Pyruvate was also determined enzymatically by recording the increase in absorbance at 340 nm (ΔA340) in the presence of excess NAD+ and lactate dehydrogenase from rabbit muscle. Formate was similarly determined enzymatically by using NAD+-dependent formate dehydrogenase from Candida boidinii. Both enzymes were obtained from Roche Diagnostics.
RESULTS
Lactate as electron donor.
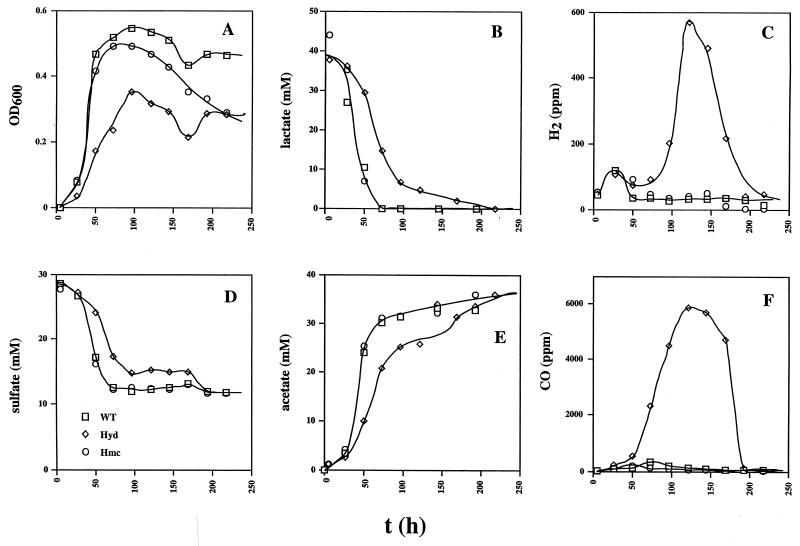
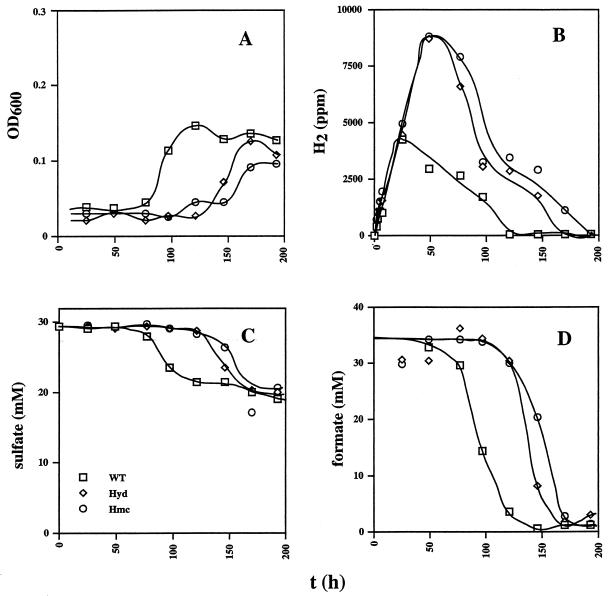
The wild-type and hmc mutant strains showed similar kinetics of growth (0 to 50 h) in cultures containing 38 mM lactate and 28 mM sulfate (Fig. 2A). The kinetics of utilization of lactate and sulfate and production of acetate (Fig. 2B, D, and E, respectively) are consistent with the use of two lactate molecules per sulfate molecule reduced and the formation of one acetate molecule per lactate molecule. A cell yield (Ysulfate) of 10.3 g/mol of sulfate can be calculated from the data in Fig. 2. H2 production by Desulfovibrio prior to growth on lactate- and sulfate-containing medium (the hydrogen burst) was seen as the small peak in hydrogen concentration to 100 ppm at 25 h in cultures of all three strains (Fig. 2C). No CO was produced during this burst period. The hmc strain maintained a lower cell density than the wild type in the stationary growth phase (Fig. 2A, 100 to 250 h). The hyd mutant grew slower and to a lower cell density under these conditions, as observed before (16). Peculiarly, no sulfate was reduced from 100 to 175 h. However, lactate continued to be metabolized during this period, leading to significant amounts of H2 (Fig. 2C, 600 ppm, 0.024 mM) and, surprisingly, even larger amounts of CO (Fig. 2F, 6,000 ppm, 0.24 mM). The H2 and CO formed, as well as the remaining lactate, were all oxidized when sulfate reduction resumed from 175 to 200 h. Although the production and subsequent consumption of trace amounts of CO has been reported before (10), the production of large amounts of CO as a result of the deletion of the genes for Fe-only hydrogenase was totally unexpected. In a duplicate experiment, the hyd mutant produced a similarly high concentration of CO (6,230 ppm at 115 h, relative to the wild-type (171 ppm) and hmc mutant (49 ppm) strains.
