Abstract
The 3-hydroxypropionate cycle is a bicyclic autotrophic CO2 fixation pathway in the phototrophic Chloroflexus aurantiacus (Bacteria), and a similar pathway is operating in autotrophic members of the Sulfolobaceae (Archaea). The proposed pathway involves in a first cycle the conversion of acetyl-coenzyme A (acetyl-CoA) and two bicarbonates to l-malyl-CoA via 3-hydroxypropionate and propionyl-CoA; l-malyl-CoA is cleaved by l-malyl-CoA lyase into acetyl-CoA and glyoxylate. In a second cycle, glyoxylate and another molecule of propionyl-CoA (derived from acetyl-CoA and bicarbonate) are condensed by a putative β-methylmalyl-CoA lyase to β-methylmalyl-CoA, which is converted to acetyl-CoA and pyruvate. The putative l-malyl-CoA lyase gene of C. aurantiacus was cloned and expressed in Escherichia coli, and the recombinant enzyme was purified and studied. β-Methylmalyl-CoA lyase was purified from cell extracts of C. aurantiacus and characterized. We show that these two enzymes are identical and that both enzymatic reactions are catalyzed by one single bifunctional enzyme, l-malyl-CoA lyase/β-methylmalyl-CoA lyase. Interestingly, this enzyme works with two different substrates in two different directions: in the first cycle of CO2 fixation, it cleaves l-malyl-CoA into acetyl-CoA and glyoxylate (lyase reaction), and in the second cycle it condenses glyoxylate with propionyl-CoA to β-methylmalyl-CoA (condensation reaction). The combination of forward and reverse directions of a reversible enzymatic reaction, using two different substrates, is rather uncommon and reduces the number of enzymes required in the pathway. In summary, l-malyl-CoA lyase/β-methylmalyl-CoA lyase catalyzes the interconversion of l-malyl-CoA plus propionyl-CoA to β-methylmalyl-CoA plus acetyl-CoA.
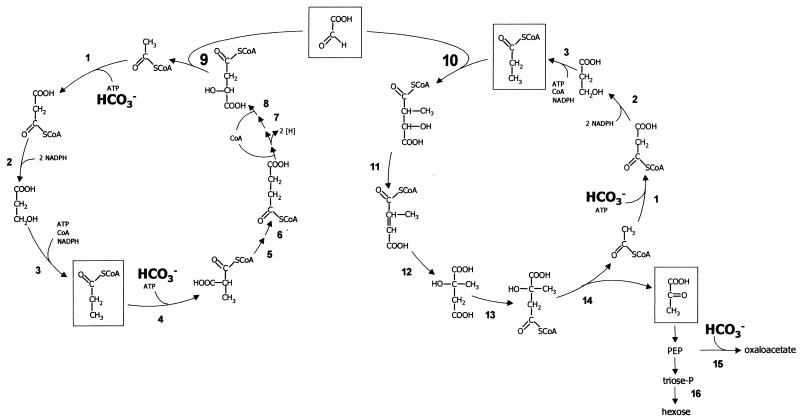
The phototrophic green nonsulfur bacterium Chloroflexus aurantiacus (Bacteria) uses a new autotrophic CO2 fixation pathway termed the 3-hydroxypropionate cycle (1, 11, 17-22, 37-38) (Fig. 1, left). This bacterium grows optimally at 55°C under heterotrophic conditions but can also grow in mineral salt medium using CO2 as sole carbon source (21, 31-32, 36). A similar, yet not identical, pathway appears to operate in CO2 fixation by autotrophic acidophilic Archaea of the family Sulfolobaceae (Crenarchaeota), such as Acidianus brierleyi, Metallosphaera sedula, and Sulfolobus metallicus (6, 23, 27). The proposed 3-hydroxypropionate cycle is actually a bicyclic pathway (18) and is shown in Fig. 1.
FIG. 1.
Proposed bicyclic pathway for autotrophic CO2 fixation in C. aurantiacus. (Left) 3-Hydroxypropionate cycle for CO2 fixation affording glyoxylate as first net CO2 fixation product. (Right) Proposed glyoxylate assimilation cycle. 1, acetyl-CoA carboxylase; 2, malonyl-CoA reductase; 3, propionyl-CoA synthase; 4, propionyl-CoA carboxylase; 5, epimerase; 6, methylmalonyl-CoA mutase; 7, citrate cycle enzymes (succinate dehydrogenase, fumarate hydratase); 8, succinyl-CoA:l-malate CoA transferase; 9, l-malyl-CoA lyase; 10, erythro-β-methylmalyl-CoA lyase; 11, postulated β-methylmalyl-CoA dehydratase; 12, postulated mesaconyl-CoA hydratase; 13, postulated citramalate activation by succinyl-CoA:l-citramalate CoA transferase; 14, postulated citramalyl-CoA lyase; 15, phosphoenolpyruvate carboxylase; 16, gluconeogenesis enzymes. erythro-β-methylmalyl-CoA, mesaconyl-CoA, and citramalate were identified as putative intermediates of the proposed glyoxylate assimilation cycle (18).
Each turn of the first cycle results in the net fixation of two molecules of bicarbonate into one molecule of glyoxylate (Fig. 1, left). Acetyl-coenzyme A (acetyl-CoA) is carboxylated to malonyl-CoA by conventional ATP-dependent acetyl-CoA carboxylase. The reduction of malonyl-CoA to propionyl-CoA requires 3 NADPH and 1 Mg-ATP (products, AMP and PPi) and proceeds via free 3-hydroxypropionate as an intermediate (1, 19, 22, 38). Propionyl-CoA is carboxylated to methylmalonyl-CoA followed by isomerization of methylmalonyl-CoA to succinyl-CoA. Succinyl-CoA is used for malate activation by CoA transfer, forming succinate and l-malyl-CoA; succinate in turn is oxidized to malate by conventional enzymes. l-Malyl-CoA is cleaved into acetyl-CoA and glyoxylate by l-malyl-CoA lyase (EC 4.1.3.24) (Fig. 1, reaction 9) (17).
In a second cycle, another molecule of acetyl-CoA and bicarbonate are converted to propionyl-CoA (Fig. 1, right). Glyoxylate and propionyl-CoA are disproportionated to acetyl-CoA and pyruvate via erythro-β-methylmalyl-CoA, mesaconyl-CoA, and citramalate (18). Hence, acetyl-CoA (the primary CO2 acceptor molecule) is reformed from glyoxylate (the primary CO2 fixation product). Propionyl-CoA gives rise to pyruvate, the central biosynthetic or gluconeogenetic precursor.
There are three characteristic processes in the proposed autotrophic CO2 fixation pathway that are not present in other autotrophs. The first one is the reduction of malonyl-CoA to propionyl-CoA. This complex reaction sequence formally involves five steps (Fig. 1, left) but is catalyzed by only two enzymes. These two enzymes, malonyl-CoA reductase (reaction 2) and propionyl-CoA synthase (reaction 3), have recently been purified and characterized (1, 22).
The second process concerns the formation of the primary CO2 acceptor acetyl-CoA from malate under release of glyoxylate. Previous work showed that malate is converted to l-malyl-CoA with succinyl-CoA as CoA donor, a reaction that is catalyzed by succinyl-CoA:l-malate CoA transferase (reaction 8). l-Malyl-CoA is cleaved into acetyl-CoA and glyoxylate by l-malyl-CoA lyase (reaction 9) (17).
