Abstract
TxeR, a sigma factor that directs Clostridium difficile RNA polymerase to recognize the promoters of two major toxin genes, was shown to stimulate its own synthesis. Whether expressed in C. difficile, Clostridium perfringens, or Escherichia coli, TxeR stimulated transcription of fusions of the txeR promoter region to reporter genes. As is the case for the tox genes, txeR expression was responsive to the cellular growth phase and the constituents of the medium. That is, the level of expression in broth culture was low during the exponential growth phase, but rapidly increased as cells approached the stationary phase. In the presence of excess glucose, expression from the txeR promoter was repressed. The results support a model for toxin gene expression in which synthesis of TxeR is induced by specific environmental signals. The increased level of TxeR then permits high-level expression of the toxin genes. The study of txeR gene regulation in C. difficile was made possible by introduction of a mobilizable, replicative plasmid via conjugation with E. coli.
Clostridium difficile, the major cause of antibiotic-associated diarrhea and pseudomembranous colitis, is also the most frequently identified cause of hospital-acquired infectious diarrhea (22). Typically, a patient subjected to high doses of antibiotics develops a severe diarrheal disease that is accompanied by the unusual presence of large numbers of C. difficile cells in the intestinal tract. Two large toxin proteins, encoded by the toxA (tcdA) and toxB (tcdB) genes, are thought to be the primary virulence factors (1, 33, 40, 43). Both toxins A and B disrupt the actin cytoskeleton of intestinal epithelial cells through modification of Rho family proteins by UDP-glucose-dependent glycosylation (17, 18, 43). Virulent strains of C. difficile vary in the level of toxin production (7, 15), but toxin synthesis seems to be coordinated inasmuch as strains that produce low levels of toxin B also produce low levels of toxin A (9, 25).
The toxA and toxB genes lie within an ∼19-kb, five-gene chromosomal cluster that has some of the properties of a pathogenicity island (3, 4, 9). It was recently shown that the open reading frame tcdE, separating the toxB and toxA genes, encodes a holin-like function that may play a role in the release of the toxin proteins (41). Avirulent strains, such as strain CD37 (38), lack this cluster completely (3, 9). The tox genes are each transcribed from their own promoters, as well as by read-through transcription from an upstream promoter (7, 10). Transcription from the gene-specific promoters is repressed during rapid exponential growth in rich medium, but becomes induced as cells enter the stationary phase (7). If a rapidly metabolizable carbon source, such as glucose, is present in the medium, however, the tox genes remain repressed even many hours after cells enter the stationary phase (7). Other physiological conditions, such as the presence of biotin (45) and certain amino acids (16, 19-21, 44), also contribute to the regulation of tox gene expression by unknown mechanisms.
Expression of the tox genes in the stationary phase is strongly dependent on TxeR, the product of the gene located immediately upstream of toxB (27, 32). TxeR is a novel RNA polymerase sigma factor that permits core RNA polymerase to recognize the promoters of the toxA and toxB genes and initiate their transcription (27). When these promoters are fused to reporter genes and cloned in Escherichia coli or Clostridium perfringens, the expression of the reporter construct depends almost completely on the presence of the txeR gene in the same cell (27, 32). Moreover, TxeR-dependent stationary-phase induction of the tox genes in C. perfringens is repressed by glucose (27). Interestingly, the toxB and toxA promoters resemble those of certain other toxin and bacteriocin genes in several Clostridium species (7, 8, 13), and TxeR is similar in sequence to proteins that regulate those genes (28, 29, 32).
We sought to understand the mechanism by which environmental conditions influence tox gene expression. One possibility is that these conditions control the synthesis of TxeR, thereby regulating tox genes indirectly. To test this hypothesis, we examined the regulation of txeR gene expression and found that it was also induced during the stationary phase, was also repressed by glucose, and was autoregulatory in vivo, in three different bacterial hosts.
MATERIALS AND METHODS
Construction of reporter fusions for E. coli.
The promoter region of txeR was amplified by PCR from pCD21 (9), a generous gift from J. L. Johnson (Department of Biochemistry, Virginia Tech, Blacksburg). The primer at the 5′ end contained a HindIII site (underlined) (5′-GCGCAAGCTTTTAGATGGTTGCAGAGT-3′) and the 3′ primer contained a BamHI site (underlined) (5′-CGGGATCCTGCATAAAATCACCCTCT-3′). The resulting 0.85-kb PCR product was subcloned in pACYC184 and named pAC-TP. The reporter gene for these experiments, the C. difficile toxin A repeating unit (ARU) coding sequence (6), was removed from plasmid pAC-BP-ARU (32) as a 3.24-kb BamHI fragment and subcloned in pAC-TP to create pAC-TP-ARU. For expression studies, pAC-TP-ARU was introduced by CaCl2-mediated transformation (5) into E. coli strain BL21(DE3) carrying either the TxeR-expressing plasmid pT7-txeR (32) or the vector pT7-7.
Growth of E. coli reporter strains and assay of ARU by ELISA.
E. coli strain BL21(DE3) carrying pAC-TP-ARU and pT7-txeR or pT7-7 was grown in Terrific broth (42). Samples of cells harvested after various times of incubation were suspended in phosphate-buffered saline (pH 7.4) containing Complete protease inhibitor cocktail (Boehringer-Mannheim) and broken by sonication or agitation with glass beads. After removal of debris by centrifugation, the supernatant fluids were stored at −20°C. Total protein concentrations were determined with the Coomassie Plus protein assay reagent (Pierce). The ToxA test (TechLab, Inc., Blacksburg, Va.) was used to determine the level of ARU in lysates following the 2-h protocol. Units of enzyme-linked immunosorbent assay (ELISA) reactivity were arbitrarily defined as the A450 multiplied by the reciprocal of the highest dilution with an absorbance of 0.2 to 2.5. The level of ARU is expressed as units per milligram of total protein in the lysate. The extracts were also analyzed by immunoblot assays. Forty micrograms of total protein from each sample at the 20-h time point was denatured by boiling for 3 min with 2.5% sodium dodecyl sulfate (SDS) and 5% 2-mercaptoethanol and subjected to electrophoresis in 7.5% separating-4% stacking SDS-polyacrylamide gel electrophoresis (PAGE) gels (23). Proteins were transferred to a nitrocellulose membrane, exposed to the anti-ARU monoclonal antibody PCG-4 (32) at a dilution of 1:1,000, and then detected by incubation with a 1:2,000 dilution of antimouse immunoglobulin G (IgG)-peroxidase conjugate.
