Abstract
The TatA and TatB proteins are essential components of the twin arginine protein translocation pathway in Escherichia coli. C-terminal truncation analysis of the TatA protein revealed that a plasmid-expressed TatA protein shortened by 40 amino acids is still fully competent to support protein translocation. Similar truncation analysis of TatB indicated that the final 30 residues of TatB are dispensable for function. Further deletion experiments with TatB indicated that removal of even 70 residues from its C terminus still allowed significant transport. These results imply that the transmembrane and amphipathic helical regions of TatA and TatB are critical for their function but that the C-terminal domains are not essential for Tat transport activity. A chimeric protein comprising the N-terminal region of TatA fused to the amphipathic and C-terminal domains of TatB supports a low level of Tat activity in a strain in which the wild-type copy of either tatA or tatB (but not both) is deleted.
Protein transport to the periplasm of most bacteria proceeds by one of two pathways. The major route of protein translocation across the cytoplasmic membrane is the Sec pathway, to which substrates are targeted by N-terminal signal peptides. Protein translocation by the Sec machinery is by an N- to C-terminal threading mechanism driven by SecA-dependent ATP hydrolysis (19, 22). However, a critical subset of proteins is transported in a Sec-independent manner. Such proteins are also synthesized with N-terminal signal peptides that, in this case, harbor a consensus SRRxFLK motif (2). These twin arginine signal peptides target precursor proteins to the twin arginine translocation (Tat) system that is structurally and mechanistically related to the ΔpH-dependent thylakoid import pathway of chloroplasts (15). Substrates of the Tat pathway are frequently cofactor-containing proteins that bind their cofactors in the cytoplasmic compartment and are thus folded prior to translocation (3).
In Escherichia coli, four proteins, TatA, TatB, TatC, and TatE, have been identified that are required for Tat-dependent protein translocation (4, 24, 26, 33). TatC is an essential component of the Tat system in bacteria and chloroplasts (4, 21). It is an integral inner membrane protein comprising six transmembrane α-helices with the N and C termini located in the cytoplasm (10, 11). TatA and TatE are more than 50% identical at the amino acid level. Genetic analysis indicates that they have similar functions in protein transport; however, a strain with a deletion of tatA generally displays much more severe transport defects than a tatE deletion strain (24). This is consistent with expression studies that indicate that tatA is transcribed and translated at a 50- to 100-fold higher level than tatE (14). TatB shares some sequence homology with TatA/E (approximately 25% identity at the amino acid level) but is considerably longer. TatB is an essential component of the E. coli Tat machinery. Thus, an in-frame deletion of the tatB gene leads to a complete block in the export of seven endogenous Tat substrates. The mutation can be complemented by extra copies of tatB supplied in trans but not by multicopy tatA/E, highlighting the distinct functions of these two classes of proteins (26). However, it has recently been demonstrated that there is still some export of the artificial Tat substrate TorA-ColV in the tatB mutant strain, suggesting that, depending upon the substrate, TatB may not always be completely essential for function of the E. coli Tat pathway (13). Moreover, on the basis of whole genome sequences, it can be inferred that a functional Tat system in some bacteria may require just one copy of a TatA/B/E-like protein in addition to TatC (3, 34). This suggests that, under some circumstances, the function of either TatA or TatB is dispensable or that the properties of TatA/E and TatB are combined in a single type of polypeptide.
TatA/B family proteins are predicted to have in common a structure that comprises an N-terminal transmembrane α-helix, followed by an adjacent amphipathic domain and a C-terminal region of variable length. Recently, it has been demonstrated that both TatA and TatB are true integral inner membrane proteins. However, upon removal of the N-terminal transmembrane domain, TatA becomes a soluble protein while TatB retains peripheral interactions with the membrane (10). These observations suggest that the amphipathic domain of TatB is able to mediate some level of interaction with the membrane. The two proteins can be further distinguished by virtue of the fact that TatB forms a tight stoichiometric complex with TatC (5). The copy numbers of the two proteins are also markedly different—by two different methods, the TatA/TatB/TatC molar ratio in the E. coli membrane has been estimated to be approximately 40:2:1 (14, 25). A complex of TatAB with an apparent molecular mass of 600 kDa containing a large molar excess of TatA over TatB has been isolated in detergent solution and forms annular structures visible by electron microscopy (25).