FIG. 2.
Growth physiology of the wild-type (WT) and hyd and hmc mutant strains in WP medium containing lactate (38 mM), sulfate (28 mM), and a headspace of 10% (vol/vol) CO2 and 90% N2. Plotted as a function of time are cell density (A), lactate concentration (B), H2 concentration in the headspace (C), sulfate concentration (D), acetate concentration (E), and CO concentration in the headspace (F). The symbol definitions in panel D apply to the entire figure.
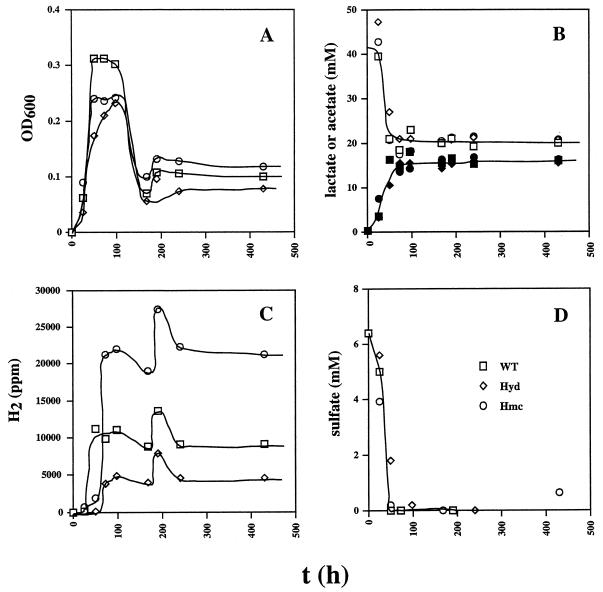
To determine if the wild-type and hmc mutant strains form H2 (and possibly CO) when the electron donor lactate is in excess, the sulfate concentration was decreased to 7 mM (Fig. 3). No growth was observed with lactate when no sulfate was added (data not shown). Complete reduction of the limiting amount of sulfate at 50 h (Fig. 3D) gave rise to cell mass (OD600 = 0.2 to 0.3), which processed some lactate fermentatively, forming high H2 headspace concentrations in cultures of all three strains (Fig. 3C). This inhibited further fermentation of lactate, which remained constant at 20 mM from 100 to 400 h. No significant amounts of CO were formed during lactate fermentation. Of the initial lactate concentration (38 mM), 14 mM was used for sulfate reduction and 20 mM remained. Hence, approximately 4 mM was used for the formation of biomass and fermentative processing. An OD600 of 0.25 corresponds to a cell dry weight of 0.08 g/liter. With an established average formula for biomass dry weight (CH1.8O0.5N0.2) of 24.6 g/mol of carbon, the biomass concentration is found to correspond to 3 mM C. Hence, approximately 1 mM lactate was assimilated into biomass and 3 mM was processed fermentatively, yielding hydrogen and pyruvate. Subsequent fermentation of pyruvate did not yield CO under these conditions. Headspace H2 concentrations produced by the hmc mutant, the wild type, and the hyd mutant were 20,000, 10,000, and 5,000 ppm (0.8, 0.4, and 0.2 mM, respectively). These were almost constant from 100 to 400 h, except for a transient hump at 200 h. This corresponded to an increase in the OD600, indicating a metabolic adjustment, not an experimental error. These differences in H2 concentration resulting from lactate fermentation were reproducible (±5%) in a duplicate experiment. Although these concentrations are significant, 10-fold higher concentrations (6 mM H2) would result if the fermentation could be described solely by the sum of reactions 1 and 2 in Table 1, suggesting that reduced organic fermentation products were also formed. The observed formation of only 16 mM acetate from 18 mM lactate (Fig. 3B) is in agreement with this suggestion. Hence, cytoplasmic reduction of organic acids may provide a sink for some of the hydrogen formed in lactate fermentation, when sulfate is absent. The higher headspace hydrogen concentration in cultures of the hmc mutant indicates that transport of electrons derived from periplasmic hydrogen oxidation to cytoplasmic organic fermentation pathways is, in part, Hmc mediated. The lower headspace hydrogen concentrations of the hyd mutant indicate that Fe-only hydrogenase may function in hydrogen production, not consumption, under these conditions.