Construction of reporter fusions for C. perfringens.
The construction of pTUM307 (a TxeR-expressing plasmid derived from pTRKL2) and plasmids pTUM181 and pTUM182, carrying toxA-gusA and toxB-gusA fusions, respectively, was described previously (27). To create a txeR-gusA fusion plasmid (pTUM183), a 504-bp fragment corresponding to positions −493 to +11 with respect to the txeR translational start codon was amplified by PCR with primers OBD15H and OBD16X (7) and cloned in the gusA reporter plasmid pTUM177 (27). Plasmids were introduced into C. perfringens strain SM101 (46) by electroporation.
Construction of a txeR-gusA fusion strain of C. difficile.
The txeR gene originally cloned in pDG1664 (27) was excised as a HindIII-EcoRV fragment and subcloned between the HindIII and EcoRV sites of plasmid pSK− (Stratagene), creating pTUM507. A HindIII fragment of plasmid pCD28 (7) carrying a txeR-gusA translational fusion was then inserted into pTUM507 and into pSK−, giving rise to plasmids pTUM525 and pTUM515, respectively. The direction of transcription of the txeR-gusA fusion is the same in pTUM515 and pTUM525. In pTUM525, the txeR gene is downstream of the fusion and in the same orientation. The appropriate fragments carrying the gusA fusions from pTUM515 and pTUM525 were subcloned with SacI and Asp718 into the corresponding sites of the mobilizable C. perfringens shuttle vector pJIR1457 (26), resulting in the construction of pJIR2126 and pJIR2127, respectively.
Plasmids pJIR1457, pJIR2126, and pJIR2127 were introduced by transformation into E. coli strain S17-1, which carries the broad-host-range plasmid RP4, which is capable of mobilizing IncP plasmids, such as pJIR1457 (37). These constructs were then transferred to the nontoxinogenic C. difficile strain CD37 (38) by conjugation. Two milliliters of an overnight culture of CD37 in BHIS medium (38) was used to inoculate 90 ml of the same medium, and this mixture was incubated at 37°C for 6 h or until the culture began to produce gas. Meanwhile, exponential-phase cultures of E. coli donor strains derived from strain S17-1 were prepared in 2× YT medium (31) supplemented with trimethoprim (10 μg/ml) and erythromycin (150 μg/ml). A 100-μl sample of each culture was then placed on a thick (ca. 25 ml) BHIS agar plate. The cultures were mixed and spread over the surface of the plate. Samples of each culture were also spread on separate plates of the same medium to serve as controls. The plates were incubated in an atmosphere of 10% H2-10% CO2-80% N2 for 20 to 24 h at 37°C. The growth from each plate was resuspended in 3 ml of diluent (0.01% peptone, 0.1% sodium thioglycolate, 0.85% sodium chloride), and 100 μl of this mixture was spread on BHIS agar supplemented with erythromycin (50 μg/ml) as an enrichment step to increase the number of C. difficile transconjugants (P. Noack, personal communication). After incubation for approximately 16 h, the growth was resuspended in 1 ml of diluent as before. Samples (100 μl) were then spread on BHIS plates supplemented with rifampin and erythromycin and incubated anaerobically for a minimum of 48 h. For each conjugation, plasmids were isolated from three independently derived C. difficile transconjugants. The restriction profiles of these plasmids were identical to those of the parent plasmid (data not shown), indicating that passage through C. difficile did not lead to any rearrangements or deletions. The C. difficile CD37 derivatives that carried pJIR1457, pJIR2126, and pJIR2127 were designated JIR8030, JIR8034, and JIR8035, respectively.
Growth of C. perfringens and C. difficile and β-glucuronidase assays.
Unless otherwise noted, C. perfringens and C. difficile strains were grown for β-glucuronidase assays in TY medium or TY medium containing 1% glucose (TYG) as described previously (7). Cultures were incubated at 37°C in an anaerobic chamber (Coy Laboratory Products, Inc.) in an atmosphere of 10% H2, 10% CO2, and 80% N2. In order to determine the expression profiles of the toxA-gusA, toxB-gusA, and txeR-gusA fusions in C. perfringens, overnight cultures in TYG medium were subcultured at a 1:100 dilution in either TY or TYG medium. Samples of cells were collected during different stages of growth to determine the GusA activity as a function of the growth phase. β-Glucuronidase from C. perfringens strains was assayed as previously described (27).
Assays of β-glucuronidase from C. difficile cultures were performed by a modification of the method of Mani and Dupuy (27). Ten-milliliter samples of growing cultures were removed at 2-h intervals for a period of 8 to 10 h. The cells were harvested by centrifugation, and the cell pellet was stored at −20°C. The pellet was thawed at room temperature, and the cells were resuspended in 0.8 ml of Z buffer (60 mM Na2HPO4 · 7H2O [pH 7.0], 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4 · 7H2O, 50 mM 2-mercaptoethanol). To achieve lysis, 200 to 250 μl of 150- to 212-μm-diameter unwashed glass beads (Sigma G-9018) was added, and the mixture was mixed and treated in a BIO101 Fast Prep FP120 bead beater (BIO101 Savant) at setting 6 for 45 s. The lysates were cooled on ice and then briefly centrifuged to remove the beads. A 0.4-ml sample was then removed, mixed with 0.4 ml of Z buffer, and incubated at 37°C for 5 min. The enzyme reaction was started by the addition of 0.16 ml of 6 mM p-nitrophenyl β-d-glucuronide (Sigma) and stopped by the addition of 0.4 ml of 1.0 M Na2CO3. β-Glucuronidase units were calculated as described before (7, 30).