The third characteristic feature is a new pathway used to assimilate glyoxylate (18). The proposed pathway shown in Fig. 1 (right) involves the condensation of glyoxylate and propionyl-CoA to erythro-β-methylmalyl-CoA (reaction 10). The putative enzyme β-methylmalyl-CoA lyase, which catalyzes the initial condensation reaction of this second cycle, was demonstrated in partially purified protein fractions from C. aurantiacus, but it was not further purified and characterized. Besides β-methylmalyl-CoA, mesaconyl-CoA, citramalate, and citramalyl-CoA are likely intermediates, the latter being cleaved into acetyl-CoA and pyruvate.
In the present work the putative l-malyl-CoA lyase (mclA) gene of C. aurantiacus was expressed in Escherichia coli, and β-methylmalyl-CoA lyase was purified from cell extracts of C. aurantiacus. We show that both reactions (Fig. 1, reactions 9 and 10) are catalyzed by one enzyme. Interestingly, the enzyme works with two different substrates at the same time in two different directions.
MATERIALS AND METHODS
Bacteria and growth conditions.
C. aurantiacus strain OK-70-fl (DSM 636) was grown anaerobically at 55°C and pH 8 in 12-liter glass fermentors under autotrophic or heterotrophic conditions as described earlier (17). Cells were stored in liquid nitrogen until use. E. coli strain XL1-Blue recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB lacIqZΔM15 Tn10 [Tetr]) was grown at 37°C in Luria-Bertani (LB) medium (34). Ampicillin was added to E. coli cultures to a final concentration of 50 μg/ml. Cell extracts were prepared aerobically as described previously (17).
Materials.
Chemicals were obtained from Fluka (Neu-Ulm, Germany), Merck (Darmstadt, Germany), Sigma-Aldrich (Deisenhofen, Germany), or Roth (Karlsruhe, Germany). Biochemicals were purchased from Roche Diagnostics (Mannheim, Germany), Applichem (Darmstadt, Germany), or Gerbu (Craiberg, Germany). Materials and equipment for protein purification were obtained from Amersham Biosciences (Freiburg, Germany) or Millipore (Eschborn, Germany). Materials for cloning and expression were purchased from MBI Fermentas (St. Leon-Rot, Germany), MWG Biotech AG (Ebersberg, Germany), or Peqlab Biotechnologie (Erlangen, Germany). [2-14C]propionate was obtained from Hartmann Analytic (Braunschweig, Germany).
Syntheses. (i) l-Malyl-CoA.
l-Malyl-CoA was chemically synthesized according to the methods of Eggerer and Grünewälder (10), with slight modification. The synthesis intermediate l-malylcaprylcysteamine [S-(β-hydroxysuccinyl)-N-caprylcysteamine] was synthesized by Richard Krieger (Institut für Organische Chemie, Freiburg, Germany) as described elsewhere (10, 28). l-Malyl-CoA was stored as a freeze-dried powder at −20°C. It contained 70% CoA-thioester and 30% free CoA as determined by high-performance liquid chromatography (HPLC) separation and detection at 260 nm (see below).
(ii) Succinyl-CoA, acetyl-CoA, and propionyl-CoA.
Succinyl-CoA, acetyl-CoA, and propionyl-CoA were synthesized as described elsewhere (17), and the dry powders were stored at −20°C.
(iii) [2-14C]propionyl-CoA.
[2-14C]propionyl-CoA was synthesized according to the protocol described previously for synthesis of [1,2-14C]acetyl-CoA (17), using [2-14C]propionate instead of [1,2-14C]acetate.
(iv) β-Methylmalyl-CoA.
β-Methylmalyl-CoA was synthesized enzymatically from propionyl-CoA and glyoxylate using a preparation of recombinant l-malyl-CoA/β-methylmalyl-CoA lyase protein. A reaction mixture (50 ml) containing 200 mM morpholinepropanesulfonic acid (MOPS)-K+ buffer (pH 7.7), 2.5 mM glyoxylate, 2 mM MgCl2, 2.5 mM propionyl-CoA, 36.7 kBq of [2-14C]propionyl-CoA as tracer, and 6.3 ml of recombinant l-malyl-CoA/β-methylmalyl-CoA lyase protein fraction (14.4 mg of protein; 2 U) was incubated at 55°C. After 60 min of incubation, the mixture was adjusted to pH 1.8 by addition of 1 M HCl. Protein was removed by centrifugation. The supernatant was extracted twice with diethyl ether to remove remaining glyoxylate. The volume of the supernatant was reduced to 5 ml by flash evaporation at 30°C (3 kPa) and subjected to a reversed-phase column (250 by 20 mm, 10 μm; Grom-Sil 120 ODS-4 HE; Crom, Herrenberg-Kayh, Germany), which was developed by a step gradient (64 ml each) of 2.9, 4.8, 5.7, 6.7, 8.6, and 10.5% acetonitrile (vol/vol) in 50 mM potassium phosphate buffer, pH 6.7, with a flow rate of 8 ml min−1. The effluent was monitored by a radiomonitor and photometrically (260 nm). A radioactive fraction that eluted at a retention volume of 200 ml was collected and its pH was adjusted to pH 1.8 by addition of 1 M HCl. The acetonitrile was evaporated by flash evaporation at 30°C (2 kPa), the sample was lyophilized, and the powder was stored at −20°C. It contained 92% CoA-thioester and 8% free CoA as determined with 5,5′-dithiobis(2-nitrobenzoate) (Ellman's reagent) and detection at 412 nm (ɛ412 = 13,600 M−1 cm−1 [9]). The reaction was also used to determine the equilibrium constant keq.
Enzyme assays.
l-Malyl-CoA/β-methylmalyl-CoA lyase was tested at 55°C in the lyase or condensation direction.
(i) Lyase reaction.
The l-malyl-CoA- or β-methylmalyl-CoA-dependent formation of glyoxylate was monitored photometrically at 324 nm with phenylhydrazine in a continuous assay as described previously (15, 17) (ɛ324 for glyoxylate phenylhydrazone = 17,000 M−1 cm−1 [33]). The assay mixture (0.5 ml) contained 200 mM MOPS-K+ buffer (pH 7.7), 4 mM MgCl2, 3.5 mM phenylhydrazinium chloride, 1 mM l-malyl-CoA or β-methylmalyl-CoA, and protein. The reaction was started by addition of CoA-thioester. Phenylhydrazine had no effect on the stability of CoA derivatives.
(ii) Condensation reaction.
The propionyl-CoA- or acetyl-CoA-dependent consumption of glyoxylate was monitored photometrically with phenylhydrazine in a discontinuous assay. The assay mixture (0.2 ml) contained 200 mM MOPS-K+ buffer [pH 7.7], 4 mM MgCl2, 2 mM glyoxylate, 4 mM concentration of propionyl-CoA or acetyl-CoA, and protein. The reaction was started by the addition of protein. After 20 min of incubation, samples (25 μl) were retrieved, diluted, and cooled down to room temperature by addition of 0.975 ml of 200 mM MOPS-K+ buffer (pH 7.4) containing 3.5 mM phenylhydrazinium chloride. After 15 min of incubation at room temperature, the formed glyoxylate phenylhydrazone was detected at 324 nm. In additional experiments, glyoxylate was replaced by pyruvate or oxaloacetate (2 mM concentration each), and acetyl-CoA or propionyl-CoA were replaced by succinyl-CoA. When the apparent Km values were to be determined, the concentration of the CoA-thioester or glyoxylate was varied between 0.05 and 20 mM; glyoxylate was either omitted (lyase reactions) or its concentration remained fixed at 2 mM, and the samples (25 μl) were developed after 5 min of incubation at 55°C (condensation reaction). Buffers used to determine the pH optimum were 200 mM MOPS-K+, pH 6.0 to 8.9, at room temperature, which corresponded to pH 5.7 to 8.6 at 55°C; the ΔpH of 0.3 was experimentally determined. The dependence of the reaction on divalent metal ions was investigated by addition of 1 mM EDTA to the reaction mixture in the absence of a divalent cation. The temperature optimum of the l-malyl-CoA lyase reaction was determined by varying the reaction temperature between 45 and 75°C.
