Abstract
In Streptomyces griseus, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) switches on aerial mycelium formation and secondary metabolite biosynthesis. An A-factor-dependent transcriptional activator, AdpA, activates multiple genes required for morphological development and secondary metabolism in a programmed manner. A region upstream of a zinc-containing metalloendopeptidase gene (sgmA) was found among the DNA fragments that had been isolated as AdpA-binding sites. The primary product of sgmA consisted of N-terminal pre, N-terminal pro, mature, and C-terminal pro regions. sgmA was transcribed in an AdpA-dependent manner, and its transcription was markedly enhanced at the timing of aerial mycelium formation. AdpA bound two sites in the region upstream of the sgmA promoter; one was at about nucleotide position −60 (A site) with respect to the transcriptional start point of sgmA, and the other was at about position −260 (B site), as determined by DNase I footprinting. Transcriptional analysis with mutated promoters showed that the A site was essential for the switching on of sgmA transcription and that the B site was necessary for the marked enhancement of transcription at the timing of aerial mycelium formation. Disruption of the chromosomal sgmA gene resulted in a delay in aerial hypha formation by half a day. SgmA is therefore suggested to be associated with the programmed morphological development of Streptomyces, in which this peptidase, perhaps together with other hydrolytic enzymes, plays a role in the degradation of proteins in substrate hyphae for reuse in aerial hypha formation.
Members of the gram-positive, soil-inhabiting, filamentous bacterial genus Streptomyces produce a wide variety of secondary metabolites and show complex morphological differentiation culminating in sporulation. These characteristics make members of this genus important as industrial microorganisms and as one of the model prokaryotes for studying multicellular differentiation (3, 4). On agar medium, one or more germ tubes formed from a germinating spore grow into substrate hyphae, which branch frequently and grow rapidly by cell wall extension at the hyphal tips. Subsequently, aerial hyphae emerge by reuse of material assimilated into the substrate mycelium, such as DNA, proteins, and storage compounds. Many cells in substrate hyphae thus lyse and die (5, 21, 31). At this stage of development, therefore, enzymes for the degradation of these substances, such as proteases, nucleases, lipases, and glucanases, are presumably required. In fact, nucleases and serine proteases are associated with the development of aerial hyphae from substrate hyphae (5, 23). When the apical growth of aerial hyphae stops, in contrast to the substrate mycelium, septa are formed at regular intervals along the hyphae to form many unigenomic compartments within a sheath. The sporulation septa consist of two membrane layers separated by a double layer of cell wall material, which permits the eventual separation of adjacent spores (7). Spore chains usually consist of many tens of spores. The spores are separated from one another by a constriction process and liberated by constriction and subsequent dissolution of the cell wall.
In Streptomyces griseus, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) at an extremely low concentration acts as a switch for aerial mycelium formation and secondary metabolite formation (9-11). An A-factor-deficient S. griseus mutant strain neither forms aerial mycelium nor produces streptomycin. We thus expected that A-factor would induce the biosynthesis and activation of hydrolytic enzymes for cannibalizing substrate hyphae to supply material to newly forming aerial mycelium. A-factor is produced in a growth-dependent manner and accumulates until the end of exponential growth. It triggers the A-factor regulatory cascade by inducing the transcription of adpA, which encodes a transcriptional activator, by binding the A-factor-specific receptor ArpA, which has bound the promoter of adpA, and dissociating ArpA from the promoter (25). AdpA induced in this way activates the transcription of strR, which encodes a pathway-specific transcriptional activator, leading to streptomycin biosynthesis. AdpA also activates adsA, which encodes an extracytoplasmic function sigma factor of RNA polymerase that is essential for aerial mycelium formation (32). Considering the A-factor regulatory cascade, we believe that the hydrolyzing enzymes for the cannibalization of substrate hyphae are under the control of AdpA and AdsA.
In the present study, we identified a gene encoding a zinc-containing metalloendopeptidase (SgmA) in the gene library containing DNA fragments bound and controlled by AdpA (12, 32). As expected, aerial mycelium formation by sgmA disruptants was delayed. We describe here the identification of the metalloendopeptidase gene as one of the targets of AdpA, transcriptional analysis of the gene in relation to morphological development, and determination of the two AdpA-binding sites upstream of the promoter by DNase I footprinting. One of the two binding sites was required for the transcriptional activation of sgmA by AdpA, and the other was necessary for the enhancement of sgmA transcription at a specific time.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. griseus IFO13350 (6) was obtained from the Institute of Fermentation, Osaka, Japan. S. griseus ΔadpA (25) and ΔadsA (32) mutants were described previously. Streptomyces strains were grown in YMPD medium (yeast extract [Difco], 0.2%; meat extract [Wako Pure Chemicals], 0.2%; Bacto Peptone [Difco], 0.4%; NaCl, 0.5%; MgSO4 · 7H2O, 0.2%; glucose, 1%). YMPD agar medium contained 2% agar. R2YE medium (8) was used for the regeneration of protoplasts. Thiostrepton (50 μg/ml) and neomycin (20 μg/ml) were added when necessary. As Streptomyces plasmids, high-copy-number plasmid pIJ702, containing the thiostrepton resistance and melanin production genes (15), with a copy number of 40 to 100 per genome, and low-copy-number plasmid pKU209, containing the ampicillin and thiostrepton resistance genes (14), with a copy number of 1 or 2 per genome, were used. Plasmid pADP10H, carrying adpA on high-copy-number vector pIJ487, was previously described (25). Escherichia coli JM109 and vectors pUC18 and pUC19 for DNA manipulation were purchased from Takara Shuzo. E. coli JM110 containing dam and dcm mutations was used for preparing nonmethylated Streptomyces DNA for gene disruption. Histidine-tagged AdpA (AdpA-H) was purified from E. coli BL21(DE3) harboring pET-adpA as described previously (32). Media and growth conditions for E. coli were described by Maniatis et al. (18). Ampicillin (50 μg/ml) and kanamycin (50 μg/ml) were used when necessary.