Gel mobility shift experiments.
txeR promoter-containing fragments of 354 (PtxeR), 298 (P1txeR), and 302 (P2txeR) bp, corresponding to positions −466 to −114, −545 to −247, and −351 to −49, respectively, with respect to the translational start codon, were amplified by PCR, with the primers 5′-GAAAAGATTTTAATTTAATGATTG-3′ and 5′-GAAAATATGCATTATGAATAC-3′ (P1 + P2), 5′-GAGAGGAGAATGTTCTAAAAT-3′ and 5′-AAAAAATCTATATGACATATTTTTA-3′ (P1), and 5′-TTACTTGAAAATTGATCTATTTT-3′ and 5′-GACAACATTGGAATTAAATCAG-3′ (P2) and end labeled with T4 polynucleotide kinase (U.S. Biochemicals, Cleveland, Ohio) and [γ-32P]ATP (3,000 Ci/mmol; Amersham) as recommended by the manufacturer. The 203-bp lacUV5 promoter fragment has been described previously (36). The labeled fragments (0.2 nM) were incubated for 60 min at room temperature in 10 μl of glutamate buffer (40 mM HEPES [pH 8], 100 mM MgCl2, 100 mM potassium glutamate, 500 μg of bovine serum albumin per ml) containing C. difficile RNA polymerase holoenzyme (N. Mani, A. L. Sonenshein, and B. Dupuy, unpublished data) or 100 to 200 nM E. coli RNA polymerase core enzyme (Epicentre) that had been preincubated with TxeR or E. coli σ70 (see below). Three microliters of a heparin-dye solution (150 μg of heparin per ml, 0.1% bromophenol blue, 50% sucrose) in glutamate buffer was added, and the mixture was loaded during electrophoresis on a 4.5% polyacrylamide gel prepared in Tris-borate-EDTA (TBE) buffer. After electrophoresis (2 h at 13 V/cm), the gel was dried and analyzed by autoradiography. During a preincubation, RNA polymerase holoenzyme was reconstituted by incubating E. coli core RNA polymerase with a fourfold molar excess of TxeR protein or σ70 for 30 min at 37°C. E. coli σ70 was a gift from A. Kolb.
Expression and purification of TxeR.
Expression of six-His-tagged TxeR (TxeR-His6 [encoded in pCD54]) was carried out with E. coli BL21-codon plus(DE3)-RIL cells (Stratagene) as described previously (27). After expression, cells were harvested by centrifugation at 20,000 × g for 20 min, resuspended in lysis buffer (20 mM Tris-HCl, 0.3 M NaCl, 10% glycerol, 10 mM imidazole [pH 7.9]), and lysed by sonication. The cell debris was removed by centrifugation as described above. The supernatant fluid was applied to a 1-ml Ni+-nitrilotriacetic acid agarose column (Qiagen), and proteins were allowed to bind for 1 h at 4°C. After three washes with 5 ml of lysis buffer, TxeR-His6 was eluted with 5 ml of elution buffer (20 mM Tris-HCl, 0.3 M NaCl, 10% glycerol [pH 7.9]) supplemented with 20, 40, 60, 100, or 250 mM imidazole. Samples of each 0.5-ml fraction were mixed with gel loading buffer and subjected to SDS-PAGE. Fractions containing highly purified TxeR protein (the 100 mM imidazole fractions) were then pooled and dialyzed overnight against a buffer containing 50 mM sodium phosphate (pH 8), 300 mM NaCl, and 50% glycerol. TxeR purity was assessed by Coomassie blue staining of an SDS-PAGE gel and by Western blot analysis (35). Mouse anti-TxeR antibody was prepared as described previously (27).
Western blot analysis of TxeR in crude extracts.
To measure the levels of TxeR protein in C. difficile, crude cell extracts were prepared by sonication of late-stationary-phase cells of C. difficile strain VPI10463 grown in TY or TYG medium. The resulting extracts were clarified by centrifugation at 27,000 × g for 20 min, and the supernatant fluids were stored at −80°C. Protein concentrations were determined by the method of Bradford (2). Eighty micrograms of total protein was subjected to electrophoresis in an SDS-PAGE (15% polyacrylamide) gel and transferred to a nitrocellulose membrane. The membrane was treated with anti-TxeR antibody (1:100 dilution) for 3 h and detected following treatment with goat anti-mouse IgG-alkaline phosphatase conjugate (1:1,000 dilution) secondary antibody for 1 h.
RESULTS
TxeR activates its own expression in E. coli.
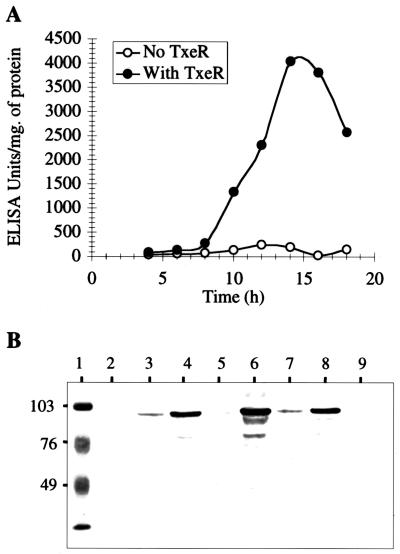
Sigma factors that regulate gene expression in response to environmental cues often regulate their own expression (12). To determine whether TxeR is autoregulatory, we measured expression from the txeR promoter in three different experimental systems. Previous work has shown that expression from the C. difficile tox promoters in E. coli depends on the presence of the txeR gene (32), implying that TxeR is functional in this gram-negative host cell. Moreover, TxeR can stimulate E. coli RNA polymerase to initiate transcription from the tox promoters in vitro (27). We therefore introduced into E. coli a plasmid (pAC-TP-ARU) that carries a fusion of a reporter (the repeat sequence of toxin A; ARU) to the txeR promoter region and a second plasmid that either does or does not encode TxeR (pT7-txeR or pT7-7, respectively). The presence of the txeR gene in trans greatly stimulated expression from the txeR promoter (Fig. 1A). At 14 h, ARU expression in the TxeR-producing strain was 20-fold higher than in the control strain. The results of the ELISAs were confirmed by Western blotting (Fig. 1B). Expression from the toxA, toxB, and txeR promoters was strongly enhanced when the txeR gene was present; a basal level of txeR-promoted ARU expression could be seen even in the absence of TxeR.