In this study, we have sought to define in more detail the structural requirements for TatA and TatB and the interrelationship between the two proteins. Our previous studies have shown that the N-terminal transmembrane domains of TatA and TatB are essential for their activities (10). Therefore, we have constructed a series of C-terminal truncations of the two proteins and assessed their abilities to support Tat-dependent transport. In addition, we have constructed chimeric proteins in which the N-terminal transmembrane regions of TatA and TatB have been swapped. Our data suggest that a chimeric protein comprising the N-terminal region of TatA fused to the amphipathic and C-terminal domains of TatB shows some properties of TatA protein function and some properties of TatB protein function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used in this study are described in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid(s) | Genotype | Source or reference |
|---|---|---|
| Bacterial strains | ||
| MC4100 | F− ΔlacU169 araD139 rpsL150 relA1 ptsF rbs flbB5301 | 7 |
| VJS5833 | Δ(lacIZYA)X74 ΔphoA532 pcnB1 zad-981::Tn10d (Kmr) | 30 |
| K38 | hfrC phoA4 pit-10 tonA22 ompF627 relA1 spoT1 λ+ | 18 |
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF) U169 | Promega |
| MC4100-P | Same as MC4100 pcnB1 zad-981::Tn10d (Kmr) | 6 |
| JARV16 | Same as MC4100 ΔtatA ΔtatE | 26 |
| JARV16-P | Same as MC4100 ΔtatA ΔtatE pcnB1 zad-981::Tn10d (Kmr) | 26 |
| BØD | Same as MC4100 ΔtatB | 26 |
| BØD-P | Same as MC4100 ΔtatB pcnB1 zad-981::Tn10d (Kmr) | 26 |
| BEAD | Same as MC4100 ΔtatAB ΔtatE | This work |
| BEAD-P | Same as MC4100 ΔtatAB ΔtatE pcnB1 zad-981::Tn10d (Kmr) | This work |
| PLAWT | Same as JARV16 attB::Ptat tatA+ | This work |
| PLBWT | Same as BØDattB::Ptat ΔtatA tatB+ | This work |
| PLAx | Same as JARV16 attB::Ptat(tatA-10x) | This work |
| PLBx | Same as BØD attB::Ptat ΔtatA(tatB-10x) | This work |
| Plasmids | ||
| pGP1-2 | Kmr encodes T7 RNA polymerase | 31 |
| pNR42 | Cmr encodes T7 RNA polymerase | 26 |
| pNR14 | pT7.5; suf1+ Ampr | 26 |
| pBluescript II KS(+) | Ampr | Stratagene |
| pFAT415 | pBluescript TatA+ | 26 |
| pPALA10, -20, -30, -40, -50, -60, -67 | Same as pFAT415 but with stepwise 3′ truncations of tatA by 10, 20, 30, 40, 50, 60, and 67 codons | This work |
| pFAT416 | pBluescript TatB+ | 26 |
| pPALB10, -20, -30, -40, -50, -60, -70, -80, -90, -100, -110, -120, -130, -140, -149 | Same as pFAT416 but with stepwise 3′ truncations of tatB by 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, 120, 130, 140, and 149 codons | This work |
| pSU18 | Cmr | 1 |
| pSUB30 | pSU18 carrying tatB truncated by 30 codons | This work |
| pACBN | pSU18 carrying tatB codons 1-20 fused to tatA codons 21-89 | This work |
| pBCAN | pSU18 carrying tatA codons 1-20 fused to tatB codons 21-171 | This work |
| pACBNKS | pBluescript carrying tatB codons 1-20 fused to tatA codons 21-89 | This work |
| pBCANKS | pBluescript carrying tatA codons 1-20 fused to tatB codons 21-171 | This work |
During all genetic manipulations, E. coli strains were grown aerobically in Luria-Bertani (LB) medium (23). Concentrations of antibiotics were as described previously (7). The growth phenotypes of mutants with trimethylamine N-oxide (TMAO) as the sole respiratory electron acceptor were determined on M9 minimal medium (23) agar plates supplemented with 0.5% glycerol and 0.4% TMAO and incubated in a gas jar under a hydrogen-carbon dioxide atmosphere. The sodium dodecyl sulfate (SDS) resistance phenotype of mutants was tested on LB agar plates containing 2% SDS (6). For TMAO reductase assays, cells were cultured in modified Cohen and Rickenberg medium (9) supplemented with 0.2% glucose and 0.4% TMAO.
Plasmid and strain construction.
The plasmids and strains constructed in this work are shown in Table 1. Plasmid pFAT415 carries the tatA gene and more than 500 bp of upstream DNA in pBluescript (26). Plasmid pFAT416 carries more than 500 bp of the DNA sequence upstream of tatA (as in pFAT415) but also the ΔtatA allele and the intact tatB gene in pBluescript (26). Successive truncations of multiples of 30 bp (10 codons) from the 3′ ends of the tatA and tatB genes down to codon 22 of each gene were constructed by PCR techniques (primer sequences are available on request) and recloned into either pFAT415 or pFAT416, as appropriate. For coexpression of truncated forms of tatA and tatB in the same strain, truncated genes together with upstream tat DNA were subcloned into plasmid pSU18 by digestion with XbaI and KpnI.
Plasmid pBCAN expresses a fusion protein comprising the first 20 amino acids (i.e., the transmembrane segment) of TatA fused to residues 21 to 171 of TatB. It was constructed as follows. Codons 21 to 171 of tatB were amplified by PCR with primers TATA4 (24) and 5′-GCGGATCCGCCGGCCCGCAACGACTGCCTGTGG-3′ with pFAT416 as the template, digested with BamHI and KpnI, and cloned into the polylinker region of pSU18 to give plasmid pTBC. The promoter region and the first 60 bp (20 codons) of tatA were amplified with primers TATA1 (24) and 5′-GCGCTTTAAAAAGCAGTACAACGATGACGGC-3′ with pFAT415 as the template, digested with KpnI, and ligated into pTBC, which had been previously digested with KpnI and NaeI.
Plasmid pACBN expresses a fusion protein comprising the first 20 amino acids (i.e., the transmembrane segment) of TatB fused to residues 21 to 89 of TatA. It was constructed as follows. Codons 21 to 89 of tatA were amplified by PCR with primers 5′-GCGGTACCTTACACCTGCTCTTTATCG-3′ and 5′-GCGGATCCGCCGGCACCAAAAAGCTCGGC-3′ with pFAT415 as the template, digested with BamHI and KpnI, and cloned into the polylinker region of pSU18 to give plasmid pTAC. The promoter region and the first 60 bp (20 codons) of tatB were amplified with primers TATA1 (7) and 5′-CAGAACGACGAGGCC-3′ with pFAT416 as the template, digested with KpnI, and ligated into pTAC, which had been previously digested with KpnI and NaeI. The chimeras were subsequently also subcloned into pBluescript by digestion with XbaI and KpnI.