General recombinant DNA studies.
Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were purchased from Takara Shuzo. [α-32P]dCTP (110 TBq/mmol) for DNA labeling with a BcaBest DNA labeling system (Takara Shuzo) and [γ-32P]ATP (220 TBq/mmol) for end labeling at 5′ ends with T4 polynucleotide kinase were purchased from Amersham Pharmacia Biotech. DNA was manipulated in Streptomyces (8) and in E. coli (1, 18) as described earlier. Nucleotide sequences were determined by the dideoxy chain termination method with a Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham) or an ABI Prism DNA sequencing kit and an automated DNA sequencer.
Cloning and nucleotide sequencing of the metalloendopeptidase gene.
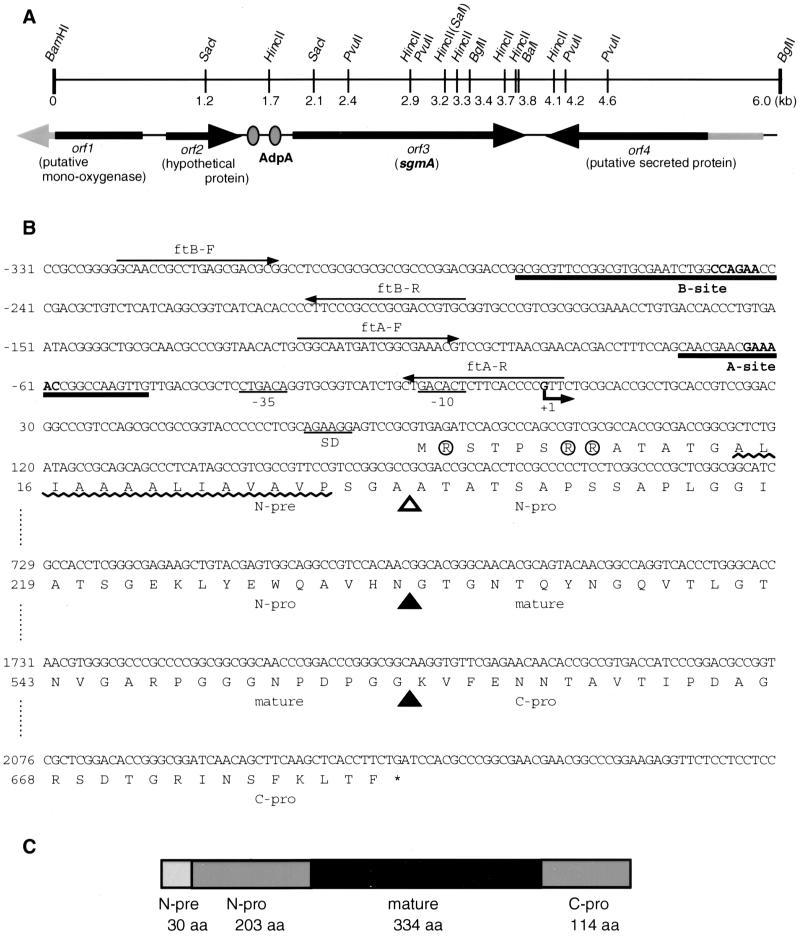
We chose one (AdBS18) of the DNA fragments that had been isolated as targets of AdpA by the gel mobility shift-PCR procedure (32). The DNA fragment was 32P labeled with the BcaBest DNA labeling system and used as a hybridization probe against restriction digests of the chromosomal DNA of S. griseus IFO13350. Several cycles of Southern hybridization and cloning by colony hybridization yielded a 6.0-kb DNA fragment upstream and downstream of the original DNA fragment (see Fig. 2A). The nucleotide sequence of the 6.0-kb fragment was determined and open reading frames (ORFs) were predicted by FRAME Plot analysis (13).
FIG. 2.