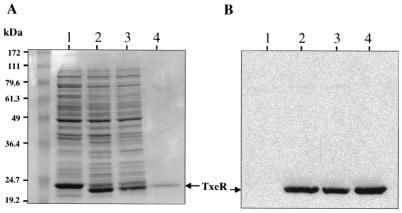
FIG. 1.
Expression of txeR in E. coli. (A) E. coli strains carrying a txeR-ARU fusion were grown in broth and assayed for ARU production by ELISA (see Materials and Methods). The cells left the rapid-exponential-growth-phase after about 5 h. Solid circles, strain containing the TxeR-expressing plasmid pT7-txeR; open circles, strain carrying the vector pT7-7. (B) E. coli strains carrying txeR-ARU, toxA-ARU, and toxB-ARU fusions were grown in broth and harvested at the 20-h time point. Extracts were analyzed by Western blots with antibody to ARU. Lanes: 1, low-molecular-mass protein standards (kilodaltons); 2, no fusion plus pT7-7 vector; 3, toxA-ARU plus pT7-7; 4, toxA-ARU plus pT7-txeR; 5, toxB-ARU plus pT7-7; 6, toxB-ARU plus pT7-txeR; 7, txeR-ARU plus pT7-7; 8, txeR-ARU plus pT7-txeR; 9, no fusion plus pT7-txeR.
TxeR also activates its own expression in C. perfringens.
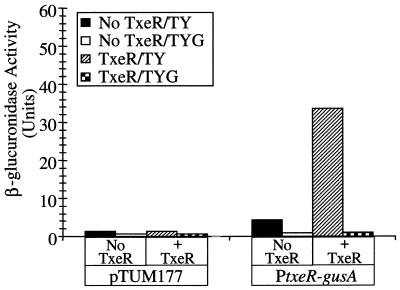
We next introduced a txeR-gusA fusion plasmid into C. perfringens strains containing either the vector pTRKL2 alone or a derivative of pTRKL2 carrying the txeR gene with its natural promoter. We have shown previously that a genetically manipulatable strain of C. perfringens, although not ideal, can be used for gene regulation experiments (7, 27). In the absence of the txeR gene, the txeR-gusA fusion showed only low-level expression in cells in TY medium (Fig. 2). This activity was greatly enhanced when the txeR gene was present in trans, indicating that txeR is subject to positive autoregulation (Fig. 2). Expression of the txeR-gusA fusion was repressed in C. perfringens cells in glucose-containing medium whether the txeR gene was present or not. The gusA reporter vector pTUM177, lacking any promoter driving gusA expression, gave little or no β-glucuronidase activity under any condition (Fig. 2).
FIG. 2.
Expression of a txeR-gusA fusion in stationary-phase cells of C. perfringens. A DNA fragment containing the promoter region of txeR gene was cloned in the reporter fusion vector pTUM177 and introduced into C. perfringens with or without the txeR gene in trans. β-Glucuronidase activity of late-stationary-phase cells grown in TY or TYG medium was determined as described previously (7). In control experiments, cells carrying the fusion vector alone were assayed. Solid bars, TY medium, no TxeR; open bars, TYG medium, no TxeR; striped bars, TY medium with TxeR; checkered bars, TYG medium with TxeR.
Growth-phase-dependent regulation of tox-gusA and txeR-gusA fusions in C. perfringens.
Expression of C. difficile toxin genes is regulated by the growth phase and by the composition of the growth medium (7). The tox mRNA levels are low during the rapid-exponential-growth phase, but increase as the cells reach the stationary phase. If a readily utilizable carbon source, such as glucose, is present in the medium, however, tox genes are repressed even in stationary-phase cells (7).
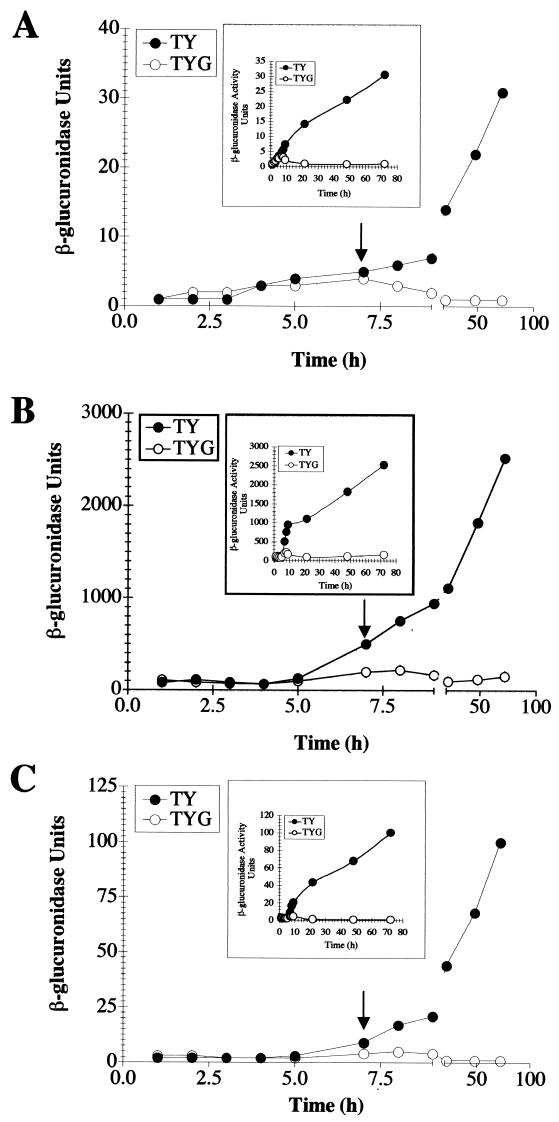
Because high-level expression of the C. difficile tox genes in C. perfringens is coordinate and depends on the presence of TxeR (27), we tested whether the txeR promoter is also subject to growth phase and carbon source regulation. As shown in Fig. 3A, TxeR-dependent expression of a txeR-gusA fusion in C. perfringens was low during the exponential growth phase, but rapidly increased as cells entered the stationary phase (6 to 8 h postinoculation). The presence of 1% glucose in the medium repressed this stationary-phase-associated increase in txeR promoter activity. This regulation correlated well with the TxeR-dependent expression of the toxA-gusA and toxB-gusA fusions in C. perfringens (Fig. 3B and C) and is consistent with the pattern of tox gene expression seen in C. difficile (7).