Formation of acetyl-CoA and β-methylmalyl-CoA from l-malyl-CoA and propionyl-CoA.
The test mixture (0.2 ml) contained 200 mM MOPS-K+ buffer (pH 7.7), 4 mM MgCl2, 2.5 mM l-malyl-CoA, 0.75 mM CoA (present as an impurity of l-malyl-CoA), 2.5 mM propionyl-CoA, and 24 μg of purified recombinant l-malyl-CoA lyase. l-Malyl-CoA was omitted in a control experiment. The addition of recombinant enzyme started the reactions. Samples of 50 and 150 μl were taken after 0 and 20 min of incubation at 55°C, and the reaction was stopped by addition of 10 and 30 μl of 1 M HCl, respectively. Protein was removed by centrifugation, and samples were analyzed for glyoxylate and for CoA thioesters by HPLC. A reversed-phase column (LiChrospher 100; end-capped; 5 μm, 125 by 4 mm; Merck, Darmstadt, Germany) was used for separation of CoA-thioesters. A gradient from 2 to 10% acetonitrile in 200 mM sodium phosphate buffer, pH 4.8, with a flow rate of 1 ml min−1 over 40 min was used. CoA-thioesters were detected at 260 nm. Retention times were 2 min (free organic acids), 8 min (l-malyl-CoA), 10 min (β-methylmalyl-CoA; free CoA), 16 min (acetyl-CoA), and 24 min (propionyl-CoA).
Cloning and expression of a putative l-malyl-CoA lyase (mlcA) gene in E. coli XL1-Blue.
Standard protocols were used for preparation, cloning, transformation, amplification, and purification of DNA (3, 34). Plasmid DNA preparation was performed according to the method of Birnboim and Doly (4). On the basis of a DNA and protein sequence alignment of the mlcA gene (l-malyl-CoA lyase protein EC 4.1.3.24; accession number AAB58884) from Methylobacterium extorquens, a region on the C. aurantiacus genome (contig 965) showed one highly conserved putative l-malyl-CoA lyase gene. Two oligonucleotides were designed: (i) 5′-GAGCATCATGAAGGGTATTC-3′ (20-mer; primer contained PagI restriction site) partially corresponding to nucleotides 7026 to 7045 of contig 965 of the genome database of C. aurantiacus (http://www.jgi.doe.gov/JGI_microbial/html/chloroflexus/chloro_homepage.html); and (ii) 5′-CTTGCTGCAGCGTCACAGACC-3′ (21-mer, primer contained PstI restriction site) partially corresponding to nucleotides 9351 to 9372. These primers were used in a PCR containing 0.5 μg of chromosomal DNA of C. aurantiacus and 2.5 U of Pwo polymerase (Peqlab Biotechnologie), and the Peqlab DNA amplification kit was used. An annealing temperature of 60°C was used to amplify a 2,347-bp genomic region which contained the putative mlcA gene. The PCR product was purified and cloned into pTrc 99A (accession number U13872; Amersham Biosciences) by using the NcoI and PstI restriction sites of the multiple cloning site. Plasmid PCR and colony PCR confirmed the correct integration of the PCR fragment into the vector. The recombinant plasmid was transformed into E. coli XL1-Blue, and the expression of the putative mlcA gene was induced at an optical density at 578 nm of 0.7 (12-liter fermentor, 37°C) by addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside to the LB-ampicillin medium. After induction, the culture was incubated for 4 h at room temperature. Cells were harvested at 4°C and stored in liquid nitrogen prior to purification of recombinant protein. Transformed E. coli XL1-Blue cells containing the pTrc-vector but no PCR fragment served as a control for the expression of the putative mlcA gene.
Computer analysis.
The BLAST program was used to search protein databases at the National Center for Biotechnology Information (Bethesda, Md.) and the local C. aurantiacus server (http://spider.jgi-psf.org/JGI_microbial/bin/psf_blast?PROJEKT_ID = 2351478) at the Department of Energy (DOE) Joint Genome Institute (Walnut Creek, Calif.).
Purification of recombinant l-malyl-CoA lyase from E. coli.
The purification was performed at 4°C, followed by measuring the l-malyl-CoA lyase reaction.
(i) Heat precipitation.
Cell extract (100,000 × g supernatant) from 25 g of cells (wet weight) was incubated at 65°C for 20 min to precipitate unwanted protein from E. coli cells, followed by ultracentrifugation (100,000 × g) at 4°C for 60 min.
(ii) DEAE-Sepharose fast flow chromatography.
The supernatant after heat precipitation (30 ml) was applied to a 50-ml DEAE-Sepharose Fast Flow column (Amersham Biosciences; flow rate, 3 ml min−1) which had been equilibrated with 20 mM MOPS-K+ buffer (pH 7.2) containing 10% (vol/vol) glycerol (buffer A). The column was washed with four bed volumes of buffer A and three bed volumes of buffer A containing 100 mM KCl and developed with a linear gradient from buffer A to buffer A plus 200 mM KCl over 250 ml. Active fractions (120 to 180 mM KCl) were pooled (150 ml) and stored at −80°C.
(iii) Size exclusion chromatography.
The volume of the active pool obtained from DEAE-Sepharose chromatography was reduced to 4 ml by ultrafiltration (Amicon YM 10 membrane; Millipore) and applied to a 120-ml HiLoad Superdex 200 16/60 column (Amersham Biosciences; flow rate, 1 ml min−1). The column was developed with 20 mM MOPS-K+ buffer (pH 7.6) containing 10% (vol/vol) glycerol and 150 mM KCl. The active protein eluted with a retention volume of 59 to 76 ml, and active fractions were pooled (18 ml) and stored at −80°C.
Purification of β-methylmalyl-CoA lyase from C. aurantiacus.
The purification was performed at 4°C or room temperature and followed by measuring the condensation reaction of glyoxylate with propionyl-CoA.
(i) Heat precipitation.
Cell extract (100,000 × g supernatant) from 25 g of cell mass (wet weight) of autotrophically grown C. aurantiacus was incubated at 65°C for 10 min to precipitate unwanted protein, carotenoids, and other colored compounds, followed by ultracentrifugation (100,000 × g) at 4°C for 30 min.
(ii) DEAE-Sepharose fast flow chromatography.
The supernatant after heat precipitation (30 ml) was applied to a 50-ml DEAE-Sepharose Fast Flow column (Amersham Biosciences; flow rate, 3 ml min−1) which had been equilibrated with 20 mM M-K+ buffer, pH 7.4, containing 10% (vol/vol) glycerol (buffer B). The column was washed with two bed volumes of buffer B and three bed volumes of buffer B containing 100 mM KCl and developed with a linear gradient from buffer B to buffer B plus 200 mM KCl over 250 ml. Active fractions (100 to 160 mM KCl) were pooled (94 ml) and stored at −80°C.