ORFs in the cloned 6.0-kb fragment and nucleotide sequence of the promoter and coding regions of sgmA. (A) In the 6.0-kb fragment, two complete ORFs (ORF 2 and sgmA) and two truncated ORFs (ORF 1 and ORF 4) are present. The two AdpA-binding sites in front of sgmA are shown (gray ovals). (B) Nucleotide sequence of sgmA, with the transcriptional start point of sgmA taken as +1. The two AdpA-H-binding sites (A and B sites) determined by DNase I footprinting (see Fig. 5) are indicated. The bold letters in the binding sites were changed so that AdpA no longer bound these sites (see Fig. 6). The positions of the ftA and ftB primers used for preparing 32P-labeled probes for DNase I footprinting are also shown. Likely −35 and −10 sequences for the sgmA promoter are underlined. A purine-rich sequence, AGAAGG, 7 nucleotides upstream of the start codon, may serve as a ribosome-binding (SD) sequence. The signal sequence of SgmA can be divided into an Arg cluster region (Arg residues are circled), a hydrophobic region (wavy line), and an Ala/Pro repeat region. During the maturation of SgmA, secondary cleavages at two sites (closed triangles), between Asn-233 and Gly-234 and between Gly-567 and Lys-568, likely occur after secretion. A likely cleavage site (open triangle) for a signal peptidase is also shown. (C) Schematic representation of the structure of SgmA. aa, amino acids.
For the construction of pIJ702-sgmA, the sgmA sequence was amplified by PCR with the following primers: 5′-GCCGCATGCGATCCACGCCCAGCCGTCGC-3′ (the underlined letters indicate an SphI site and represent Met-1 [the ATG in the SphI site; originally GTG] to Arg-8) and 5′-GCCGGATCCCTTCCGGGCCGTTCGTTCGC-3′ (the underlined letters indicate a BamHI site and represent the region 11 to 30 nucleotides downstream of the stop codon of sgmA). After no errors during PCR had been verified, the amplified fragment was inserted between the SphI and the BglII sites of pIJ702 to construct pIJ702-sgmA. The ATG in the SphI site in pIJ702 serves as the start codon of melC1 (15). pIJ702-sgmA thus constructed had the sgmA sequence just downstream of the melC1 promoter. As a control plasmid, pIJ702-d, in which the small SphI-BglII fragment was deleted from pIJ702, was constructed. This plasmid does not direct the synthesis of tyrosinase; we have observed that this synthesis slightly affects the morphogenesis of S. griseus.
For the construction of pSGML, the 4.2-kb BamHI-PvuII fragment containing a 5′ portion of ORF 1, the whole ORF 2, the whole sgmA gene, and a 3′ portion of ORF 4 (see Fig. 2A) was cloned between the BamHI and the SmaI sites of pUC19. The sgmA sequence was excised as a 3.0-kb SacI fragment and inserted into the SacI site of pUC18. The sgmA sequence was then excised as an EcoRI-SalI fragment and inserted between the EcoRI and the XhoI sites of pKU209, resulting in pSGML.
Gel mobility shift assay.
Purification of AdpA-H from E. coli BL21(DE3) and the gel mobility shift assay were described previously (32). AdBS18 (441 bp; corresponding to positions −249 to +192; see Fig. 2B), originally cloned as a target of AdpA, was present in the EcoRI site of pUC19. This fragment was amplified with commercial primers M4 and RV (Takara Shuzo), 32P labeled with T4 polynucleotide kinase, and used as a probe. A 533-bp fragment and a 507-bp fragment upstream and downstream, respectively, of AdBS18 were also examined by the gel mobility shift assay. For amplification of the upstream region, 5′-CGGCGGAGCGATGCCTGGAG-3′ (corresponding to nucleotide positions −741 to −722; see Fig. 2B) and 5′-GGGGTGTGATGACCGCCTGA-3′ (positions −209 to −228) were used as primers, and the 5′ ends of the amplified fragment were 32P labeled. For the downstream region, 5′-CTCATAGCCGTCGCCGTTCC-3′ (positions +135 to +154) and 5′-GCCCAGCCAGACCACCTTGC-3′ (positions +641 to +622) were similarly used.
S1 nuclease mapping.
Methods for RNA isolation from cells grown on cellophane on the surface of agar medium and S1 nuclease mapping were as described by Kelemen et al. (16). Hybridization probes were prepared by PCR with a pair of 32P-labeled and nonlabeled primers. For high-resolution S1 nuclease mapping, 5′-GCGGCAATGATCGGCGAAAC-3′ (nucleotide positions −121 to −102, with the transcriptional start point of sgmA taken as +1 [determined later]; see Fig. 2B) and 5′-CGCGGACTCCTTCTGCGAGG-3′ (positions +75 to +56) were used. For low-resolution S1 nuclease mapping, 5′-CGACGCTGTCTCATCAGGCG-3′ (positions −241 to −222) and 5′-GGAGGTGGCGGTCGCGGCGC-3′ (positions +179 to +160) were used. hrdB, encoding a principal σ factor of RNA polymerase (28), was used to check the purity and the amount of the RNA used as described previously (32). Protected fragments were analyzed on 6% polyacrylamide DNA sequencing gels by the method of Maxam and Gilbert (19).
DNase I footprinting.
The method of DNase I footprinting was described by Yamazaki et al. (32). For analysis of the AdpA-binding site in AdBS18, a 32P-labeled fragment was prepared by PCR with ftA-F and ftA-R, covering positions −120 to +3, as the primers (see Fig. 2B). For determination of an additional AdpA-binding site, ftB-F and ftB-R, covering positions −322 to −190, were similarly used.