FIG. 3.
Growth-phase-dependent expression of txeR-gusA and tox-gusA fusions in C. perfringens. C. perfringens cells carrying txeR in trans and gusA fusions to the txeR (A), toxA (B), or toxB (C) promoters were grown anaerobically in TY or TYG medium, and the β-glucuronidase activities of cells at different stages of growth were determined over a period of 72 h. The data have been plotted so as to magnify the expression pattern during the exponential phase of growth and entry into stationary phase by introducing a break in the x axis. The arrows indicate the times at which the cultures left the rapid-exponential-growth phase. The graph inset shows the plot of all data points. Solid circles, TY medium; open circles, TYG medium.
TxeR autoregulation in C. difficile.
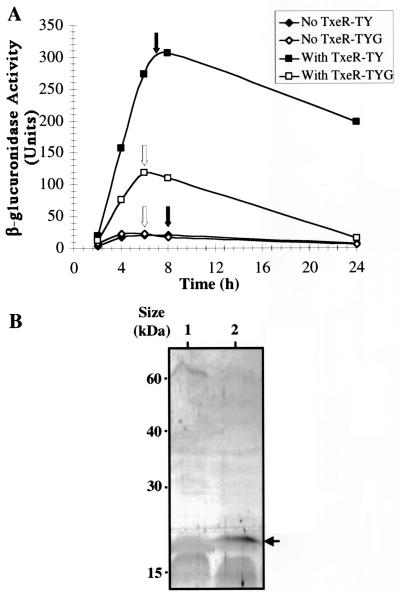
To study txeR autoregulation in C. difficile, we constructed a txeR-gusA fusion plasmid that has the txeR gene (with its natural promoter) in cis, but downstream of the fusion. This plasmid (pJIR2127) was transferred from E. coli to a nontoxinogenic strain of C. difficile by conjugation, and the resultant strain was tested for β-glucuronidase activity. A control plasmid (pJIR2126) lacked the txeR gene; a second control plasmid (pJIR1457) lacked both the fusion and the txeR gene. Consistent with the results seen in C. perfringens and E. coli, the txeR-gusA fusion showed low-level activity in the absence of txeR, and this activity was significantly increased in the presence of txeR (Fig. 4A). The increased expression, however, only occurred when the culture reached the end of the exponential growth phase (Fig. 4A). The TxeR-dependent increase in txeR-gusA expression was partially inhibited when glucose was present in the medium. TxeR-independent expression in C. difficile, however, was not affected by the presence of glucose (Fig. 4A). To verify that the levels of TxeR protein in C. difficile are indeed responsive to the environment, immunoblots of crude extracts of late-stationary-phase cells of C. difficile grown in either TY or TYG medium were probed with anti-TxeR antibody (Fig. 4B). C. difficile cells grown in TY medium contained TxeR, but cells grown in TYG medium had no detectable TxeR.
FIG. 4.
Expression of txeR in C. difficile. (A) Derivatives of C. difficile strain CD37 carrying a plasmid-borne PtxeR-gusA fusion (with or without the txeR gene) were grown in either TY or TYG medium, and β-glucuronidase activity was determined as described in Materials and Methods. The arrows indicate the times at which the cultures left the rapid-exponential-growth phase. (B) Western blot of TxeR protein in C. difficile. Crude extracts of late-stationary-phase cells of C. difficile strain VPI10463 grown in either TYG (lane 1) or TY (lane 2) medium were analyzed by immunoblotting with anti-TxeR antibody as described in Materials and Methods. The arrow shows the position of TxeR.
Overexpression and purification of TxeR.
Our previous preparation of TxeR purified after overexpression in E. coli contained some contaminating proteins, leaving open the possibility that TxeR acts on tox expression in conjunction with another protein present in E. coli extracts (27). We have modified the purification protocol, as described in Materials and Methods, and have now prepared TxeR with a purity greater than 95% (Fig. 5). The new batch of TxeR permitted E. coli core RNA polymerase to bind to and activate transcription from the toxA promoter (data not shown).
FIG. 5.
Purification of TxeR. (A) SDS-PAGE analysis of protein extracts from E. coli −codon plus(DE3)-RIL carrying either pET28-b or pCD54. Lanes: 1, crude cell extract from E. coli carrying the vector pET28-b; 2, crude cell extract from E. coli carrying pCD54 (expressing TxeR); 3, soluble fraction of cell extract from E. coli carrying pCD54; 4, TxeR-His6 purified. GIBCO/BRL low-molecular-mass standards (kilodaltons) are shown adjacent to the gel. (B) Immunodetection of TxeR by using anti-TxeR antibody. The protein samples in each lane correspond to those in panel A.
TxeR enables RNA polymerase to bind to the txeR promoter region.