(iii) Phenyl-Sepharose chromatography.
The DEAE fraction was adjusted to a final concentration of 1 M ammonium sulfate by using a 2 M ammonium sulfate solution. The protein fraction was then centrifuged and the supernatant was directly applied to a 20-ml phenyl-Sepharose column (Amersham Biosciences; flow rate, 1 ml min−1). The column had been equilibrated with 20 mM MOPS-K+ buffer (pH 7.9) containing 1 M (NH4)2SO4 and 10% (vol/vol) glycerol. After washing the column with one bed volume of this buffer, the column was developed with 120 ml of a decreasing linear gradient of 1 to 0 M ammonium sulfate, at a flow rate 1 ml min−1. The activity eluted with 500 to 250 mM salt and the pooled fractions (35 ml) were concentrated and desalted immediately by ultrafiltration to a final volume of 2.2 ml.
(iv) Size exclusion chromatography.
The sample obtained from phenyl-Sepharose chromatography and ultrafiltration was applied to a 120-ml HiLoad Superdex 200 16/60 column, with a flow rate of 1 ml min−1. The column was developed with 20 mM MOPS-K+ buffer (pH 7.6) containing 10% (vol/vol) glycerol and 150 mM KCl. The active protein eluted with a retention volume of 62 to 72 ml, and fractions were pooled (11 ml) and stored at −80°C.
(v) Resource Q chromatography.
The sample obtained from size exclusion chromatography was reduced to 1.9 ml and desalted by ultrafiltration using 20 mM MOPS-K+ buffer (pH 7.6) containing 10% glycerol (vol/vol) (buffer C). The protein solution was applied in two runs each onto a 1-ml Resource Q column (Amersham Biosciences; flow rate, 5 ml min−1) which had been equilibrated with buffer C. The column was washed with two bed volumes of buffer C and developed with a gradient from buffer C to buffer C plus 1 M KCl over 20 ml. Active fractions (130 to 250 mM KCl) were pooled (4 ml) and stored at −20°C.
Determination of molecular mass.
The native molecular masses of the purified recombinant protein and the purified protein from C. aurantiacus were estimated using a 120-ml HiLoad Superdex 200 16/60 column (Amersham Biosciences; flow rate, 1 ml min−1). The column was developed with 20 mM MOPS-K+ buffer (pH 7.6) containing 10% (vol/vol) glycerol and 150 mM KCl and calibrated with the following molecular mass standards: ferritin (450 kDa), bovine serum albumin (67 kDa), and ovalbumin (45 kDa). A native gel (8% polyacrylamide) was performed to obtain further information about the molecular masses of the proteins. Bovine serum albumin served as a molecular standard as follows: monomer, 67 kDa; dimer, 134 kDa; trimer, 201 kDa; tetramer, 268 kDa.
Other methods.
Cell extracts were prepared as described previously (17). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12.5% polyacrylamide) was performed as described by Laemmli (26). Proteins were stained with Coomassie blue according to the method of Zehr et al. (39). Protein levels were determined by the method of Bradford (5), using bovine serum albumin as the standard. Determination of the N-terminal amino acid sequence of the purified β-methylmalyl-CoA lyase protein from C. aurantiacus after blotting on polyvinylidene difluoride membrane was performed by TopLab (Martinsried, Germany) using an Applied Biosystems Procise 492 sequencer (Weiterstadt, Germany). The phenylthiohydantoin derivatives were identified with an online Applied Biosystems Analyzer 140 C. Optical absorption spectra of the purified β-methylmalyl-CoA lyase from C. aurantiacus extract (0.44 mg of protein ml−1) and of the purified recombinant l-malyl-CoA lyase from E. coli (2.4 mg of protein ml−1) were collected at room temperature using a Perkin Elmer Life Science Lambda 2S spectrometer and the same buffer as a blank.
RESULTS
Identification of a putative l-malyl-CoA lyase gene of C. aurantiacus and expression in E. coli.
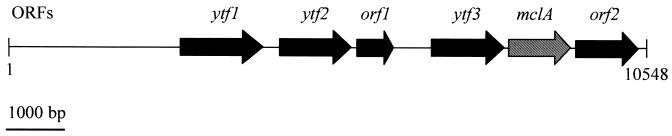
The l-malyl-CoA lyases from M. extorquens (formerly Pseudomonas sp. strain AM1) and Aminobacter aminovorans (formerly Pseudomonas sp. strain MA) have been studied extensively (2, 8, 12-13, 15-16, 33). The enzyme catalyzes a fully reversible Claisen condensation-retrocondensation reaction, with equilibrium constants keq {([acetyl-CoA] × [glyoxylate])/[l-malyl-CoA]} of 4.7 × 10−4 M (as determined for the M. extorquens enzyme) (13) and ∼3 × 10−3 M (as determined for the A. aminovorans enzyme) (15). Screening of the incomplete C. aurantiacus genome for a malyl-CoA lyase-like gene gave one hit on contig 965, with an E-value of 3e − 42 (Fig. 2, mclA). The gene coded for a 38-kDa protein and was flanked by a putative CoA transferase gene coding for a 45-kDa protein (Fig. 2, ytf3), which may function in the formation of l-malyl-CoA from malate and succinyl-CoA. Such an enzyme has been proposed for the 3-hydroxypropionate cycle (Fig. 1, reaction 8). The DNA fragment encompassing the putative CoA transferase gene and the malyl-CoA lyase gene was amplified by PCR, cloned in E. coli, and expressed. E. coli cell extract was heat precipitated, since the l-malyl-CoA lyase activity tolerates 20 min of incubation at 65°C. The supernatant of the heat-treated recombinant E. coli extract showed l-malyl-CoA lyase activity (lyase reaction, 1.5 μmol min−1 mg of protein−1 at 55°C), and this activity was missing (<0.002 μmol min−1 mg of protein−1) in heat-treated cell extract from recombinant E. coli cells lacking the DNA insert. The reverse reaction (condensation reaction of glyoxylate with acetyl-CoA) was catalyzed at 55°C with a specific activity of 0.32 μmol min−1 mg of protein−1. The ratio of the rates of lyase activity over condensation activity at pH 7.4 was 4.6 (Table 1). SDS-PAGE revealed a major protein band of 38 kDa (Fig. 3A).
FIG. 2.
Organization of putative protein coding sequences (ORFs) on contig 965 (10.5 kb) of the C. aurantiacus strain J-10-fl genome. ytf1, putative CoA transferase (1.44 kb, 479 aa, 53 kDa); ytf2, putative CoA transferase (1.22 kb, 405 aa, 45 kDa); orf1, hypothetical protein of unknown function (0.64 kb, 231 aa, 23 kDa); ytf3, putative CoA transferase (1.23 kb, 409 aa, 45 kDa); mclA, putative malyl-CoA lyase gene (1.05 kb, 348 aa, 38 kDa); orf2, hypothetical protein of unknown function (1.06 kb, 352 aa, 38 kDa).
TABLE 1.