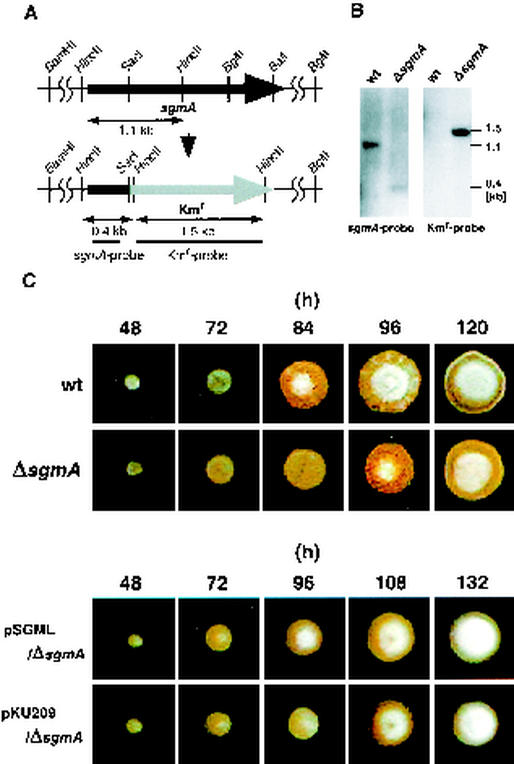
Gene disruption.
The cloned 6.0-kb BamHI-BglII fragment was placed in the BamHI site of pUC19. Between the SacI and BalI sites (corresponding to Leu-83 to Ala-655), a 1.5-kb SacI-Aor51HI fragment carrying the kanamycin resistance gene from Tn5 (2) was inserted (see Fig. 7A). The pUC19 plasmid was digested with DraI, alkali denatured with 0.1 M NaOH, and introduced by protoplast transformation into S. griseus IFO13350 as described previously (24). Correct replacement of the disrupted sgmA sequence with the intact chromosomal sgmA sequence, as a result of double crossover, was checked by Southern hybridization with the 1.5-kb kanamycin resistance gene and a 220-bp fragment containing a 5′ region of sgmA against HincII-digested chromosomal DNA.
FIG. 7.
Phenotypes of sgmA disruptants. (A) Schematic representation of disrupted sgmA on the chromosome of S. griseus IFO13350. (B) Southern hybridization to check the correct disruption of the chromosomal sgmA gene. Results obtained with the DNA fragment (nucleotide positions −28 to +192) used as the sgmA probe and the HincII fragment containing the kanamycin resistance gene and used as the Kmr probe are shown. wt, wild type. (C) Spore suspensions prepared from the wild type and mutant ΔsgmA were spread on YMPD agar medium and incubated at 28°C. The wild type formed apparent aerial hyphae at 84 h, whereas mutant ΔsgmA formed aerial hyphae at 96 h. Mutant ΔsgmA harboring sgmA on a low-copy-number plasmid (plasmid pSGML) formed aerial hyphae earlier than that harboring vector pKU209.
Alterations of the two AdpA-binding sequences by PCR.
The GAAAAC sequence in the middle of the A site (see Fig. 2B) was changed to a PstI recognition sequence, CTGCAG, as follows. The fragment (nucleotide positions −867 to +54) containing the A and B sites was amplified with primers PA-F (5′-GGCGAATTCCCATCGAGGTGCTGGCCAGG-3′ [the underlining indicates an EcoRI site]) and PA-R (5′-GGCAAGCTTGGGTACCGGCGGCGCTGGAC-3′ [the underlining indicates a HindIII site]) and placed between the EcoRI and the HindIII sites of pUC19. With the cloned fragment as the template, the EcoRI site to the A site and the A site to the HindIII site were separately amplified with primer pairs PA-F and PA-AR (5′-GGCCTGCAGGTTCGTTGCTGGAAAGGTCG-3′ [positions −66 to −85; the underlining indicates a PstI site]) and PA-AF (5′-GGGCTGCAGCGGCCAAGTTGTTGACGCGC-3′ [positions −59 to −40; the underlining indicates a PstI site]) and PA-R, respectively. The two amplified fragments were connected via the PstI site, generating the EcoRI-HindIII fragment containing nucleotide changes from GAAAAC to CTGCAG. Similarly, the CCAGAA sequence in the B site was changed to a BamHI recognition sequence, GGATCC. For this purpose, primer PA-BR (5′-GGGGGATCCCCAGATTCGCACGCCGGAAC-3′ [positions −250 to −269; the underlining indicates a BamHI site]), instead of PA-AR, and primer PA-BF (5′-GGCGGATCCCCCGACGCTGTCTCATCAGG-3′ [positions −243 to −224; the underlining indicates a BamHI site]), instead of PA-AF, were used. For generation of the EcoRI-HindIII fragment containing nucleotide changes at both A and B sites, the procedure for generating the A-site mutation was applied to the EcoRI-HindIII fragment containing the mutation at the B site. The absence of undesirable errors was checked by nucleotide sequencing.