To demonstrate that TxeR permits RNA polymerase to interact directly with the txeR promoter in vitro, gel mobility shift assays were performed with a DNA fragment encompassing the txeR promoter region (Fig. 6). C. difficile RNA polymerase was unable to bind to the txeR promoter-containing fragment in the absence of added TxeR, even though it could bind well to the promoter region of the gdh gene, a gene expressed during the exponential growth phase (27). Addition of purified TxeR enabled C. difficile RNA polymerase to form heparin-resistant complexes at the txeR promoter (Fig. 6A). TxeR was also able to activate the interaction of E. coli core RNA polymerase with the txeR promoter (Fig. 6B). (We used E. coli core RNA polymerase, because, as previously noted [27], we have been unable to purify C. difficile core polymerase totally free of associated sigma subunits.) These results provide direct biochemical evidence that TxeR acts by interacting with RNA polymerase core enzyme and stimulating binding to the promoter region of txeR (27). As a negative control, we showed that TxeR did not stimulate binding of core polymerase to the lacUV5 promoter, whereas addition of E. coli σ70 did stimulate binding (Fig. 6C). As previously shown (27), TxeR is also unable to stimulate binding of core enzyme to the C. difficile gdh promoter.
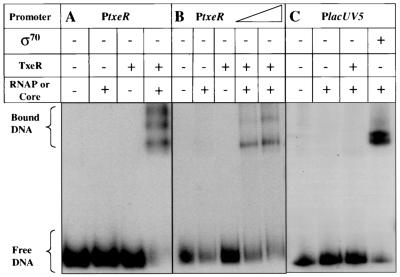
FIG. 6.
Gel mobility shift assays of the txeR promoter. A DNA fragment containing the txeR promoter region (P1+P2) was incubated with C. difficile RNA polymerase (A) or E. coli RNA polymerase (B) core enzyme alone or core enzyme that had been preincubated with TxeR (see Materials and Methods). The triangle in panel B indicates the use of increasing amounts of TxeR-containing holoenzyme (100 and 200 nM). (C) A DNA fragment containing the E. coli lacUV5 promoter was incubated with E. coli core enzyme alone or core enzyme that had been preincubated with TxeR or E. coli σ70.
The txeR promoter region contains two sets of sequences with similarity to the putative −10 and −35 regions recognized by TxeR (Fig. 7) (27). Labeled DNA fragments containing one or the other of these sites were tested separately in gel mobility shift assays. TxeR stimulated binding of RNA polymerase to both DNA fragments (Fig. 8).
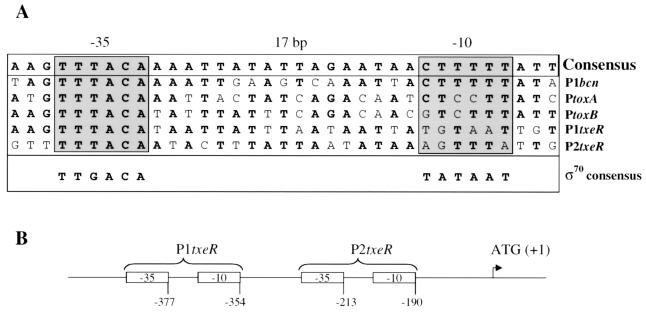
FIG. 7.
Comparison of TxeR-recognized promoters. (A) Two putative promoter elements for txeR that share similarity to the bcn and tox promoters are located upstream of the txeR open reading frame. (B) Schematic of the positions of these putative promoters with respect to the start codon of txeR.
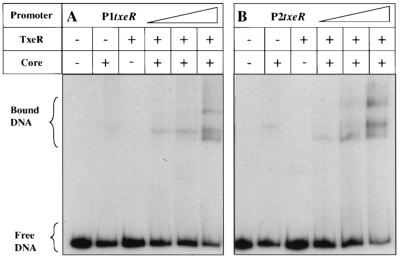
FIG. 8.
Gel mobility shift assays of txeR-recognized promoters. DNA fragments containing the putative P1txeR promoter (A) or the putative P2txeR promoter (B) were incubated with E. coli RNA polymerase core enzyme alone or in the presence of TxeR. The triangles indicate the use of increasing amounts of TxeR-containing holoenzyme (50, 100, and 200 nM).
The ability of TxeR to stimulate transcription from the txeR promoter was tested with in vitro transcription assays. Even though addition of TxeR to E. coli core RNA polymerase activated transcription from tox promoters, TxeR mixed with C. difficile RNA polymerase or E. coli core RNA polymerase failed to stimulate transcription from the txeR promoter when either or both of the potential sets of recognition sequences were present (data not shown).
DISCUSSION
Toxin synthesis in C. difficile is dependent on the growth phase (7, 15) and is responsive to several kinds of environmental signals, including the amino acid composition of the medium (16, 19, 20, 21, 44), the level of biotin (45), the nature of the carbon source (7, 21), and the temperature of cultivation (S. Karlsson, B. Dupuy, K. Mukherjee, E. Norin, L.G. Burman, and T. Åkerlund, unpublished data). However, the underlying molecular mechanisms that mediate these influences on toxin synthesis are unclear.
We had previously demonstrated that toxin synthesis in C. difficile is under the control of TxeR, an alternative sigma factor (27). Since both toxA and toxB gene-specific promoters show coordinate regulation, we hypothesized that the regulatory effects observed on toxin expression might be mediated via modulation of TxeR expression or activity. Our present results show that TxeR positively regulates its own expression in a manner analogous to its effect on the tox promoters and suggest a model for the response of the tox genes to cell growth conditions. During rapid exponential growth, the synthesis of TxeR is mostly repressed and the tox genes are silent. When cells enter the stationary phase, repression of txeR is relieved, synthesis of TxeR increases, and transcription of the tox genes is induced. Since synthesis of TxeR is autocatalytic, this model also explains the rapid amplification of toxin mRNA synthesis when cells make the transition from exponential to stationary phases. This model is consistent with the notion that the txeR gene has at least two promoters, one of which is relatively weak and independent of TxeR. This promoter provides a basal level of TxeR expression during exponential growth and is insensitive to glucose. TxeR-dependent transcription is subject to growth phase and carbon source-mediated regulation. Moreover, TxeR is active as a positive autoregulator at 37°C, but less so at other temperatures (Karlsson et al., unpublished data). It remains to be seen whether TxeR-dependent transcription is also influenced by the availability of biotin and various amino acids (16, 19, 20, 44, 45). It should be noted that the effect of glucose on txeR expression does not exclude the possibility that glucose also acts independently at the tox promoters.