Strain BEAD (MC4100 ΔtatAB ΔtatE) was constructed from strain BAD (MC4100 ΔtatAB) (13) by transfer of the mutant tatE allele present on plasmid pFAT44 (24) by standard procedures (12). The Tn10d-linked pcnB1 allele was subsequently moved into this strain by P1 transduction as described previously (26). Strains PLA1 and PLA3-5 carry chromosomal deletions of the tatA and tatE genes, with either the full-length form or truncated forms of tatA (full-length tatA, PLAWT; tatA truncated by 10 codons, PLA1; tatA truncated by 20 codons, PLA2; tatA truncated by 30 codons, PLA3; tatA truncated by 40 codons, PLA4; tatA truncated by 50 codons, PLA5) together with the tat promoter region present at the att site. Analogous strains PLB1 and PLB3-11 have chromosomal deletions of the tatB gene, with either the full-length form or a truncated form of tatB (full-length tatB, PLBWT; tatB truncated by 30 to 120 codons, PLB3-12) together with the tat promoter region present at the att site. The full-length and truncated alleles of tatA and tatB were amplified with the primers 5′-GCGCAGATCTCGGCCAGTGAATTGTAATACG-3′ and 5′-GCGCAGATCTGCTCGAAATTAACCCTCACTAAAGG-3′ with the appropriate pBluescript-encoded truncated tatA or tatB gene as the template. The PCR products were digested with BglII and cloned into the BamHI site of plasmid pRS552 (28). The tat alleles were delivered onto the chromosome of either JARV16 (MC4100 ΔtatA ΔtatE) or BØD (MC4100 ΔtatB) as described elsewhere (28).
All clones obtained from PCR-amplified DNA were sequenced to ensure that no mutations had been introduced.
Protein methods.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting were carried out as described previously (16, 32), with affinity-purified TatA and TatB antisera (25), and immunoreactive bands were visualized with the ECL detection system (Amersham Pharmacia Biotech).
Expression and stability of some truncated forms of TatA and TatB was further monitored by radiolabeled pulse-chase analysis in strain K38(pGP1-2) as described previously (26). Tat substrate SufI from plasmid pNR14 was expressed under control of the T7 promoter. Strains MC4100(pNR42), PLAWT(pNR42), PLA1-5(pNR42), PLBWT(pNR42), and PLB3-12(pNR42) were transformed with pNR14, and radiolabeled pulse-chase analysis was performed as previously described (26).
Subcellular fractions for TMAO reductase activity measurements were prepared from small (30-ml) cultures with the cold osmotic shock protocol (29). TMAO:benzyl viologen oxidoreductase activity was measured as described previously (27). Protein concentrations were estimated by the method of Lowry et al. (17).
RESULTS
Truncation analysis of TatA.
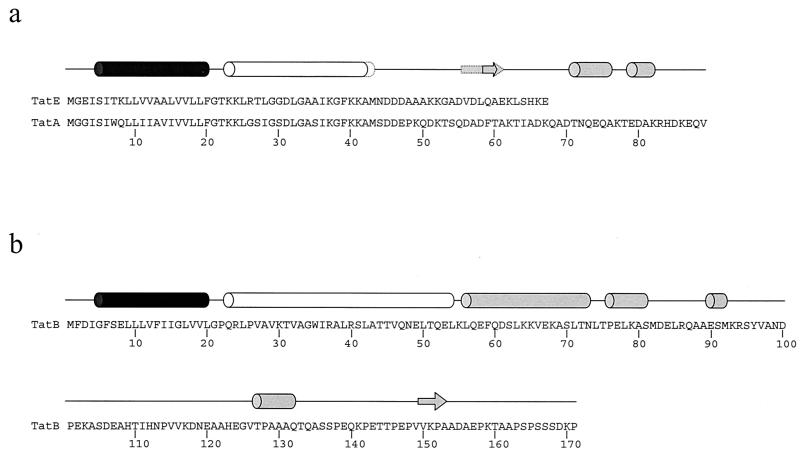
The E. coli tatA gene encodes a 9.6-kDa polypeptide of 89 amino acid residues. Structural predictions (Fig. 1A) suggest an N-terminal hydrophobic α-helix to residue 20, followed by a short hinge region and a longer amphipathic α-helix extending to amino acid 42. The tatE gene encodes a protein of 67 residues with a predicted mass of 7.0 kDa. It shares almost 60% amino acid identity with TatA along its entire length and can functionally substitute for TatA. In keeping with the high percentage of amino acid identity, the positions of the predicted hydrophobic and amphipathic α-helical regions in TatE are indistinguishable from those in TatA (Fig. 1A). Both proteins have C-terminal regions that are predicted to be mainly random coil. Because of the high sequence and structural similarity between the two proteins, coupled with our previous observations that TatE is poorly expressed but functionally equivalent to TatA (14, 24, 26), in the following experiments, we chose to study TatA alone.
FIG. 1.
Structural predictions for TatA, TatB, and TatE. Predicted structures were determined with the PSIPRED program at http://bioinf.cs.ucl.ac.uk/psipred/. (a) Predictions for TatA and TatE. Secondary structural elements common to both proteins are shown with a solid line, and those unique to TatE are shown with a dotted line. (b) Structural prediction for TatB. α-Helical regions are represented as cylinders, and β-sheets are represented as arrows. The transmembrane α-helical domains of TatA, TatB, and TatE are black, and the strongly amphipathic α-helical domains are white. Further predicted helical regions are grey.