FIG. 3.
Growth physiology of the wild-type (WT) and hyd and hmc mutant strains in WP medium containing lactate (38 mM), sulfate (7 mM), and a headspace of 10% (vol/vol) CO2 and 90% N2. Plotted as a function of time are cell density (A), lactate (open symbols) and acetate (filled symbols) concentrations (B), H2 concentration in the headspace (C), and sulfate concentration (D). The symbol definitions in panel D apply to the entire figure.
TABLE 1.
Standard free-energy changes of relevant reactions
| Reaction | ΔG0′ (kJ/mol)a |
|---|---|
| 1. Lactate → pyruvate + H2 | +43.2 |
| 2. Pyruvate + 2 H2O → acetate + HCO3− + H2 + H+ | −47.1 |
| 3. Pyruvate → acetate + CO | −32.0 |
| 4. CO + 2 H2O → HCO3− + H2 + H+ | −15.1 |
| 5. Pyruvate + H2O → acetate + formate + H+ | −48.5 |
| 6. H2O + formate → HCO3− + H2 | +1.4 |
Gibbs free energies of formation from elements were used for the compounds indicated (20). These were then combined to calculate ΔG0′.
Pyruvate as electron donor.
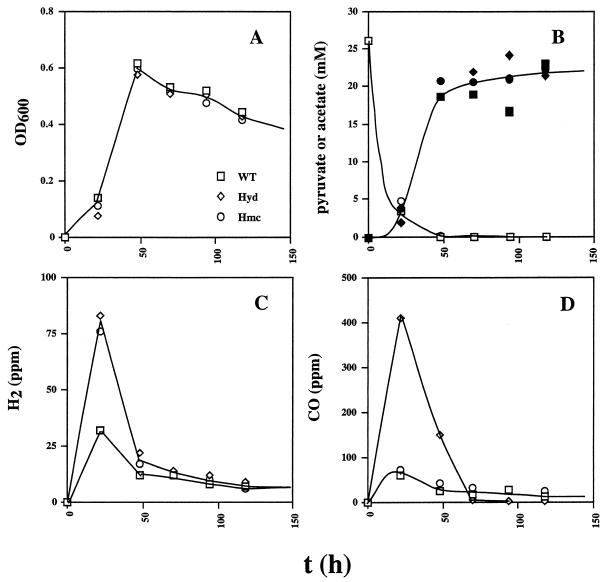
With pyruvate (30 mM) as the electron donor for sulfate reduction (28 mM), all three strains grew identically (Fig. 4A) and had similar kinetics of pyruvate consumption and acetate production (Fig. 4B). The strains also consumed similar amounts of sulfate with identical kinetics (data not shown), consistent with a 4:1 stoichiometry of reaction between pyruvate and sulfate and with formation of one acetate molecule for each pyruvate molecule oxidized. The data indicated a Ysulfate of 23.8 g/mol of sulfate under these conditions. At the start of growth, a small burst of H2 and CO was observed (Fig. 4C and D). The H2 burst was more pronounced in the hyd and hmc mutants (Fig. 4C), whereas the CO burst was only more prominent in the hyd mutant strain. (Fig. 4D). In contrast to the use of lactate, no temporary halt in the reduction of sulfate by the hyd mutant strain with associated production of H2 and/or CO was observed later in the growth curve.
FIG. 4.
Growth physiology of the wild-type (WT) and hyd and hmc mutant strains in WP medium containing pyruvate (30 mM), sulfate (28 mM), and a headspace of 10% (vol/vol) CO2 and 90% N2. Plotted as a function of time are cell density (A), pyruvate concentration determined enzymatically (open symbols) and acetate concentration (filled symbols) (B), H2 concentration in the headspace (C), and CO concentration in the headspace (D). The symbol definitions in panel A apply to the entire figure.