Purification of recombinant l-malyl-CoA lyase from E. colia
| Purification step | Total l-malyl-CoA lyase activity (μmol min−1) | Total protein (mg) | Specific l-malyl-CoA lyase activity (μmol min−1 mg of protein−1) | Recovery of l-malyl-CoA lyase activity (%) | Purification of l-malyl-CoA lyase (fold) | Specific condensation activityb (lyase/condensation activity ratio)
|
|
|---|---|---|---|---|---|---|---|
| Glyoxylate + propionyl-CoA | Glyoxylate + acetyl-CoA | ||||||
| Heat precipitationc (65°C, 20 min) | 208.5 | 135.0 | 1.5 | 100 | (1) | 0.419 (3.6) | 0.323 (4.6) |
| DEAE-Sepharose | 149.8 | 109.5 | 1.4 | 71.8 | 0.88 | 0.525 (2.7) | 0.416 (3.4) |
| Gel filtration | 100.0 | 43.2 | 2.3 | 47.9 | 1.5 | 0.501 (4.6) | 0.477 (4.8) |
l-Malyl-CoA lyase was purified from 25 g (wet weight) of E. coli cells, and the l-malyl-CoA lyase reaction was followed at 55°C.
Specific activities (in micromoles per minute per milligram of protein) are shown for the condensation reaction of glyoxylate with propionyl-CoA or acetyl-CoA; values in parentheses are ratios of specific activities for the l-malyl-CoA lyase reaction and the condensation reaction with propionyl-CoA or acetyl-CoA.
100,000 × g supernatant.
FIG. 3.
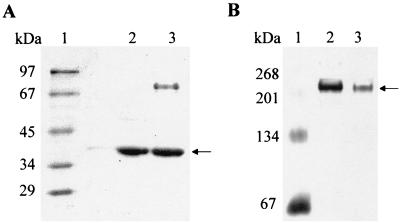
(A) Denaturing PAGE (12.5%) of purified recombinant l-malyl-CoA lyase of C. aurantiacus compared to purified β-methylmalyl-CoA lyase of C. aurantiacus (5 μg each). Lanes: 1, molecular mass marker (rabbit phosphorylase b, 97 kDa; bovine serum albumin, 67 kDa; egg ovalbumin, 45 kDa; lactate dehydrogenase, 34 kDa; carboanhydrase, 29 kDa; lysozyme, 14 kDa); 2, recombinant l-malyl-CoA lyase (size exclusion fraction); 3, purified β-methylmalyl-CoA lyase (Resource Q fraction) for comparison. The arrow indicates the corresponding molecular mass of around 38 kDa of both enzyme preparations. (B) Native PAGE (8%) of both enzyme preparations (5 to 10 μg of protein). Lanes: 1, molecular mass marker (bovine serum albumin monomer, 67 kDa; dimer, 134 kDa; trimer, 201 kDa; tetramer, 268 kDa); 2, recombinant l-malyl-CoA lyase (size exclusion fraction); 3, purified β-methylmalyl-CoA lyase (Resource Q fraction) for comparison. The arrow indicates a molecular mass of ≥210 kDa for both enzymes. The gels were stained with Coomassie brilliant blue R-250.
Purification and properties of the recombinant l-malyl-CoA lyase.
The recombinant l-malyl-CoA lyase was purified in two steps from heat-treated extract of 25 g of E. coli cells with a yield of 48% (Table 1). The specific lyase activity at 55°C was 2.3 μmol of l-malyl-CoA cleaved min−1 mg of protein−1. The ratio of lyase activity/condensation activity remained virtually unchanged during purification. SDS-PAGE showed a 38-kDa protein and three additional very faint protein bands (<5%) (Fig. 3A) which were also present in E. coli cells lacking the DNA insert (data not shown). The properties of the recombinant enzyme are summarized in Table 2. The protein was colorless and exhibited an absorption maximum near 280 nm, indicating that it contained no UV-visible light-absorbing cofactor. Determination of the native molecular mass by size exclusion chromatography indicated a native molecular mass of approximately 210 ± 20 kDa. In native PAGE gels, the protein migrated with an estimated molecular mass of ≥210 kDa (Fig. 3B). This indicates that the native enzyme has an α5 or α6 composition. The catalytic number (55°C, pH 7.1) for l-malyl-CoA consumption was 8.8 s−1, assuming an α6 native enzyme of the calculated 228 kDa, and the apparent Km for l-malyl-CoA was 64 μM. The optimal pH for enzyme activity at 55°C was 7.1 (corresponding to 7.4 at room temperature). The enzyme was stable at room temperature for days and could be stored at −80°C. Enzyme activity required the presence of Mg2+, which could partially be replaced by Mn2+ or Co2+. Consequently, complexing agents such as EDTA completely inhibited the enzyme, and activity could be restored by adding excess Mg2+. Pyruvate was not condensed with acetyl-CoA, whereas the rate of oxaloacetate condensation was 40% of the rate observed with glyoxylate; the product of that reaction may be citryl-CoA. The optimal temperature for activity was 70°C, as determined in the continuous spectrophotometric lyase reaction assay using l-malyl-CoA as substrate.
TABLE 2.
Molecular and catalytic properties of recombinant l-malyl-CoA lyase and β-methylmalyl-CoA lyase from C. aurantiacus
| Property | Recombinant l-malyl-CoA lyasea/β-methylmalyl-CoA lyase | |
|---|---|---|
| Substrates | erythro-β-Methylmalyl-CoA, l-malyl-CoA, glyoxylate, propionyl-CoA, acetyl-CoA | |
| Products | Glyoxylate, propionyl-CoA, acetyl-CoA, erythro-β-methylmalyl-CoA, l-malyl-CoA | |
| Specific activitiesa (μmol min−1 mg of protein−1) | 6.5 ± 0.1 (β-methylmalyl-CoA lyase); 2.3 ± 0.2 (l-malyl-CoA lyase); 0.50 ± 0.01 (propionyl-CoA condensation); 0.48 ± 0.01 (acetyl-CoA condensation) | |
| Apparent Km values (mM) | 0.089 ± 0.02 (β-methylmalyl-CoA); 0.061 ± 0.01 (l-malyl-CoA); 1.2 ± 0.6 (propionyl-CoA); 0.36 ± 0.15 (acetyl-CoA); 2.0 ± 0.5 (glyoxylate) | |
| pH optimum | 7.1 (55°C) | |
| Temperature optimum | 70°C | |
| Native molecular mass | 210 ± 20 kDa | |
| Subunit molecular mass | 38 kDa | |
| Suggested composition | α5 or α6 | |
| Catalytic numbersa (s−1; per hexamer) | 24.5 ± 0.4 (β-methylmalyl-CoA consumption); 8.8 ± 0.8 (l-malyl-CoA consumption); 1.9 ± 0.02 (glyoxylate consumption + propionyl-CoA); 1.8 ± 0.04 (glyoxylate consumption + acetyl-CoA) | |
| Specificity | erythro-β-Methylmalyl-CoA, 100%; l-malyl-CoA, 35 ± 1%; propionyl-CoA, 100%; acetyl-CoA, 96 ± 2%; succinyl-CoA, <1%; glyoxylate, 100%; oxaloacetate + propionyl-CoA, 11 ± 3%; oxaloacetate + acetyl-CoA, 22 ± 18%; pyruvate, <1% | |
| Influence of divalent cations (concn [mM]) | Mg2+ (4), 100%; Mg2+ (0), <2%; Mn2+ (4), 31%; Co2+ (4), 31%; Ni2+ (4), 12%; Ca2+ (4), 4% | |
| Inhibitor (concn [mM]) | EDTA (1), <2% |
Data obtained for recombinant l-malyl-CoA lyase.
Purification and properties of β-methylmalyl-CoA lyase from C. aurantiacus.