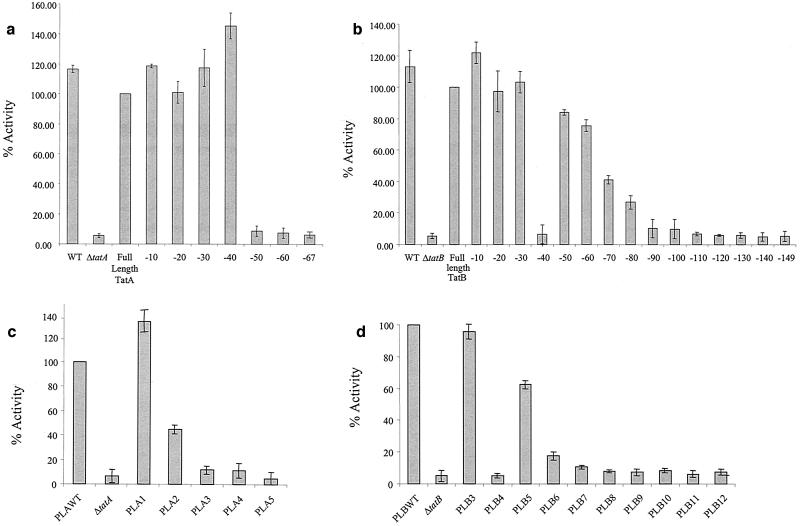
We have constructed a series of truncations of TatA by the sequential removal of blocks of 10 amino acids from the C-terminal end. Initially, these fusions were expressed on a multicopy plasmid vector in a strain with the chromosomal copies of both tatA and tatE deleted. In addition, the strain carried the pcnB allele to restrict the plasmid copy number. The ability of the TatA truncations to complement the Tat− phenotype of the tatA/E deletion strain was assessed by measuring the levels of TMAO reductase activity (due to the Tat substrate protein TorA) in the periplasm of strains expressing the truncated proteins. As shown in Fig. 2A, C-terminal truncations of TatA by up to 40 residues still supported full export of TorA to the periplasm. These results are consistent with our previous observation that plasmid-expressed TatE (which is 22 amino acids shorter than TatA) can fully restore Tat transport activity to a strain with tatA and tatE deleted (26) but suggest that a TatA protein of only 49 amino acids retains full functionality. However, deletions of 50 residues or more from the C terminus of TatA resulted in very low levels of periplasmic TorA activity that were indistinguishable from those of the strain with tatA and tatE deleted.
FIG. 2.
Periplasmic TMAO reductase activities assayed from tat mutant strains expressing C-terminally truncated TatA and TatB proteins. Panels a and b show periplasmic TMAO reductase activities determined from strains harboring plasmid-borne truncation alleles. (a) Activities determined for ΔtatA/E pcnB mutant strain JARV16-P expressing C-terminally truncated TatA proteins. One hundred percent activity is that determined from the periplasmic fraction of JARV16-P carrying full-length tatA. (b) Activities determined for ΔtatB pcnB mutant strain BØD-P expressing C-terminally truncated TatB proteins. One hundred percent activity is that determined from the periplasmic fraction of BØD-P carrying full-length tatB. Panels c and d show periplasmic TMAO reductase activities from strains carrying chromosomally encoded truncation alleles integrated at the att site. (c) Activities determined for the ΔtatA/E attB::Ptat (tatA-10x) series of strains. One hundred percent activity is that determined from the periplasmic fraction of the tatA/E mutant expressing the full-length tatA allele: (d) Activities determined for the ΔtatB attB::Ptat ΔtatA (tatB-10x) series of strains. One hundred percent activity is that determined from the periplasmic fraction of the tatB mutant expressing the full-length tatB allele. WT, wild type. Cells were fractionated, and TMAO reductase activity was assayed as described in the text.
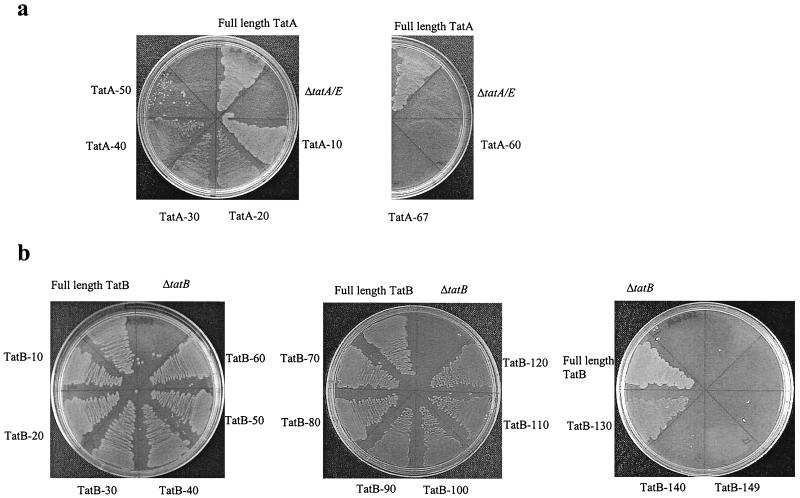
Since the TorA activity assay cannot distinguish between strains that show a low level of TorA export and those that are completely blocked for Tat transport, we employed two growth tests to further analyze strains expressing the truncated TatA proteins. In the first, we tested the ability of the strain lacking tatA and tatE and expressing the truncated TatA protein to grow anaerobically with TMAO as the sole electron acceptor. In agreement with the results in Fig. 2A, truncations of up to 40 residues supported strong growth on TMAO. The truncated protein lacking 50 residues permitted weak growth, suggesting that a very low but nonzero quantity of TorA was still exported. The strains with the remaining truncations failed to grow (results not shown). We have shown previously that tat deletion mutants are sensitive to growth in the presence of the ionic detergent SDS because of a defect in the barrier of the cell envelope. Therefore, as a second growth test, we tested the ability of the strain lacking tatA and tatE and expressing the truncated TatA protein to form colonies on LB plates containing 2% SDS. In full agreement with the results of Fig. 2A and of the TMAO plate tests, truncations of up to 40 residues supported strong growth in the presence of the detergent while TatA lacking 50 residues still permitted weak growth (Fig. 3A). Truncations of greater than 50 residues did not allow growth on SDS-containing medium.