For determination of the binding of AdpA to the mutated A and B sites by gel mobility shift assays, 32P-labeled probes of 303 bp for the A site and 300 bp for the B site were prepared by PCR with primer pairs 5′-TCAGGCGGTCATCACACCCC-3′ (positions −228 to −209) and 5′-CGCGGACTCCTTCTGCGAGG-3′ (positions +75 to +56) for the A site and 5′-CCACGCTCGCGGCGTCGGGG-3′ (positions −399 to −380) and 5′-ACGTTTCGCCGATCATTGCC-3′ (positions −100 to −119) for the B site. A 32P-labeled probe of 453 bp containing both A and B sites was also prepared with the primers 5′-TGGACGACCCGGACCGCTTC-3′ (positions −378 to −359) and 5′-CGCGGACTCCTTCTGCGAGG-3′ (positions +75 to +56).
The SacI-HincII fragment containing the mutation at the A or B site or both was excised from the EcoRI-HindIII fragment. Each of these fragments was replaced with the corresponding SacI-HincII fragment within the BamHI-SalI fragment on pUC19. The kanamycin resistance gene on a HindIII fragment was then inserted into the HindIII site, generating a plasmid used for gene replacement. The mutations at the A and B sites were thus inserted into the corresponding region on the chromosome of wild-type S. griseus by two rounds of single crossover. Correct insertion was checked by direct sequencing of the 0.9-kb SacI-SacI region covering the A and B sites (see Fig. 2A).
Nucleotide sequence accession number.
The nucleotide sequence reported in this article has been deposited in the DDBJ, EMBL, and GenBank DNA databases under accession no. AB085745.
RESULTS AND DISCUSSION
Isolation of a DNA fragment as a target of AdpA.
We established a simple and rapid procedure for isolation of the chromosomal DNA fragments from S. griseus IFO13350 that were recognized and bound by AdpA (12, 32). Our repeated experiments yielded more than 60 DNA fragments as candidates for the targets of AdpA. We chose one (AdBS18) of these candidates for further study. AdBS18 in the EcoRI site of pUC19 was excised by EcoRI digestion and subjected to a gel mobility shift assay with AdpA-H. As shown in Fig. 1A, AdBS18 was retarded by AdpA. An excess amount of cold AdBS18 abolished the binding (data not shown), indicating that the binding was specific. AdBS18 consisted of 441 bp (nucleotide positions −249 to +192) (Fig. 2), as indicated by nucleotide sequencing. As described below, this fragment contained one complete AdpA-binding site (A site) and one incomplete AdpA-binding site (B site).
FIG. 1.
Binding of AdpA-H to AdBS18 and to regions upstream and downstream of AdBS18, as determined by a gel mobility shift assay. (A) AdBS18 was 32P labeled and used in a gel mobility shift assay. The amounts of AdpA-H were 0.02 μg (lane 2), 0.2 μg (lane 3), and 0.4 μg (lane 4). Lane 1 is a control lane in which no AdpA-H was present. The positions of AdpA-H-bound (closed triangle) and free (open triangle) probes are shown. The position of the gel well is also indicated by an arrow. (B) Fragments of approximately 500 bp upstream (left; nucleotide positions −741 to −209 [see Fig. 2B]) and downstream (right; positions +135 to +641) of AdBS18 were separately 32P labeled by PCR and used as probes in a gel mobility shift assay. Lane 1 is a control lane in which no AdpA-H was present. The amounts of AdpA-H were 0.2 μg (lane 2) and 0.4 μg (lane 3).
A large BamHI-BglII DNA fragment covering AdBS18, in a total of 6.0 kb, was cloned by standard DNA manipulation (Fig. 2A). During the gel mobility shift assay, we noted that an additional region located upstream of AdBS18 was bound by AdpA-H. We prepared a ca. 500-bp fragment upstream of AdBS18 and subjected it to a gel mobility shift assay (Fig. 1B). The upstream region was also retarded by AdpA-H, and an excess amount of the cold probe abolished the binding. On the other hand, a ca. 500-bp fragment downstream of AdBS18, used as a control, was not retarded. It was thus apparent that AdpA-H recognized and bound a second site in addition to AdBS18. We compared the approximate binding affinities of the two binding sites by changing the amounts of AdpA-H in the gel mobility shift assays. No significant difference in binding affinities between the two sites was observed.
Cloning and nucleotide sequencing of the metalloendopeptidase gene.
We determined the nucleotide sequence of the cloned 6.0-kb fragment and predicted the extents and directions of ORFs by FRAME Plot analysis (13). AdBS18 (441 bp) was found to be located in front of an ORF encoding a protein of 681 amino acids, suggesting that AdpA bound the promoter region of this gene and activated its expression. The amino acid sequence from Gly-234 to Gly-567 was found to match completely that of the metalloendopeptidase (SGMP II for S. griseus metalloendopeptidase II) purified from S. griseus K-1 (17). The homologies of the three other ORFs with those on the Streptomyces coelicolor A3(2) genome (http://www.sanger .ac.uk/Projects/S_coelicolor) were as follows: ORF 1, putative mono-oxygenase (DNA database accession no. 2SCG61.20C); ORF 2, hypothetical protein (SCD95A.38C); and ORF 4, putative secreted protein (SCBAC17A6.09C). Although S. coelicolor A3(2) contains two sgmA homologues in tandem (SC3D11.03C and SC3D11.04C), the gene organization in the neighbors is totally different from that in S. griseus.