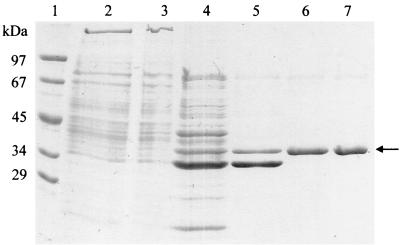
Extract of autotrophically grown C. aurantiacus cells contained a propionyl-CoA-dependent glyoxylate-consuming activity of 0.017 μmol min−1 mg of protein−1, which was down-regulated in heterotrophically grown cells by a factor of 4. This β-methylmalyl-CoA lyase was purified 30-fold from 25 g of autotrophically grown cells of C. aurantiacus in five steps, with a yield of 7% (Table 3). SDS-PAGE revealed one strong band corresponding to 38 kDa and two additional protein bands (Fig. 4). The UV-visible spectrum, native molecular mass (Fig. 3B), and enzyme stability properties were indistinguishable from those of recombinant l-malyl-CoA lyase. β-Methylmalyl-CoA synthesis required glyoxylate and propionyl-CoA, the reaction depended on Mg2+, and EDTA reversibly inhibited the enzyme (Table 2). The specific condensation activity was 0.51 μmol min−1 mg of protein−1 (55°C, pH 7.4), corresponding to a catalytic number of 1.9 s−1 (assuming a 228-kDa α6 holoenzyme). The determined N-terminal amino acid sequence (MRKLAHNFYK) was identical to the deduced N-terminal sequence of recombinant l-malyl-CoA lyase, and the apparent Km value for l-malyl-CoA (57 μM) was similar to that of recombinant l-malyl-CoA lyase. The apparent Km values for propionyl-CoA and acetyl-CoA were 1.8 and 0.5 mM, respectively, and 2.0 mM for glyoxylate. In the condensation reaction with propionyl-CoA, pyruvate was inactive, whereas oxaloacetate was transformed at 14% of the rate observed with glyoxylate; the product of that reaction may be methylcitryl-CoA.
TABLE 3.
Purification of β-methylmalyl-CoA lyase from autotrophically grown cells of C. aurantiacusa
| Purification step | Total condensation reaction activity (μmol min−1)b | Total protein (mg) | Specific condensation reaction activity (μmol min−1 mg of protein−1)b | Recovery of condensation reaction activity (%)b | Purification of condensation reaction activity (fold)b | Total l-malyl-CoA cleavage activity (μmol min−1) | Specific l-malyl-CoA cleavage activity (μmol min−1 mg of protein−1) | Recovery of l-malyl-CoA cleavage activity (%) | Purification of l-malyl-CoA cleavage activity (fold) |
|---|---|---|---|---|---|---|---|---|---|
| Cell extractc | 13.1 (18.6) | 768 | 0.017 (0.024) | 100 (100) | 1 (1) | 36.3 | 0.047 | 100 | 1 |
| Heat precipitation (65°C, 10 min) | 14.4 (15.8) | 375 | 0.038 (0.042) | 109 (85.1) | 2.2 (1.7) | ND | ND | ND | ND |
| DEAE-Sepharose | 11.1 (11.6) | 63.9 | 0.174 (0.182) | 84.5 (62.7) | 10.2 (7.5) | ND | ND | ND | ND |
| Phenyl-Sepharose | 5.2 (ND) | 12.6 | 0.414 (ND) | 39.7 (ND) | 24.2 (ND) | ND | ND | ND | ND |
| Gel filtration | 2.1 (ND) | 2.6 | 0.805 (ND) | 16.0 (ND) | 47.1 (ND) | ND | ND | ND | ND |
| Resource Q | 0.92 (0.91) | 1.8 | 0.512 (0.503) | 7.0 (6.9) | 29.9 (21.0) | 2.2 | 1.2 | 6.0 | 26.1 |
β-Methylmalyl-CoA lyase was purified from 25 g (wet weight) of C. aurantiacus cells; the glyoxylate reaction was followed at 55°C.
The first value refers to the condensation of glyoxylate with propionyl-CoA, and the value in parentheses refers to the condensation of glyoxylate with acetyl-CoA.
100,000 × g supernatant.
ND, not determined.
FIG. 4.
Denaturing PAGE (12.5%) of cell extract of autotrophically grown C. aurantiacus and β-methylmalyl-CoA lyase at various steps of purification (20 μg each). Lanes: 1, molecular mass marker (rabbit phosphorylase b, 97 kDa; bovine serum albumin, 67 kDa; egg ovalbumin, 45 kDa; lactate dehydrogenase, 34 kDa; carboanhydrase, 29 kDa); 2, cell extract; 3, cell extract after heat precipitation; 4, DEAE-Sepharose fraction; 5, phenyl-Sepharose fraction; 6, size exclusion fraction; 7, Resource Q fraction. The gel was stained with Coomassie brilliant blue R-250. The arrow indicates a 38-kDa protein band that corresponds to β-methylmalyl-CoA lyase.
Further evidence for the bifunctional enzyme l-malyl-CoA lyase/β-methylmalyl-CoA lyase.
The comparison of the properties of recombinant l-malyl-CoA lyase and β-methylmalyl-CoA lyase strongly suggests that these two enzymes are identical. To corroborate this conclusion, each of the two enzyme preparations was tested with the respective substrates of the other enzyme. Indeed, recombinant l-malyl-CoA lyase not only condensed glyoxylate with acetyl-CoA (0.48 μmol min−1 mg of protein−1) to l-malyl-CoA, but also with propionyl-CoA (0.50 μmol min−1 mg protein−1) to β-methylmalyl-CoA (Table 1). The ratio l-malyl-CoA lyase activity/condensation activity with both acetyl-CoA and propionyl-CoA was near 4 throughout purification.
The recombinant enzyme was, therefore, used for enzymatic synthesis of β-methylmalyl-CoA. We assume that the product was the erythro stereoisomer of β-methylmalyl-CoA, since this form was identified before by nuclear magnetic resonance spectroscopy of products formed by the cell extract or partially purified protein fractions from [1,2,3-13C]propionyl-CoA and glyoxylate (18). When tested in the lyase direction, both enzyme preparations showed higher specific activities with β-methylmalyl-CoA (100%; 6.5 and 3.2 μmol min−1 mg−1 for recombinant enzyme and purified enzyme from C. aurantiacus, respectively) than with l-malyl-CoA (35%). The apparent Km values for β-methylmalyl-CoA of the two enzyme preparations were similar, 89 ± 21 μM (mean ± standard deviation).
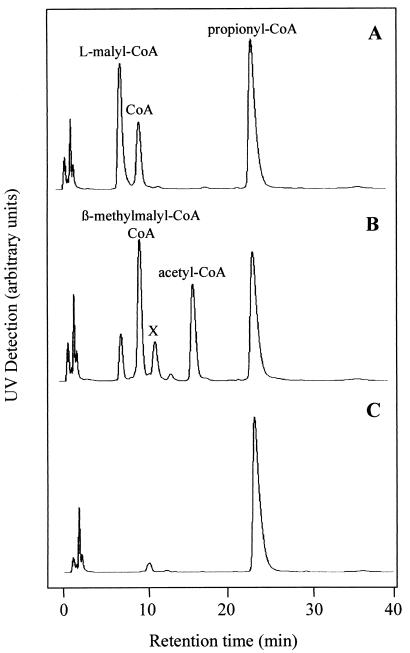
The β-methylmalyl-CoA synthesis reaction was used to determine the equilibrium constant keq {([propionyl-CoA] × [glyoxylate])/[β-methylmalyl-CoA]} of 7.1 × 10−4 M (55°C, pH 7.1), which corresponds to a ΔG′ of +19.6 kJ (55°C). At millimolar concentrations of substrates, the enzyme reaction is expected to be fully reversible, as was observed experimentally. When l-malyl-CoA and propionyl-CoA (2.5 mM, each) were added to the recombinant enzyme, acetyl-CoA and β-methylmalyl-CoA were formed, which was shown by HPLC (Fig. 5). In this assay, 0.3 mM glyoxylate was formed at equilibrium. When propionyl-CoA was omitted, the glyoxylate concentration at equilibrium was 0.6 mM, which corresponds to an equilibrium constant of 2 × 10−4 M for the l-malyl-CoA lyase reaction. This value is close to the reported value of 4.7 × 10−4 M (13). The combined glyoxylate formation and consumption by one enzyme results in the observed lower glyoxylate concentration when both l-malyl-CoA and propionyl-CoA are present.