FIG. 3.
Growth of tat mutant strains expressing TatA and TatB truncations on medium containing SDS. (a) Growth of ΔtatA/E pcnB mutant strain JARV16-P expressing either full-length TatA or a range of C-terminally truncated proteins. (b) Growth of ΔtatB pcnB strain BØD-P expressing full-length TatB or a range of C-terminally truncated proteins.
Inactive truncated TatA did not exhibit a dominant-negative phenotype when expressed in a Tat+ strain. In addition, none of the truncated TatA proteins was able to complement a ΔtatB strain for growth on either TMAO or SDS, which is consistent with the differing functions of TatA and TatB.
Truncation analysis of TatB.
The E. coli tatB gene encodes a protein of 171 amino acids with a predicted molecular mass of 18.4 kDa. Structural prediction analysis of TatB (Fig. 1B) indicates an N-terminal hydrophobic α-helix, similar to those of TatA and TatE, extending to residue 20. This is followed by a helical region extending to amino acid 81, of which at least the first section is predicted to be strongly amphipathic. The C-terminal portion is predicted to be mainly random coil.
To define the minimal functional unit of TatB, we constructed a parallel series of truncations of TatB, again by the sequential removal of groups of 10 amino acids from the C terminus. We expressed these fusions in a strain carrying a chromosomal deletion in tatB and, in addition, the pcnB allele to restrict the plasmid copy number. The truncated proteins were initially assessed for the ability to support Tat transport activity by assaying the levels of periplasmic TMAO reductase activity in a strain in which they were expressed. The results are shown in Fig. 2B. TatB proteins lacking up to 30 amino acids from the C termini behaved essentially the same way as the wild-type protein for the transport of TMAO reductase. Proteins truncated by 50 to 100 residues showed a progressively diminishing but significant level of periplasmic TMAO reductase activity. An interesting anomaly was seen with the TatB −40 construct, which was indistinguishable from the tatB deletion strain in terms of periplasmic TorA activity. Strains expressing proteins C-terminally truncated by 110 residues or more had only background levels of periplasmic TMAO reductase activity.
The TatB truncations were also tested for the ability to support growth of the ΔtatB strain on minimal medium containing TMAO. In agreement with the assay results shown in Fig. 2B, strains carrying truncations lacking up to 100 amino acids from the C terminus were able to grow with TMAO as the sole electron acceptor. Furthermore, proteins truncated by 110 and 120 amino acids also supported growth, consistent with the export of a very low, but significant, level of TMAO reductase. The TatB −40 construct supported only very weak growth on TMAO, consistent with the TorA activity measurements (results not shown). A further test of growth in the presence of SDS (Fig. 3B) fully supported the results observed with TMAO. However, in this case, some protection from the killing effect of SDS was even afforded to the tatB strain expressing a TatB protein of only 41 residues (truncated by 130 amino acids from the C terminus). The inactive truncated TatB forms did not show a dominant-negative phenotype when expressed in a Tat+ strain. In addition, none of them were able to complement the ΔtatA/E strain for growth on either TMAO or SDS.
Stability of truncated TatB.
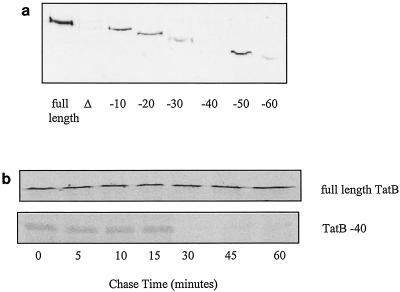
A surprisingly low level of Tat transport activity was found in the tatB mutant strain expressing the TatB −40 truncation. To check the stability of the truncated protein, we examined the levels of protein by Western blotting. As shown in Fig. 4A, the TatB −40 truncated protein could not be detected after Western blotting although we were able to detect TatB proteins truncated by at least 60 amino acids with our TatB antiserum. These observations suggest that the TatB −40 protein may be unstable. This is confirmed in Fig. 4B (bottom), where pulse-chase analysis of expression of TatB −40 shows that although the protein is synthesized, it is subjected to rapid degradation. In contrast, the full-length protein (Fig. 4B, top) is stable over the time period of the experiment.
FIG. 4.
The TatB protein that is C-terminally truncated by 40 residues is unstable. (a) Western blot analysis of whole cells of ΔtatB mutant strain BØD either devoid of plasmids (lane Δ) or carrying plasmids expressing full-length TatB or C-terminally truncated TatB protein, as indicated. Approximately 50 μg of protein was loaded in each lane. (b) Full-length TatB (upper gel) or TatB truncated by 40 residues from the C terminus (lower gel) expressed in strain K38(pGP1-2) was pulse-labeled for 5 min by the addition of [35S]methionine and then chased from time zero with unlabeled methionine. Protein synthesis and stability were assessed by SDS-PAGE of whole cells, followed by autoradiography.