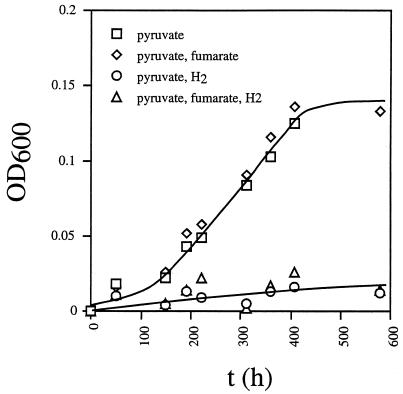
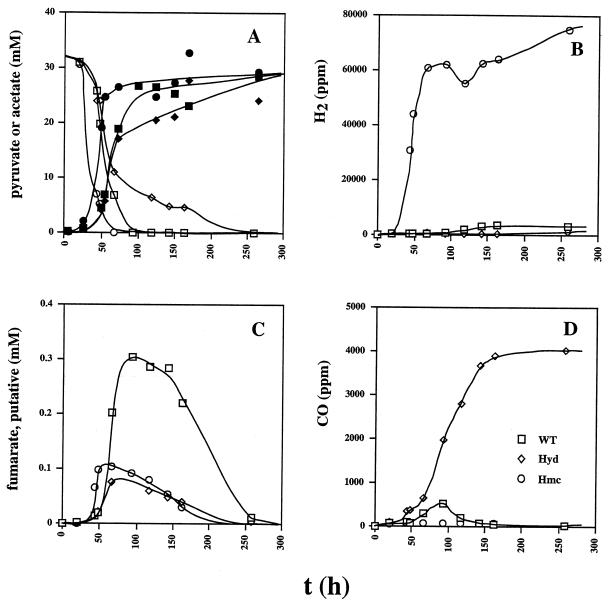
D. vulgaris can grow fermentatively on pyruvate. However, the doubling time was 100 to 130 h and cultures reached a maximal OD600 of 0.15 (Fig. 5). Hydrogen inhibited fermentative growth of wild-type D. vulgaris on pyruvate. Fermentative metabolism of pyruvate was therefore investigated with cultures containing 30 mM pyruvate and a limiting amount of sulfate (3 mM). As in Fig. 3D, all of the sulfate was fully reduced at 50 to 75 h (data not shown), giving similar amounts of biomass for all three strains (OD600 = 0.4 to 0.6). The cell density then declined to an OD600 of 0.2 at 250 h in all three strains (data not shown). Pyruvate was completely metabolized fermentatively (Fig. 6A), giving rise to ca. 27 mM acetate (Fig. 6A). As in Fig. 2, use of the organic acid and corresponding production of acetate by the hyd mutant were slower than by the other two strains. In terms of reducing gas production, the three strains were all different. The hmc mutant produced up to 75,000 ppm (7.5% [vol/vol] of the gas phase; 3 mM) of H2. In contrast, the hyd mutant produced less H2 than the wild-type strain (Fig. 6B). However, it produced 4,000 ppm (0.16 mM) of CO, which remained as a stable fermentation product (Fig. 6D). Although these concentrations are significant, they are small relative to the concentration of acetate, indicating formation of other nongaseous fermentation products, i.e., fumarate, succinate, and isobutyrate, as indicated below.
FIG. 5.
Effect of fumarate and/or hydrogen on fermentative growth of wild-type D. vulgaris on pyruvate. Growth was in WP medium containing pyruvate (30 mM) and no sulfate. Fumarate (34 mM) was added where indicated. Addition of hydrogen was done by changing the headspace gas from 90% (vol/vol) N2-10% CO2 to 80% (vol/vol) H2-20% CO2. Cell density is plotted as a function of time.
FIG. 6.
Growth physiology of the wild-type (WT) and hyd and hmc mutant strains in WP medium containing pyruvate (30 mM), sulfate (3 mM), and a headspace of 10% (vol/vol) CO2 and 90% N2. Plotted as a function of time are concentrations of pyruvate (open symbols) and acetate (filled symbols) (A), H2 concentration in the headspace (B), concentration of a putative fumarate intermediate (C), and CO concentration in the headspace (D). The symbol definitions in panel D apply to the entire figure.
Formate as electron donor.