FIG. 5.
Formation of acetyl-CoA and β-methylmalyl-CoA from l-malyl-CoA and propionyl-CoA by recombinant l-malyl-CoA lyase at 55°C. Results are shown for HPLC detection of CoA-thioesters after 0 min of incubation (A), after 20 min of incubation (B), or in a control experiment where l-malyl-CoA was omitted after 20 min of incubation (C). Retention times: 8 min (l-malyl-CoA), 10 min (β-methylmalyl-CoA; CoA), 16 min (acetyl-CoA), 24 min (propionyl-CoA). The reaction mixture (0.2 ml) contained 200 mM MOPS-K+ buffer (pH 7.7), 4 mM MgCl2, 2.5 mM l-malyl-CoA, 2.5 mM propionyl-CoA, and 24 μg of protein. Note that the l-malyl-CoA preparation contained 30% CoA as an impurity. Therefore, besides 2.5 mM l-malyl-CoA, 0.75 mM CoA was also initially present in the corresponding assay mixture. An unidentified product is indicated by the X.
DISCUSSION
Evidence for a bifunctional l-malyl-CoA/β-methylmalyl-CoA lyase.
The following arguments strongly suggest that only one enzyme, which is encoded by the l-malyl-CoA lyase gene (mclA), catalyzes the l-malyl-CoA lyase and β-methylmalyl-CoA lyase reaction (1). Only one l-malyl-CoA lyase-like gene was identified so far in the C. aurantiacus genome (2). The N-terminal amino acid sequence of β-methylmalyl-CoA lyase was identical to that of l-malyl-CoA lyase deduced from the gene (3). The sizes of subunit and native enzyme of both preparations and all other molecular and stability properties studied were identical within the range of accuracy of the methods (4). Both enzyme activities required Mg2+ and were inactivated by EDTA (5). The Vmax values and the apparent Km values for various substrates were virtually indistinguishable (6). The ratios of the rates of the two enzyme activities in cell extracts and in purified enzymes were virtually identical.
Role of the enzyme, required enzyme activity, and regulation.
We propose that in vivo the role of the enzyme is to catalyze the following reactions: (i) l-malyl-CoA ⇄ acetyl-CoA + glyoxylate; and (ii) propionyl-CoA + glyoxylate ⇄ β-methylmalyl-CoA.
The overall reaction (Fig. 5) is l-malyl-CoA + propionyl-CoA ⇄ acetyl-CoA + β-methylmalyl-CoA. This reaction involves glyoxylate as an intermediate. The enzyme represents the last enzyme of the first (carbon fixation) cycle and the starting enzyme of the second (glyoxylate assimilation) cycle (Fig. 1, reactions 9 and 10). The simultaneous operation of forward and reverse reactions of an enzyme with ambiguous substrate specificity resulting in such an intriguing transformation reaction is rather uncommon and reduces the number of enzymes required in the autotrophic pathway.
The specific activities of l-malyl-CoA cleavage and glyoxylate condensation with propionyl-CoA to β-methylmalyl-CoA in cell extract amounted to 0.047 and 0.017 μmol min−1 mg of protein−1, respectively. These in vitro rates must be at least as high as the minimal rates that can explain the growth rate under autotrophic conditions. The specific rate of CO2 fixation of cells growing with a generation time of 20 h is approximately 0.035 μmol min−1 mg of cell protein−1. Since this CO2 fixation cycle fixes three molecules of bicarbonate, the minimal specific rate of the enzymes of the bicyclic pathway should be on the order of 0.012 μmol min−1mg of protein−1. The specific enzyme activities observed meet these criteria. Also, the fourfold up-regulation of enzymatic activity under autotrophic conditions is in line with this function.
Genes adjacent to the malyl-CoA lyase gene and presence of related genes on the chromosome of C. aurantiacus.
Contig 965 of the C. aurantiacus genome (Fig. 2) contains some other open reading frames (ORFs), besides the one representing the l-malyl-CoA lyase gene studied here (mclA), that may play a role in autotrophic carbon metabolism. Three ORFs (ytf1, ytf2, and ytf3) likely represent succinyl-CoA-dependent CoA transferases, and ORF ytf3 is possibly succinyl-CoA:l-malate CoA transferase (for a review, see reference 14). Unfortunately, expression of ORF ytf3 in E. coli did not result in formation of a functional protein and the expected CoA transferase activity could not be shown (unpublished results). Searching for enzymes catalyzing Claisen-type condensation or retrocondensation reactions revealed an ORF on contig 967, which is similar to 3-hydroxy-3-methylglutaryl-CoA lyase (EC 4.1.3.4) and is next to another putative CoA transferase. We are currently testing the possibility that these two ORFs actually represent citramalyl-CoA lyase and succinyl-CoA:citramalate CoA transferase, respectively. These two enzymes have been postulated to play a role in the glyoxylate assimilation cycle (Fig. 1, reactions 13 and 14).
Comparison with l-malyl-CoA lyase from M. extorquens, A. aminovorans, and other bacteria.
l-Malyl-CoA/β-methylmalyl-CoA lyase from C. aurantiacus is similar to l-malyl-CoA lyase from M. extorquens (13) and A. aminovorans (15-16) with respect to the molecular mass of the subunit (35 to 39 kDa) (2, 7), native molecular mass (190 to 290 kDa) (8, 13), subunit composition (5-6), sequence homology (M. extorquens, 324 amino acids [aa], 35.4 kDa; C. aurantiacus, 348 aa, 38 kDa; 37% aa identity, 51% nucleotide identity), substrate specificity, kinetics constants, and the Mg2+ dependency of the reaction. These enzymes also slowly condensed glyoxylate with propionyl-CoA at 10% of the rate of acetyl-CoA condensation; the product of that reaction was not identified (13), but it was probably β-methylmalyl-CoA. The enzyme from C. aurantiacus differs mainly in two aspects, which reflect the growth properties and metabolism of the organism. The enzyme is more heat stabile, and it catalyzes the propionyl-CoA condensation reaction at higher rates.
In principle, the condensation of propionyl-CoA and glyoxylate with formation of β-methylmalate (25) has been reported earlier in Bacillus spp. (29, 35), but its physiological role remained unclear. A similar reverse (lyase) reaction was proposed for the generation of glyoxylate from acetate in acetate-grown Rhodospirillum rubrum cells lacking isocitrate lyase (24, 30).
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany, and the Fonds der Chemischen Industrie, Frankfurt, Germany.