Coexpression of the shortest fully functional alleles of TatA and TatB gives an active Tat system.
The shortest forms of plasmid-expressed TatA and TatB that separately support full Tat transport activity are a TatA protein lacking the C-terminal 40 amino acids and a TatB protein missing the final 30 amino acids. To confirm that these truncated proteins were, indeed, fully active, we coexpressed the plasmid-encoded TatA −40 and TatB −30 proteins in an E. coli strain with the chromosomal copies of tatA, tatB, and tatE deleted. As shown in Table 2, the periplasmic TorA activity in the strain with the chromosomal copies of tatA, tatB, and tatE deleted and expressing the two truncated proteins was comparable to that of the wild type, indicating that the two proteins had full activity.
TABLE 2.
Periplasmic TMAO reductase activities of strains expressing wild-type and truncated forms of TatA and TatB
| Strain | Relevant genotype | Periplasmic TMAO reductase activity (μmol/min/mg of periplasmic protein) |
|---|---|---|
| MC4100-P | Tat+ | 0.98 |
| BEAD-P | ΔtatAB ΔtatE | 0.09 |
| BEAD-P(pFAT405/pFAT416) | ΔtatAB ΔtatE pTatA+ pTatB+ | 0.94 |
| BEAD-P(pALA30/pSUB40) | ΔtatAB ΔtatE pTatA(−40) pTatB(−30) | 1.04 |
Chromosomally encoded truncated TatA and TatB at the att site.
We sought to extend our analysis of the phenotype of selected TatA and TatB truncations by assessing their effects on the kinetics of transport through the Tat pathway. We therefore investigated the effects of some of the truncated alleles on the rate of export of the Tat substrate SufI. It was not possible to undertake these experiments with the plasmid-encoded truncations, since the pre-SufI substrate and the T7 polymerase were also plasmid encoded. To carry out these experiments, we therefore introduced the truncated tatA and tatB alleles onto the chromosome of the ΔtatA/E and ΔtatB mutants. The mutant alleles, together with tat promoter DNA, were recombined onto the chromosome of the tat mutant strains at the lambda phage attachment (att) site. We have shown previously that although the presence of the pcnB mutation does greatly reduce the level of plasmid-borne Tat proteins, there is still an approximately 25-fold overexpression of protein relative to the chromosomally encoded level (6). Therefore, we first examined the effects of the presence of the single-copy full-length and truncated forms of tatA and tatB on Tat activity by assessing the levels of periplasmic TMAO reductase. Introduction of the wild-type tatA and tatB genes into the att site of, respectively, the ΔtatA/E and ΔtatB strains resulted in levels of periplasmic TorA activity comparable to that of the parental strain MC4100 (Fig. 2C and D). The recombinant strains also exported plasmid-expressed SufI with the same kinetics as the wild type (data not shown). Moreover, Western blotting showed that the full-length TatA and TatB proteins expressed from the att site were produced at levels similar to those of the proteins expressed from their native chromosomal location (results not shown). Thus, the att-integrated alleles behaved identically to the native chromosomal alleles.
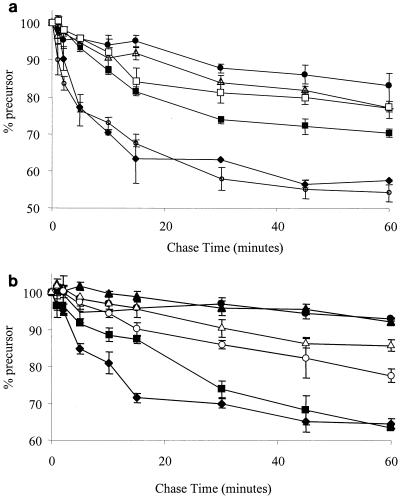
The chromosomally encoded tatB truncation series (Fig. 2D) behaved in a way similar to that of the plasmid-encoded truncations. The TatB −30 truncation restored full periplasmic TorA activity to the ΔtatB strain (Fig. 2D). TatB truncated by 50 to 120 amino acids produced a progressive decrease in the ability to export TorA but allowed growth on both SDS and TMAO plates (data not shown), mirroring the results obtained with the plasmid-borne truncations. As with the plasmid-encoded TatB −40 truncation, the strain with the corresponding chromosomally encoded protein showed hardly any Tat activity. These observations were supported by pulse-chase analysis of SufI export (Fig. 5B). The ΔtatB attB::Ptat ΔtatA(tatB-30) mutant strain appeared to export SufI at the same rate as the ΔtatB attB::Ptat ΔtatA tatB+ mutant strain. The tatB mutant expressing TatB truncations from positions −30, −60, and −90 showed a progressively decreased, although still detectable, rate of SufI export. In contrast, the rate of export seen with the tatB mutant strain expressing the −40 truncation was indistinguishable from that of the strain with the tatB mutation alone.
FIG. 5.
Pulse-chase analysis of SufI export in tat mutant strains expressing C-terminally truncated TatA and TatB proteins. SufI precursor protein, expressed in tatA/E or tatB mutant strains carrying tatA or tatB truncation alleles integrated at the att site, was pulse-labeled for 5 min by the addition of [35S]methionine and then chased from time zero with unlabeled methionine. Precursor synthesis and processing were assessed by SDS-PAGE of whole cells, followed by autoradiography. The mean percentage of total SufI protein remaining in the precursor form in whole cells at each of the indicated time points is plotted (n = 3 to 6). The bars represent the standard error of the mean. (a) Export of SufI in strains expressing full-length TatA and truncated forms of TatA. Symbols: ⧫, PLAWT (full-length TatA); ○, PLA1 (TatA −10); □, PLA2 (TatA −20); ▪, PLA3 (TatA −30); ▵, PLA4 (TatA −40); •, PLA5 (TatA-50). (b) Export of SufI in strains expressing full-length TatB and truncated forms of TatB. Symbols: ⧫, PLBWT (full-length TatB); ▪, PLB3 (TatB −30); ▴, PLB4 (TatB −40); ○, PLB6 (TatB −60); ▵, PLB9 (TatB −90); •, BØD (ΔtatB).