Like hydrogen, formate is metabolized by a periplasmic dehydrogenase, yielding periplasmic protons when oxidizing formate (Fig. 1, reaction 2). Inoculation of medium containing 35 mM formate and 28 mM sulfate gave a rapid production of H2 that proceeded linearly with time (Fig. 7B). This hydrogen resulted from formate fermentation (Table 1, reaction 6). The observed hydrogen burst is again caused by an imbalance of formate oxidation and sulfate reduction reactions prior to growth. H2 consumption, leading to a decrease in the headspace hydrogen concentration, coincided with the onset of sulfate reduction and started first in the wild type and then in the two mutant strains. The use of 35 mM formate (Fig. 7D) by all three strains gave rise to a reduction of ca. 8 mM sulfate (Fig. 7C), in agreement with the expected 4:1 stoichiometry. Little cell mass was formed. The data in Fig. 7A indicated a Ysulfate of 3.8 g/mol of sulfate for all three strains. No CO was produced under these culture conditions. Acetate, included at a concentration of 3 mM for use as a possible carbon source, was not detectably metabolized. A hydrogen uptake role in formate oxidation is indicated for both Fe-only hydrogenase and the Hmc complex under these conditions, because absence of either leads to higher headspace hydrogen concentrations.
FIG. 7.
Growth physiology of the wild-type (WT) and hyd and hmc mutant strains in WP medium containing formate (35 mM), sulfate (28 mM), and a headspace of 10% (vol/vol) CO2 and 90% N2. Plotted as a function of time are cell density (A), H2 concentration in the headspace (B), sulfate concentration (C), and formate concentration (D). The symbol definitions in panel C apply to the entire figure.
Detection of an organic acid intermediate.
Analysis of organic acid concentrations by high-pressure liquid chromatography (HPLC) indicated the following retention times for the standards (10 mM) that were routinely used: succinate, 12.2 min; lactate, 13.3 min; formate, 14.3 min; acetate, 15.4 min; propionate, 17.6 min; butyrate, 20.7 min. A metabolic intermediate with a maximal concentration at 75 to 100 h was observed at 14.2 min whenever lactate or pyruvate was used as the electron donor. Despite its similar retention time, the intermediate was not formate. It was easily seen with UV but not with refractive-index detection, indicating that it has a much higher molar absorption coefficient at 210 nm than formate. Samples with a high concentration of the intermediate did not have significant formate concentrations when formate was analyzed enzymatically. Determination of the retention times of a variety of organic acids indicated that the intermediate was likely fumarate, which has a high UV absorbance and a retention time similar to that of formate. Transient fumarate concentrations were especially high during pyruvate fermentation by the wild-type strain (Fig. 6C). Approximately 10-fold lower maximal concentrations were observed in the cultures described in Fig. 2 to 4. Formation of succinate (up to 1.5 mM) and isobutyrate (putative, up to 3 mM) as possible end products of pyruvate fermentation was observed by HPLC (results not shown). In view of these results, it was determined if fumarate addition to D. vulgaris, growing by pyruvate fermentation, can increase cell yield. Although cell densities were slightly higher (Fig. 5), the difference was too small to allow the conclusion that D. vulgaris can use fumarate effectively as an acceptor of electrons originating from pyruvate, as shown also by others (7, 12).
DISCUSSION
The hydrogen burst of D. vulgaris was first described in detail by Tsuji and Yagi (22) as the production of H2 prior to the production of H2S and growth. Use of a sensitive reducing gas detector in the present study allowed the demonstration that D. vulgaris can also produce CO under these conditions. The hydrogen burst is therefore more appropriately referred to as the fermentation burst. Nongaseous fermentation products are also likely formed during the burst period. However, these would be difficult to detect with standard HPLC methods in view of the sensitivity required. When grown with excess sulfate, D. vulgaris exhibited a fermentation burst with all three of the organic acids tested (Fig. 2C, 4C and D, and 7B). The burst was particularly long, up to 50 h, yielding in excess of 8,000 ppm (0.32 mM) of headspace hydrogen, when formate was the electron acceptor (Fig. 7). The fermentation burst likely results in the generation of a sufficiently low redox potential and of sufficient ATP to start sulfate reduction.