Special thanks to Richard Krieger, Organische Chemie, Universität Freiburg, for the synthesis of l-malylcaprylcysteamine. We thank Nasser Gad'on, Universität Freiburg, for assistance in growing cells. Preliminary sequence data were obtained from the local C. aurantiacus server (http://spider.jgi-psf.org/JGI_microbial/bin/psf_blast?PROJEKT_ID = 2351478) at the DOE Joint Genome Institute.
REFERENCES
- 1.Alber, B. E., and G. Fuchs. 2002. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 277:12137-12143. [DOI] [PubMed] [Google Scholar]
- 2.Arps, P. J., G. F. Fulton, E. C. Minnich, and M. E. Lidstrom. 1993. Genetics of serine pathway enzymes in Methylobacterium extorquens AM1: phosphoenolpyruvate carboxylase and malyl coenzyme A lyase. J. Bacteriol. 175:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology, John Wiley and Sons, New York, N.Y.
- 4.Birnboim, H., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Burton, N. P., T. D. Williams, and P. R. Norris. 1999. Carboxylase genes of Sulfolobus metallicus. Arch. Microbiol. 172:349-353. [DOI] [PubMed] [Google Scholar]
- 7.Chistoserdova, L., and M. E. Lidstrom 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM 1. Microbiology 143:1729-1736. [DOI] [PubMed] [Google Scholar]
- 8.Cox, R. B., and J. R. Quayle. 1976. Synthesis and hydrolysis of malyl-coenzyme A by Pseudomonas AM1: an apparent malate synthase activity. J. Gen. Microbiol. 95:121-133. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, R. M. C., D. C. Elliot, W. H. Elliot, and K. M. Jones. 1986. Data for biochemical research, 3rd ed. Clarendon Press, Oxford, United Kingdom.
- 10.Eggerer, H., and C. H. Grünewälder. 1964. Zum Mechanismus der Biologischen Umwandlung von Citronensäure. IV. Synthese von Malyl-Coenzym A und seiner Diastereoisomeren. Liebigs Ann. Chem. 677:200-208. [Google Scholar]
- 11.Eisenreich, W., G. Strauss, U. Werz, G. Fuchs, and A. Bacher. 1993. Retrobiosynthetic analysis of carbon fixation in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 215:619-632. [DOI] [PubMed] [Google Scholar]
- 12.Fulton, G. L., D. N. Nunn, and M. E. Lidstrom. 1984. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J. Bacteriol. 160:718-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacking, A. J., and J. R. Quayle. 1974. Purification and properties of malyl-CoA lyase from Pseudomonas AM1. Biochem. J. 139:399-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heider, J. 2001. A new family of CoA-transferases. FEBS Lett. 509:345-349. [DOI] [PubMed] [Google Scholar]
- 15.Hersh, L. B. 1973. Malate adenosine triphosphate lyase. Separation of the reaction into a malate thiokinase and malyl coenzyme A lyase. J. Biol. Chem. 248:7295-7303. [PubMed] [Google Scholar]
- 16.Hersh, L. B. 1974. Malyl coenzyme A lyase. Mechanism of action as deduced by kinetic analysis. J. Biol. Chem. 249:5208-5212. [PubMed] [Google Scholar]
- 17.Herter, S., J. Farfsing, N. Gad'on, C. Rieder, W. Eisenreich, A. Bacher, and G. Fuchs. 2001. Autotrophic CO2 fixation in Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J. Bacteriol. 183:4305-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herter, S., G. Fuchs, A. Bacher, and W. Eisenreich. 2002.. A bicyclic autotrophic CO2 fixation pathway in Chloroflexus aurantiacus. J. Biol. Chem. 277:20277-20283. [DOI] [PubMed]
- 19.Holo, H. 1989. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch. Microbiol. 151:252-256. [Google Scholar]
- 20.Holo, H., and D. Grace. 1987. Polyglucose synthesis in Chloroflexus aurantiacus studied by 13C-NMR. Arch. Microbiol. 148:292-297. [Google Scholar]
- 21.Holo, H., and R. Sirevåg. 1986. Autotrophic growth and CO2 fixation in Chloroflexus aurantiacus. Arch. Microbiol. 145:173-180. [Google Scholar]
- 22.Hügler, M., C. Ménendez, H. Schägger, and G. Fuchs. 2002. Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Bacteriol. 184:2404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii, M., T. Miyake, T. Satoh, H. Sugiyama, Y. Oshima, T. Kodama, and Y. Igarashi. 1997. Autotrophic carbon dioxide fixation in Acidianus brierleyi. Arch. Microbiol. 166:368-371. [DOI] [PubMed] [Google Scholar]
- 24.Ivanovsky, R. N., E. N. Krasilnikova, and I. Berg. 1997. A proposed cycle for acetate assimilation in the purple non-sulfur bacterium Rhodospirillum rubrum. FEMS Microbiol. Lett. 153:399-404. [Google Scholar]
- 25.Korkes, S. 1956. Variations on the citric acid cycle. Annu. Rev. Biochem. 25:723-728. [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Ménendez, C., Z. Bauer, H. Huber, N. Gad'on, K. O. Stetter, and G. Fuchs. 1999. Presence of acetyl-coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for the operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181:1088-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohrhauer, H., K. Christiansen, M. Gan, M. Deubig, and R. T. Holman. 1968. Improved method for the preparation of malonyl coenzyme A. J. Lipid Res. 9:398-399. [PubMed] [Google Scholar]
- 29.Nanako, H., Y. Takagi, and H. Katsuki. 1970. Formation of β-methylmalate from propionate and glyoxylate. Biochem. Biophys. Res. Commun. 41:1605-1611. [DOI] [PubMed] [Google Scholar]
- 30.Osumi, T., T. Ebisuno, H. Nakano, and H. Katsuki. 1975. Formation of β-methylmalate and its conversion to citramalate in Rhodospirillum rubrum. J. Biochem. 78:763-772. [DOI] [PubMed] [Google Scholar]
- 31.Pierson, B. K., and R. W. Castenholz. 1974. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus gen. and sp. nov. Arch. Microbiol. 100:5-24. [DOI] [PubMed] [Google Scholar]
- 32.Pierson, B. K., and R. W. Castenholz. 1974. Studies of pigments and growth in Chloroflexus aurantiacus, a phototrophic filamentous bacterium. Arch. Microbiol. 100:283-305. [DOI] [PubMed] [Google Scholar]
- 33.Salem, A. R., A. J. Hacking, and J. R. Quayle. 1973. Cleavage of malyl-CoA into acetyl-coenzyme A and glyoxylate by Pseudomonas AM1 and other C1-unit-utilizing bacteria. Biochem. J. 136:89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Sasaki, K., H. Nanako, and H. Katsuki. 1971. Enzymatic isomerization of β-methylmalate to (−)citramalate by a soil bacterium. J. Biochem. 70:441-449. [DOI] [PubMed] [Google Scholar]
- 36.Sirevåg, R., and R. Castenholz. 1979. Aspects of carbon metabolism in Chloroflexus. Arch. Microbiol. 120:151-153. [Google Scholar]
- 37.Strauss, G., W. Eisenreich, A. Bacher, and G. Fuchs. 1992. 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing archaebacterium Thermoproteus neutrophilus and in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 205:853-866. [DOI] [PubMed] [Google Scholar]
- 38.Strauss, G., and G. Fuchs. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur. J. Biochem. 215:633-643. [DOI] [PubMed] [Google Scholar]
- 39.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low-background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182:157-159. [DOI] [PubMed] [Google Scholar]