Interestingly, at least some of the environmental signals that control TxeR synthesis seem to be conserved in C. perfringens and C. difficile. The proteins (repressors or activators) that mediate environmental control of txeR transcription are completely unknown. Possible candidates include the C. difficile homologs of proteins such as CcpA (14), CodY (34), and AbrB (39), which control similar responses in Bacillus subtilis.
The extent of TxeR-independent expression of txeR that we see in C. perfringens (Fig. 2) and C. difficile (Fig. 4) may be exaggerated in our experiments, because we are measuring expression from multicopy plasmids. Moreover, glucose-dependent negative regulation may be underestimated if the regulatory protein is not present in sufficient excess. Nonetheless, one must reconcile the existence of any expression of a positive autoregulator with the failure to stimulate its own synthesis in rapidly growing cells and with the absence of tox gene expression under such conditions. Such results might be obtained if additional factors participated in the regulatory system. For example, some TxeR protein might be synthesized in cells growing in rich medium, but be turned over rapidly. As a result, the concentration of TxeR would be very low (Fig. 4B). Alternatively, an additional regulatory protein might contribute to transcriptional control of txeR and the tox genes. Although TxeR enables RNA polymerase to form stable (heparin resistant), open complexes at the txeR promoter, no detectable txeR transcript was observed in in vitro runoff transcription assays. After formation of a stable, open complex, RNA polymerase must initiate phosphodiester bond formation and break contact with the promoter to create a productive elongation complex. An additional factor may be required to facilitate these steps at the txeR promoter.
Our ability to introduce an engineered construct into C. difficile permitted the first described use of a reporter gene fusion in this organism. Conjugation between E. coli and C. difficile was reported by Liyanage et al. (24), but in that case, a nonreplicating DNA was transferred and integrated into the chromosome. Moreover, the integration event led to a lethal phenotype. In our experiments, a shuttle plasmid that replicates in both E. coli and Clostridium and carries an oriT site recognized by the IncP transfer system was mobilized and stably maintained in the C. difficile recipient cells. This advance in C. difficile genetics should facilitate a large variety of expression and complementation studies that were previously impossible.
The C. difficile tox promoters recognized by TxeR share striking similarities with each other and with the UV-inducible promoters of the bacteriocin (bcn) gene of C. perfringens (7). Since TxeR regulates its own expression, its promoter should also be similar. In fact, the region upstream of the txeR gene contains two potential candidates for TxeR-dependent promoters (Fig. 7). These promoters show a good match to the −35 consensus sequence for toxA, toxB, and bcn P1 and P3 promoters. However, the −10 regions appear to be divergent. The −10 region of P1txeR resembles more the −10 region of σ70 promoters (11). This promoter region could in principle be responsible for both the basal-level expression of a txeR-gusA fusion seen in the absence of the txeR gene and for TxeR-stimulated expression. These notions remain speculative, because our repeated efforts to experimentally identify txeR transcription start points in vivo have been unsuccessful, presumably because of the low level of expression of the txeR gene in C. difficile. We are currently creating a series of txeR-gusA fusions with successively shorter txeR-derived elements as a means of narrowing down the location of sequences necessary for txeR expression.
TxeR shares significant similarities with C. perfringens UviA, the putative positive regulator of bcn (8); with BotR, a positive regulator of Clostridium botulinum toxin genes (29); and with TetR, a positive regulator of the Clostridium tetani tetanus toxin gene (28). We suggest that all four regulators act as sigma factors of RNA polymerase, directing the core polymerase to specific promoter sites. In fact, in vivo functional complementarity has already been demonstrated for TetR and BotR (28) and for TxeR and UviA (B. Dupuy, N. Mani, and A. L. Sonenshein, unpublished data). Indeed, we have observed that addition of TxeR allows C. difficile and E. coli RNA polymerases to bind to the bcn, botulism toxin, and tetanus toxin promoters in gel mobility shift assays and to recognize the P1 and P3 promoters of the bcn gene in in vitro runoff transcription assays (Dupuy et al., unpublished data). These data lead us to suggest that the regulatory proteins controlling expression of toxins and a bacteriocin in four Clostridium spp. are derived from a common evolutionary ancestor.
Acknowledgments
We thank S. Cole for critical review of the manuscript, A. Kolb for the gift of E. coli σ70, D. Acheson for help in preparing antibody to TxeR, S. B. Melville for providing strain SM101, and S. Karlsson and colleagues for allowing us to cite their unpublished results.
This work was supported by a postdoctoral fellowship to N. Mani from the Charles A. King Trust, a research grant from the Public Health Service (GM42219) to A. L. Sonenshein, a grant from the Australian National Health and Medical Research Council to J. I. Rood, and funds from the Institut Pasteur (Paris).