A comparison of the amino acid sequence of the mature metalloendopeptidase (17) and its nucleotide sequence revealed that the metalloendopeptidase, named SgmA (for S. griseus metalloendopeptidase), is primarily translated in a pre-pro form (Met-1 to Phe-681), with a large pre-pro region in its NH2-terminal portion (Met-1 to Asn-233) and a large pro region in its COOH-terminal region (Lys-568 to Phe-681) (Fig. 2C). The NH2-terminal portion of SgmA has a constitution somewhat similar to those of the leader peptides of a cellulase from a Streptomyces sp. (22), an endoglucanase H from Streptomyces plicatus (27), and an exoglucanase from Cellulomonas fimi (26); the leader peptides can be divided into an NH2-terminal region, an Arg cluster region, a hydrophobic region, and an Ala/Pro repeat region (22, 27). SgmA contains Arg cluster, hydrophobic, and Ala/Pro repeat regions, although it lacks a stretch of an NH2-terminal region. SgmA is therefore believed to be a protease with this type of signal peptide but not with a signal peptide typical of other prokaryotic extracellular enzymes. By analogy with the secretion of the endoglucanase of S. plicatus (27) and the cellulase of the Streptomyces sp. (22), we believe that the cleavage of the signal sequence of SgmA occurs within the Ala/Pro repeat region and that a second cleavage between Asn-233 and Gly-234 occurs after its secretion. PSORT software (http://sort.ims.u-tokyo.ac.jp/) predicted the pre-pro boundary to be between Ala-30 and Ala-31. Cleavage between Gly-567 and Lys-568 also occurs, probably after SgmA has been secreted.
Transcriptional analysis of the metalloendopeptidase gene.
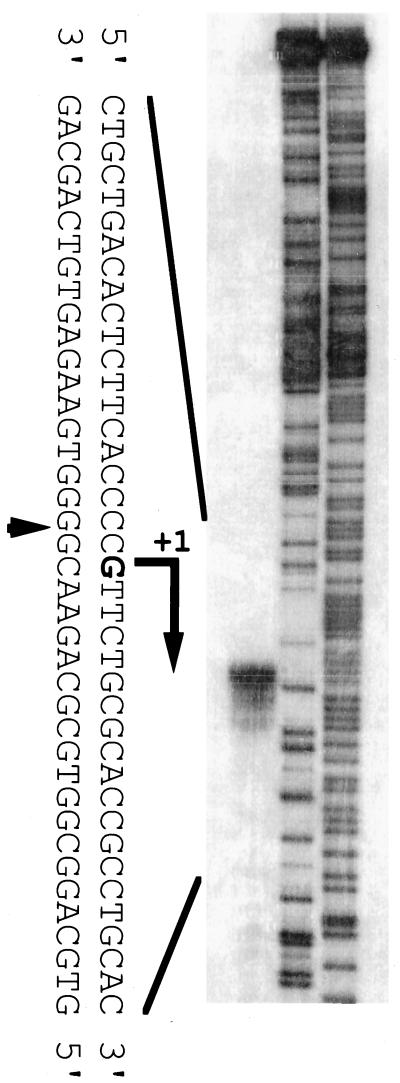
The transcriptional start point of sgmA was determined by S1 nuclease mapping with RNA prepared from cells grown on cellophane on the surface of YMPD agar medium. We searched for approximate transcriptional start points by low-resolution S1 nuclease mapping with AdBS18. A single start point was detected about 70 nucleotides upstream of the start codon, and no apparent read-through from the upstream region was detected. We next prepared an appropriate 32P-labeled probe and carried out high-resolution S1 nuclease mapping (Fig. 3). The transcriptional start point was assigned to the G residue 74 nucleotides upstream of the start codon. In front of the transcriptional start point, two sequences, CTGACA for −35 and GACACT for −10, were present. The putative −35 sequence is similar to the consensus −35 sequence (TTGACA) of the housekeeping genes in Streptomyces (29), but the −10 sequence deviates from the −10 consensus sequence (TAGRRT; R: A or G). In addition, the space between the −35 and −10 sequences is 16 bp, while the standard space in the Streptomyces genes is 17 or 18 bp.
FIG. 3.
Transcriptional start point of sgmA, as determined by high-resolution S1 nuclease mapping. RNA prepared from wild-type cells grown for 3 days on solid medium was used. The arrowhead indicates the position of the S1 nuclease-protected fragment. The 5′ terminus of the transcript was assigned to the indicated G residue because the fragments generated by the chemical sequencing reactions migrate 1.5 nucleotides further than the corresponding fragments generated by S1 nuclease digestion of the DNA-RNA hybrids (half a residue from the presence of the 3′-terminal phosphate group and one residue from the elimination of the 3′-terminal nucleotide).