The results presented in this paper suggest that, in addition to the cycling of hydrogen (Fig. 1, production through reactions 7, 8-i, and 11 and consumption through reactions 1, 3, 4, and 5), D. vulgaris Hildenborough can produce and consume CO during sulfate reduction. The suggested pathway for formation of CO from pyruvate and its subsequent conversion to CO2 and H2 through the action of cytoplasmic CO dehydrogenase (CODH) and CO-dependent hydrogenase are indicated in Fig. 1 (reactions 8-ii, 12, and 13, respectively). The genome sequence of D. vulgaris indicates the presence of a CODH very similar to that of Rhodospirillum rubrum (9). As in R. rubrum, the CODH-encoding gene is preceded by a gene for a CO-responsive transcriptional activator, CooA. In R. rubrum, CooA ensures transcription of the CODH-encoding gene only when the CO concentration reaches an appropriate threshold (8, 18). Together with a CO-dependent hydrogenase, CODH catalyzes CO oxidation to bicarbonate and hydrogen (Table 1, reaction 4). D. vulgaris also has the genes for a membrane-bound, CO-dependent hydrogenase. Unlike those of R. rubrum, these genes are not adjacent to those for CODH. Nevertheless, the presence of a CooA-regulated CODH-encoding gene in D. vulgaris makes it likely that this enzyme, together with the CO-dependent hydrogenase, functions in CO oxidation because it is synthesized only when the CO concentration reaches an appropriate threshold, as in R. rubrum.
With respect to the suggested synthesis of CO from pyruvate (Fig. 1, reaction 8-ii), it should be noted that microorganisms like Clostridium thermaceticum can transfer CO2 formed in reaction 2 of Table 1 directly to CODH, which converts the bound CO2 to CO with electrons derived from hydrogen (24). Production of CO from CO2 by this enzyme is energetically unfavorable (10), as indicated in Table 1 (reaction 4 in reverse: +15.1 kJ/mol), but this can be driven by the larger negative free-energy change of reaction 2 of Table 1. Thus, formation of CO from reactions 2 and 4 (in reverse) is, in principle, possible. However, because Desulfovibrio does not have the acetyl coenzyme A pathway for the synthesis of acetate and because its CODH is likely to catalyze CO uptake, as already discussed, it should be stated that the nature of the enzyme system that synthesizes CO from pyruvate in D. vulgaris is unknown.
D. vulgaris’ need for metabolic cycling to obtain net ATP synthesis from sulfate reduction by lactate has been explained in the introduction. Three possible mechanisms of metabolic cycling have been indicated in Fig. 1: cycling of hydrogen, as proposed previously (14); cycling of CO, for which evidence has been presented here; and cycling of formate. The latter requires the presence of a pyruvate-formate lyase (Fig. 1, reaction 8-iii). Evidence for formate cycling could not be obtained in the present study because a highly UV-absorbing organic acid tentatively identified as fumarate coeluted with formate in the HPLC system used. However, the D. vulgaris genome sequence indicates multiple pyruvate or oxo-organic acid oxidoreductases in addition to the pyruvate:ferredoxin oxidoreductase (Fig. 1, enzyme 8-i). One of these has strong homology to pyruvate-formate lyase from Escherichia coli and could represent enzyme 8-iii (Fig. 1). The fact that four different periplasmic formate dehydrogenases (Fig. 1, reaction 2), identical to the number of periplasmic hydrogenases (Fig. 1, reaction 1), are indicated by the genome sequence also provides evidence for possible formate cycling. With respect to H2 and CO, it should be stated that their steady-state concentrations are extremely low (∼4 μM in the gas phase) when wild-type D. vulgaris grows with lactate as the electron donor for sulfate reduction. However, these low steady-state values could result as the balance between significant production and consumption fluxes. The magnitude of these fluxes cannot be derived from the steady-state concentrations. Another way to approach the problem of estimating the flux involved in metabolic cycling of hydrogen and/or CO is to consider that the cells do not achieve net ATP synthesis without it. Assuming, as for hydrogen (1), a net synthesis of one ATP molecule per mol of sulfate reduced, three periplasmic protons must be formed for every two lactate molecules oxidized. Hence, the cycling flux needs to be at least 40% of the metabolic flux that converts lactate into acetate and CO2.