Surprisingly, the chromosomally expressed TatA series of truncations (Fig. 2C) differed significantly from the analogous plasmid-expressed truncated forms. Truncations of TatA by 20, 30, or 40 amino acids that supported full export of TorA when expressed in multicopy showed much lower rates when expressed in the ΔtatA/E strain in single copy. These observations were also borne out by analysis of SufI export (Fig. 5A). This could be accounted for if the truncated TatA proteins were markedly less stable than the full-length form of TatA. In the case in which these proteins are overproduced from plasmids, one could argue that, despite instability, sufficient TatA is produced to give a fully active Tat system. Unfortunately, it was not possible to test this possibility because TatA proteins truncated by more than 10 amino acids did not cross-react significantly with our TatA antiserum.
Domain swapping of TatA and TatB.
Figure 1 shows that TatA and TatB are similar in basic structural organization. To further probe the domain structure and function of TatA and TatB, we constructed two complementary genetic fusions. In one construct, we fused DNA encoding the N-terminal transmembrane domain of TatA (amino acids 1 to 20) with DNA encoding the amphipathic helix and C-terminal region of TatB (amino acids 21 to 171), giving plasmid pBCAN. The complementary construct, pACBN, comprises DNA encoding the N-terminal transmembrane domain of TatB (amino acids 1 to 20) joined to the region encoding amino acids 21 to 89 of TatA. These constructs were introduced separately into the ΔtatA/E and ΔtatB mutant strains and both individually and in combination into the ΔtatAB/E mutant strain.
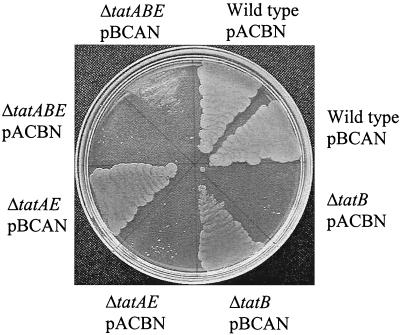
Upon subcellular fractionation, none of the strain-plasmid combinations resulted in significant accumulation of periplasmic TorA activity (data not shown). Interestingly, however, the ΔtatA/E mutant strain and the ΔtatB mutant carrying plasmid pBCAN were both capable of growth in the presence of SDS (Fig. 6). These results indicate that a Tat system comprising either TatA or TatB, along with the N-terminal domain of TatA fused to the amphipathic and C-terminal domains of TatB, has a low level of Tat function. It was, however, necessary to have a wild-type copy of either TatA or TatB present since the plasmid could not complement a strain with all of tatA, tatB, and tatE deleted (Fig. 6). In stark contrast, the complementary TatB N-terminal-TatA C-terminal fusion did not support growth of either the ΔtatA/E or the ΔtatB mutant strain with SDS. The ΔtatAB/E mutant strain also failed to grow on SDS, even when both of the fusion plasmids, pACBN and pBCAN, were present (results not shown). Taken together, these observations suggest that a function(s) unique to TatA resides in the N-terminal transmembrane helix domain whereas the function(s) unique to TatB is found in the cytoplasmic domain.
FIG. 6.
Growth of strains expressing hybrid TatA and TatB proteins on medium containing SDS. The strains and plasmids are as described in the text.
DISCUSSION
We have shown previously that, despite sharing 25% sequence identity, TatA and TatB from E. coli form two separate classes of proteins with distinct roles in protein translocation (26). Consistent with this, TatB, but not TatA, forms a tight stoichiometric complex with the TatC protein (5). In many other bacterial systems, distinct TatA and TatB homologues can be identified on the basis of amino acid sequence, suggesting that two functionally diverse proteins are necessary for Tat activity. This is also true of the analogous ΔpH system of plant thylakoids, where the Hcf106 protein (the TatB ortholog) forms a tight complex with cpTatC, which is involved in the initial recognition of precursors. Tha4, (the chloroplast TatA ortholog) functions at a step after precursor recognition in the translocation process (8, 20). Interestingly, however, some bacteria appear to have lost the requirement for separate TatA and TatB proteins. For example, the genomes of the bacterium Rickettsia prowazekii and the archaeon Aeropyrum pernix encode only one copy of a TatA/TatB-like protein. Furthermore, although Bacillus subtilis encodes three copies of TatA/TatB, analysis of the amino acid sequence shows that the proteins have features of both the TatA and TatB classes of proteins (3, 34).
In this report, we have sought to further define the features of TatA and TatB proteins. Our truncation analysis suggests that although the C-terminal domain of each protein is not strictly necessary for activity, the transmembrane and adjacent helical regions play a critical role in their function. A notable difference between the two proteins is their overall lengths; E. coli TatB is 82 amino acids longer than TatA, by virtue of a longer predicted α-helical region and an extended C terminus. However, since a TatB protein truncated from the C terminus by up to 100 amino acids still retains some TatB function, total length is not a discriminating factor. Rather, it is the length of the helical cytoplasmic domain that is of importance—the transmembrane helix-adjacent helical domain of TatB is predicted to be much longer than that of TatA, and truncation analysis shows that this reflects a functional requirement.