We determined the course of sgmA transcription by low-resolution S1 nuclease mapping. Under the culture conditions used, wild-type S. griseus strain IFO13350 grew as substrate mycelium at day 1, as a mixture of substrate mycelium and aerial mycelium at day 2, and as a mixture of aerial mycelium and spores at day 3. Mutant ΔadpA grew as substrate mycelium for 3 days. In the wild-type strain, the transcription of sgmA was detectable at days 1 and 2 and was markedly enhanced at day 3 (Fig. 4). As described below, the marked enhancement of sgmA transcription at day 3 is not required for normal aerial hypha formation. In mutant ΔadpA, however, almost no sgmA transcripts were detected, indicating that AdpA serves as a transcriptional activator for sgmA. This finding is in agreement with the observation that multiple copies of adpA (plasmid pADP10H) caused markedly enhanced transcription of sgmA, in comparison with the transcripts in the wild type.
FIG. 4.
Time course of sgmA transcription, as determined by low-resolution S1 nuclease mapping. The transcription of hrdB, which is transcribed throughout growth, was used to check the purity and amount of the mRNA used. (Upper panel) No sgmA transcript is detected in mutant ΔadpA, whereas the transcript of sgmA is detected during growth for 3 days and markedly enhanced at day 3 in the wild type (wt). (Lower panel) The transcription of sgmA is enhanced in wild-type S. griseus IFO13350 harboring pADP10H, and enhancement of sgmA transcription at day 3 also occurs.
adsA, another target of AdpA, is also switched on by AdpA (32). However, the transcription of adsA is gradually increased, reaching its peak at day 3 under these conditions, and no abrupt enhancement of adsA transcription occurs at day 3 (32). This difference will be discussed below in relation to the presence of two AdpA-binding sites for sgmA.
The pattern of sgmA transcription and the amounts of the transcripts in mutant ΔadsA are similar to those in the wild type (data not shown). This means that sgmA is not transcribed by the RNA polymerase containing σAdsA. sgmA is thus suggested to be directly controlled by AdpA.
Determination of two AdpA-binding sites by DNase I footprinting.
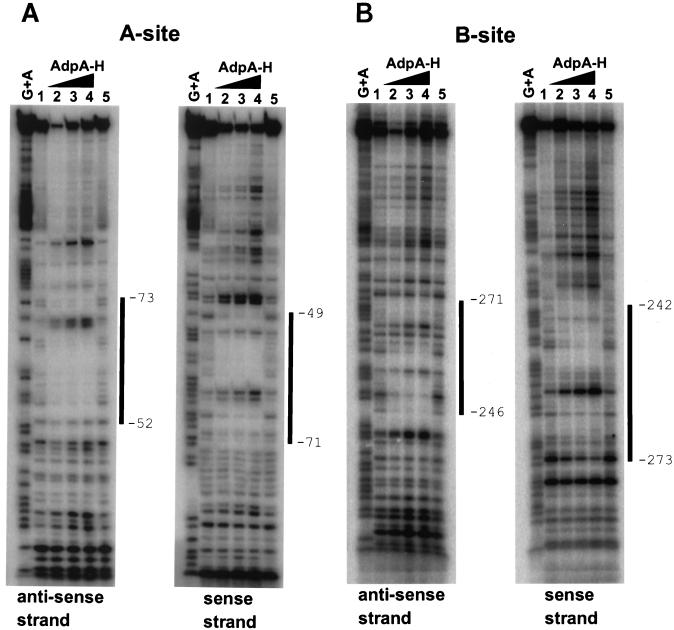
The two AdpA-binding sites upstream of sgmA were determined by DNase I footprinting with 32P-labeled ftA and ftB probes. The positions of the probes are shown in Fig. 2. The AdpA-binding site closer to the transcriptional start point, named the A site, was protected from DNase I digestion at nucleotide positions −52 to −73, with respect to the transcriptional start point, of the antisense strand and positions −49 to −71 of the sense strand (Fig. 5A). We used an image analyzer to determine the protected region where the intensities of the 32P-labeled ladders in the presence of AdpA were lower than those in the absence of AdpA. A similar DNase I footprinting assay for the additional AdpA-binding site, named the B site, showed that nucleotide positions −246 to −271 of the antisense strand and positions −242 to −273 of the sense strand were covered by AdpA (Fig. 5B).
FIG. 5.
Analysis of AdpA binding to two sites in front of the sgmA promoter by a DNase I protection assay. The DNase I digests were run with the same probes that were chemically cleaved (G+A lanes). The amounts of AdpA-H were 0.2 μg (lane 2), 0.4 μg (lane 3), and 0.8 μg (lane 4). Lanes 1 and 5 are control lanes without AdpA-H. The boundaries between the protected and unprotected nucleotides were determined with the aid of an image analyzer.
Alignment of the two binding sequences and the binding site upstream of adsA (32) does not predict an apparent consensus sequence for AdpA binding. Our unpublished results obtained from a study of the adsA promoter suggest that a sequence, 5′-AAX3G-3′ (X: any nucleotide), located at the vicinity of one of the 5′ ends of the AdpA-binding site, is important for AdpA binding. There are several positions with this sequence in both the A and the B sites.