It is not possible to estimate how much of this cycling flux follows suggested path 8-i, 8-ii, or 8-iii (Fig. 1) from the data presented here. To make an estimate, one would have to temporarily halt sulfate reduction and very rapidly (i.e., on the second, rather than the hour, time scale) measure the accumulation of potentially cycling metabolites. A temporary pause in sulfate reduction, the reasons for which are not clear, was observed in cultures of the hyd mutant with lactate and excess sulfate at the start of stationary phase (Fig. 2D, 100 h). This pause led to accumulation of hydrogen and CO (Fig. 2C and F), suggesting that these are potential cycling intermediates, a conclusion that can also be drawn from the observation of hydrogen and CO accumulation during the fermentation burst. It is not known why, when lactate was the substrate, little CO was seen during the fermentation burst (despite the fact that pyruvate is formed from lactate, which may subsequently yield CO), compared to later in the growth curve. Apparently, the physiological state (log phase or stationary phase) influences the use of the three paths suggested in Fig. 1. Likewise, one cannot draw firm conclusions with respect to the importance of path 8-i or 8-ii (Fig. 1) from the final concentrations of H2 or CO in the fermentations represented in Fig. 3 and 6. As explained already, these were often small in comparison to the amount of acetate formed.
Although the Fe-only hydrogenase mutant does not produce large amounts of CO under all of the metabolic conditions tested, it does produce much more CO than either the wild type or the hmc mutant. This increased production should help in identifying the metabolic pathway or mechanism involved. It is known that Fe-only hydrogenase is inhibited by 0.1 μM CO in aqueous solution (6). Ironically, the concentrations of CO formed by the hyd mutant (up to 0.2 mM in the gas phase and 5 μM in the aqueous phase) would be more than sufficient to inhibit Fe-only hydrogenase, had it been present. Fe-only hydrogenase may thus be required for the fermentative production of H2 from pyruvate (Table 1, reaction 2 or 4), making CO accumulate in its absence. A role for Fe-only hydrogenase in hydrogen production under fermentative conditions is also suggested by the lower hydrogen headspace concentrations in cultures of the hyd mutant strain during lactate fermentation (Fig. 3C). This role is in addition to its function in hydrogen uptake in cultures grown on medium containing only hydrogen or lactate and hydrogen as electron donor for sulfate reduction (16). In contrast, the role of the Hmc complex appears to be always hydrogen uptake (Fig. 1, reaction 4) because its absence leads to hydrogen headspace concentrations greater than or equal to those observed for the wild-type strain under all of the culture conditions tested. In the absence of sulfate, the Hmc complex may channel electrons to cytoplasmic organic acid electron acceptors (Fig. 1, reaction data not shown), because hydrogen concentrations higher than that obtained with the wild type accumulate in the absence of the Hmc complex (Fig. 3C and 6B).
CO production may also be important for D. vulgaris during growth on lactate or pyruvate because most hydrogenases have CO as a ligand in their active site (13, 23), requiring continuous availability of CO for the synthesis of active enzyme. Also, the production of small, gaseous, rapidly diffusing fermentation products, like H2 and CO, may allow methanogens or other anaerobes to grow syntrophically with D. vulgaris (11, 15).
Acknowledgments
The work reported here was done during a sabbatical leave at the Max Planck Institute for Marine Microbiology in Bremen, Germany. It was supported by a research grant from the Natural Science and Engineering Research Council of Canada and by the Max Planck Society of Germany. I acknowledge the support of a sabbatical fellowship from the University of Calgary; of a fellowship from the Hanse Wissenschaftkolleg in Delmenhorst, Germany; and of a visiting professorship from the CNRS in Marseille, France. A database of the D. vulgaris Hildenborough genome was searched at the website of The Institute for Genomic Research at http://www.tigr.org. Sequencing of this genome is financially supported by the U.S. Department of Energy.
I am very grateful to Alain Dolla, Marie-Claire Durand, Jens Harder, Daniela Lange, Ramona Appel, Christina Probian, Ralf Rabus, Johanna Voordouw, and Fritz Widdel for discussions and technical assistance.