A chimeric protein that comprises only the transmembrane domain of TatA fused to the amphipathic and C-terminal regions of TatB apparently retained a low level of Tat function, while the complementary fusion (the transmembrane domain of TatB fused to the amphipathic and C-terminal regions of TatA) was not active. Interestingly, the partially functional TatA/TatB fusion protein showed hybrid properties since it was able to partially substitute for the functions of both TatA and TatB. This points toward the transmembrane domain of TatA as being critical for the function of this protein, possibly for the formation of channel-like structures seen in electron micrographs of a purified TatA/TatB complex (25). It also indicates that the helical cytoplasmic domain performs the unique function(s) of TatB. It can be speculated that it is this region that interacts with TatC, with the transmembrane domain playing a membrane-anchoring role. This would be consistent with the proposal of Cline and Mori (8) that the twin arginine signal peptide binding site in the thylakoid Tat system was formed from an interface between cpTatC and Hcf106 (TatB).
Acknowledgments
We thank Erik de Leeuw for providing purified TatA and TatB antibodies. We acknowledge Frank Sargent for helpful discussion.
This work was supported by a BBSRC grant-in-aid to the John Innes Centre and a BBSRC-funded Ph.D. studentship to P.A.L. B.C.B. is an R. J. P. Williams Senior Research Fellow at Wadham College, Oxford, and T.P. is a Royal Society Research Fellow.
REFERENCES
- 1.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 2.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 4.Bogsch, E., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 5.Bolhuis, A., J. E. Mathers, J. D. Thomas, C. M. Barrett, and C. Robinson. 2001. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276:20213-20219. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, G., E. de Leeuw, N. R. Stanley, M. Wexler, B. C. Berks, F. Sargent, and T. Palmer. 2002. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 43:1457-1470. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cline, K., and H. Mori. 2001. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154:719-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, G. N., and H. W. Rickenberg. 1956. Concentration specifique reversible des amino acids chez Escherichia coli. Ann. Inst. Pasteur (Paris) 91:693-720. [PubMed] [Google Scholar]
- 10.de Leeuw, E., I. Porcelli, F. Sargent, T. Palmer, and B. C. Berks. 2001. Membrane interactions and self-association of the TatA and TatB components of the twin-arginine translocation pathway. FEBS Lett. 506:143-148. [DOI] [PubMed] [Google Scholar]
- 11.Drew, D., D. Sjostrand, J. Nilsson, T. Urbig, C. N. Chin Cn, J. W. de Gier, and G. von Heijne. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA 99:2690-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ize, B., F. Gérard, M. Zhang, A. Chanal, R. Volhoux, T. Palmer, A. Filloux, and L.-F. Wu. 2002. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 317:327-335. [DOI] [PubMed] [Google Scholar]
- 14.Jack, R. L., F. Sargent, B. C. Berks, G. Sawers, and T. Palmer. 2001. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 183:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keegstra, K., and K. Cline. 1999. Protein import and routing systems of chloroplasts. Plant Cell 11:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 18.Lyons, L. B., and N. D. Zinder. 1972. The genetic map of the filamentous bacteriophage f1. Virology 49:45-60. [DOI] [PubMed] [Google Scholar]
- 19.Manting, E. H., and A. J. M. Driessen. 2000. Escherichia coli translocase: the unravelling of a molecular machine. Mol. Microbiol. 37:226-238. [DOI] [PubMed] [Google Scholar]
- 20.Mori, H., and K. Cline. 2002. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell Biol. 157:205-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori, H., E. J. Summer, and K. Cline. 2001. Chloroplast TatC plays a direct role in thylakoid ΔpH-dependent protein transport. FEBS Lett. 501:65-68. [DOI] [PubMed] [Google Scholar]
- 22.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sargent, F., E. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sargent, F., U. Gohlke, E. de Leeuw, N. R. Stanley, T. Palmer, H. R. Saibil, and B. C. Berks. 2001. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur. J. Biochem. 268:3361-3367. [DOI] [PubMed] [Google Scholar]
- 26.Sargent, F., N. R. Stanley, B. C. Berks, and T. Palmer. 1999. Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J. Biol. Chem. 274:36073-36083. [DOI] [PubMed] [Google Scholar]
- 27.Silvestro, A., J. Pommier, and G. Giordano. 1988. The inducible trimethylamine-N-oxide reductase of Escherichia coli K12: biochemical and immunological studies. Biochim. Biophys. Acta 954:1-13. [DOI] [PubMed] [Google Scholar]
- 28.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based protein and operon fusion cloning tools. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 29.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 257:11591-11596. [DOI] [PubMed] [Google Scholar]
- 30.Stanley, N. R., F. Sargent, G. Buchanan, J. Shi, V. Stewart, T. Palmer, and B. C. Berks. 2002. Behaviour of topological marker proteins targeted to the Tat protein transport pathway. Mol. Microbiol. 43:1005-1021. [DOI] [PubMed] [Google Scholar]
- 31.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiner, J. H., P. T. Bilous, G. M. Shaw, S. P. Lubitz, L. Frost, G. H. Thomas, J. A. Cole, and R. J. Turner. 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93:93-101. [DOI] [PubMed] [Google Scholar]
- 34.Wu, L.-F., B. Ize, A. Chanal, Y. Quentin, and G. Fichant. 2000. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol. 2:179-189. [PubMed] [Google Scholar]