REFERENCES
- 1.Banno, Y., T. Kobayashi, H. Kono, K. Watanabe, K. Ueno, and Y. Nozawa. 1984. Biochemical characterization and biologic actions of two toxins (D-1 and D-2) from Clostridium difficile. Rev. Infect. Dis. 6(Suppl. 1):S11-S20. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, S. H., Y. J. Tang, and J. Silva, Jr. 2000. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 181:659-663. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, S. N., A. C. Y. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dove, C. H., S.-Z. Wang, S. B. Price, C. J. Phelps, D. M. Lyerly, T. D. Wilkins, and J. L. Johnson. 1990. Molecular characterization of the Clostridium difficile toxin A gene. Infect. Immun. 58:480-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuy, B., and A. L. Sonenshein. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27:107-120. [DOI] [PubMed] [Google Scholar]
- 8.Garnier, T., and S. T. Cole. 1988. Studies of UV-inducible promoters from Clostridium perfringens in vivo and in vitro. Mol. Microbiol. 2:607-614. [DOI] [PubMed] [Google Scholar]
- 9.Hammond, G. A., and J. L. Johnson. 1995. The toxigenic element of Clostridium difficile strain 10463. Microb. Pathog. 19:203-213. [DOI] [PubMed] [Google Scholar]
- 10.Hammond, G. A., D. M. Lyerly, and J. L. Johnson. 1997. Transcriptional analysis of the toxigenic element of Clostridium difficile. Microb. Pathog. 22:143-154. [DOI] [PubMed] [Google Scholar]
- 11.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, I., S. M. Whelan, T. O. Davis, and N. P. Minton. 1996. Genetic characterisation of the botulinum toxin complex of Clostridium botulinum strain NCTC 2916. FEMS Microbiol. Lett. 140:151-158. [DOI] [PubMed] [Google Scholar]
- 14.Henkin, T. M. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 15.Hundsberger, T., V. Braun, M. Weidmann, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1997. Transcriptional analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 244:735-742. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, D., T. Karasawa, R. Tanaka, M. Namiki, and S. Nakamura. 1998. Effect of isoleucine on toxin production by Clostridium difficile in a defined medium. Zentbl. Bakteriol. 287:375-386. [DOI] [PubMed] [Google Scholar]
- 17.Just, I., J. Selzer, M. Wilm, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500-503. [DOI] [PubMed] [Google Scholar]
- 18.Just, I., M. Wilm, J. Selzer, G. Rex, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J. Biol. Chem. 270:13932-13936. [DOI] [PubMed] [Google Scholar]
- 19.Karasawa, T., T. Maegawa, T. Nojiri, K. Yamakawa, and S. Nakamura. 1997. Effect of arginine on toxin production by Clostridium difficile in defined medium. Microbiol. Immunol. 41:581-585. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson, S., L. G. Burman, and T. Åkerlund. 1999. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 145:1683-1693. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson, S., A. Lindberg, E. Norin, L. G. Burman, and T. Åkerlund. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect. Immun. 68:5881-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, C. P., and J. T. LaMont. 1998. Clostridium difficile infection. Annu. Rev. Med. 49:375-390. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Liyanage, H., S. Kashket, M. Young, and E. R. Kashket. 2001. Clostridium beijerinckii and Clostridium difficile detoxify methylglyoxal by a novel mechanism involving glycerol dehydrogenase. Appl. Environ. Microbiol. 67:2004-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyerly, D. M., and T. D. Wilkins. 1995. Clostridium difficile, p.867-891. In M. J. Blaser, P. D. Smith, J. I. Ravin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, Ltd., New York, N.Y.
- 26.Lyras, D., and J. I. Rood. 1998. Conjugative transfer of RP4-oriT shuttle vectors from Escherichia coli to Clostridium perfringens. Plasmid 39:160-164. [DOI] [PubMed] [Google Scholar]
- 27.Mani, N., and B. Dupuy. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. USA 98:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marvaud, J.-C., U. Eisel, T. Binz, H. Niemann, and M. R. Popoff. 1998. TetR is a positive regulator of the tetanus toxin gene in Clostridium tetani and is homologous to BotR. Infect. Immun. 66:5698-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marvaud, J.-C., M. Gibert, K. Inoue, Y. Fujinaga, K. Oguma, and M. R. Popoff. 1998. botR/A is a positive regulator of botulinum neurotoxin and associated non-toxin protein genes in Clostridium botulinum A. Mol. Microbiol. 29:1009-1018. [DOI] [PubMed] [Google Scholar]
- 30.Melville, S. B., R. Labbe, and A. L. Sonenshein. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 62:5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Moncrief, J. S., L. A. Barroso, and T. D. Wilkins. 1997. Positive regulation of Clostridium difficile toxins. Infect. Immun. 65:1105-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moncrief, J. S., D. M. Lyerly, and T. D. Wilkins. 1999. Molecular biology of large clostridial toxins, p. 333-359. In K. Aktories and I. Just (ed.), Bacterial protein toxins. Handbook of experimental pharmacology, vol. 145. Springer-Verlag, Berlin, Germany.
- 34.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schaeffer, F., A. Kolb, and H. Buc. 1982. Point mutations change the thermal denaturation profile of a short DNA fragment containing the lactose control elements. Comparison between experiment and theory. EMBO J. 1:99-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:37-45. [Google Scholar]
- 38.Smith, C. J., S. M. Markowitz, and F. L. Macrina. 1981. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother. 19:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan, N. M., S. Pellett, and T. D. Wilkins. 1982. Purification and characterization of toxins A and B of Clostridium difficile. Infect. Immun. 35:1032-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan, K. S., B. Y. Wee, and K. P. Song. 2001. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J. Med. Microbiol. 50:613-619. [DOI] [PubMed] [Google Scholar]
- 42.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmid and cosmid clones. Focus 9:12. [Google Scholar]
- 43.von Eichel-Streiber, C., P. Bouquet, M. Sauerborn, and M. Thelestam. 1996. Large clostridial cytotoxins—a family of glucosyltransferases modifying small GTP-binding proteins. Trends Microbiol. 4:375-382. [DOI] [PubMed] [Google Scholar]
- 44.Yamakawa, K., S. Kamiya, X. Q. Meng, T. Karasawa, and S. Nakamura. 1994. Toxin production by Clostridium difficile in a defined medium with limited amino acids. J. Med. Microbiol. 41:319-323. [DOI] [PubMed] [Google Scholar]
- 45.Yamakawa, K., T. Karasawa, T. Ohta, H. Hayashi, and S. Nakamura. 1998. Inhibition of enhanced toxin production by Clostridium difficile in biotin-limited conditions. J. Med. Microbiol. 47:767-771. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]