REFERENCES
- 1.Badziong, W., and R. K. Thauer. 1978. Growth yields and growth rates of Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate and hydrogen plus thiosulfate as the sole energy sources. Arch. Microbiol. 117:209-214. [DOI] [PubMed] [Google Scholar]
- 2.Badziong, W., and R. K. Thauer. 1980. Vectorial electron transport in Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate as sole energy source. Arch. Microbiol. 125:167-174. [Google Scholar]
- 3.Bak, F., G. Scheff, and K. H. A. Jansen. 1991. Rapid and sensitive ion chromatographic technique for the determination for sulfate and sulfate reduction rates in fresh water lake sediments. FEMS Microbiol. Ecol. 85:23-30. [Google Scholar]
- 4.Cornish-Bowden, A., and M. L. Cardenas. 2001. Silent genes given voice. Nature 409:571-572. [DOI] [PubMed] [Google Scholar]
- 5.Dolla, A., B. K. J. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc-operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox potential niche establishment. Arch. Microbiol. 174:143-151. [DOI] [PubMed] [Google Scholar]
- 6.Fauque, G., H. D. Peck, Jr., J. J. Moura, B. H. Huynh, Y. Berlier, D. V. DerVartanian, M. Texeira, A. E. Przybyla, P. A. Lespinat, I. Moura, and J. LeGall. 1988. The three classes of hydrogenases from sulfate-reducing bacteria of the genus Desulfovibrio. FEMS Microbiol. Rev. 4:299-344. [DOI] [PubMed] [Google Scholar]
- 7.Grossman, J. P., and J. R. Postgate. 1955. The metabolism of malate and certain other compounds by Desulphovibrio desulphuricans. J. Gen. Microbiol. 12:429-445. [DOI] [PubMed] [Google Scholar]
- 8.He, Y., D. Shelver, R. L. Kerby, and G. P. Roberts. 1996. Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J. Biol. Chem. 271:120-123. [DOI] [PubMed] [Google Scholar]
- 9.Kerby, R. L., S. S. Hong, S. A. Ensign, L. J. Coppoc, P. W. Ludden, and G. P. Roberts. 1992. Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J. Bacteriol. 174:5284-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupton, F. S., R. Conrad, and J. G. Zeikus. 1984. CO metabolism of Desulfovibrio vulgaris strain Madison: physiological function in the absence or presence of exogenous substrates. FEMS Microbiol. Lett. 23:263-268. [Google Scholar]
- 11.McInerney, M. J., and M. P. Bryant. 1981. Anaerobic degradation of lactate by syntrophic associations of Methanosarcina barkeri and Desulfovibrio species and effect of H2 on acetate degradation. Appl. Environ. Microbiol. 41:346-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, J. D. A., and D. S. Wakerley. 1966. Growth of sulphate-reducing bacteria by fumarate dismutation. J. Gen. Microbiol. 43:101-107. [DOI] [PubMed] [Google Scholar]
- 13.Nicolet, Y., C. Piras, P. Legrand, C. E. Hatchikian, and J. C. Fontecilla-Camps. 1999. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure 7:13-23. [DOI] [PubMed] [Google Scholar]
- 14.Odom, J. M., and H. D. Peck, Jr. 1981. Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria Desulfovibrio sp. FEMS Microbiol. Lett. 12:47-50. [Google Scholar]
- 15.Phelps, T. J., R. Conrad, and J. G. Zeikus. 1985. Sulfate-dependent interspecies H2 transfer between Methanosarcina barkeri and Desulfovibrio vulgaris during coculture metabolism of acetate and methanol. Appl. Environ. Microbiol. 50:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pohorelic, B. K. J., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postgate, J. R. 1984. The sulphate-reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 18.Shelver, D., R. L. Kerby, Y. He, and G. P. Roberts. 1997. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl. Acad. Sci. USA 94:11216-11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stams, A. J. M., and T. A. Hansen. 1982. Oxygen-labile l(+)-lactate dehydrogenase activity in Desulfovibrio HL21. FEMS Microbiol. Lett. 13:389-394. [Google Scholar]
- 20.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traore, A. S., C. E. Hatchikian, J.-P. Belaich, and J. LeGall. 1981. Microcalorimetric studies of the growth of sulfate-reducing bacteria: energetics of Desulfovibrio vulgaris growth. J. Bacteriol. 145:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuji, K., and T. Yagi. 1980. Significance of hydrogen burst from growing cultures of Desulfovibrio vulgaris Miyazaki, and the role of hydrogenase, and cytochrome c3 in energy production system. Arch. Microbiol. 125:35-42. [Google Scholar]
- 23.Volbeda, A., M.-H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 24.White, D. 2000. The physiology and biochemistry of prokaryotes, 2nd edition, p. 318. Oxford University Press, New York, N.Y.
- 25.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 4. Springer Verlag, New York, N.Y.
- 26.Widdel, F., and T. A. Hansen. 1991. The dissimilatory sulfate- and sulfur-reducing bacteria, p. 585-624. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 1. Springer Verlag, New York, N.Y.