Contribution of the A site to the transcriptional activation of sgmA by AdpA.
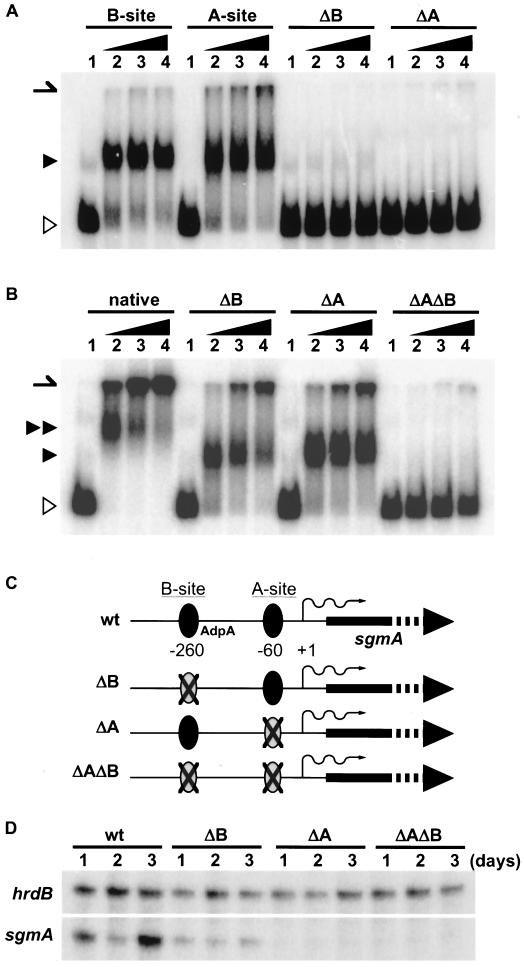
The presence of two AdpA-binding sites upstream of the sgmA promoter raised a question: are one or both of the binding sites required for transcriptional activation of the sgmA promoter by AdpA? We generated nucleotide changes by PCR so that the GAAAAC sequence in the middle of the A site was replaced by a PstI recognition sequence, CTGCAG, and inserted the mutated sequence into the corresponding chromosomal locus of wild-type S. griseus (Fig. 6A), generating mutant ΔA. The sequence changed is in one of the AAX3G sequences. The mutated sequence was no longer bound by AdpA when a ca. 300-bp fragment containing the mutated A site at the middle was assayed by a gel mobility shift assay (Fig. 6B). Similarly, we generated mutant ΔB, containing a mutated B site in the chromosome. The mutation in the B site contained nucleotide changes from CCAGAA to a BamHI recognition sequence, GGATCC, and this mutated site was not bound by AdpA. The sequence changed is also in an AAX3G sequence. We also generated mutant ΔAΔB, which contained mutations at both the A and the B sites on the chromosome. A gel mobility shift assay with the 32P-labeled probe of 453 bp containing both the A and the B mutations showed that AdpA failed to bind this fragment, whereas the 453-bp fragment containing the mutation at either the A or the B site was still bound by AdpA (Fig. 6C). The retarded signal observed for the 453-bp fragment containing intact A and B sites showed a low mobility, indicating that AdpA bound the two AdpA-binding sites in the original 453-bp fragment and that it bound the intact one in the 453-bp fragment containing a mutation at one of the two sites.
FIG. 6.
Generation of mutations at A and B sites and transcription of sgmA in S. griseus containing the mutations. (A) A DNA fragment (nucleotide positions −399 to −100) containing the B site and a DNA fragment (positions −228 to +56) containing the A site were 32P labeled and used as controls in the gel mobility shift assays. The DNA fragment containing the mutated A site (ΔA) or B site (ΔB) is not bound by AdpA-H. The positions of AdpA-H-bound (closed triangle) and free (open triangle) probes are shown. The position of the gel well is shown by an arrow. The amounts of AdpA-H were 0.2 μg (lane 2), 0.4 μg (lane 3), and 0.8 μg (lane 4). Lane 1 is a control lane without AdpA-H. (B) A DNA fragment (positions −378 to +75) containing both the A and the B sites was used. The amount of AdpA-H used for each lane was the same as that used in panel A. The mobility of the retarded original fragment (native; double closed triangles) containing the two AdpA-binding sites is slower than that of the DNA fragment containing either of the two original binding sites (ΔA or ΔB; single closed triangle). The DNA fragment containing no original binding sites (ΔAΔB) is not bound by AdpA-H. (C) Schematic representation of structures of the region upstream of the sgmA promoter on the chromosome. For example, mutant ΔB contains the intact A site and the mutated B site. wt, wild type. (D) Transcription of sgmA in the wild type and mutants ΔB, ΔA, and ΔAΔB, as determined by low-resolution S1 nuclease mapping. The 32P-labeled probe was the same as that used for Fig. 4. The ratios of the amounts of sgmA transcripts relative to those of hrdB transcripts at days 1 and day 2 in mutant ΔB were similar to those in the wild type, although no enhancement of sgmA transcription at day 3 was observed for ΔB. Almost no transcription of sgmA in mutants ΔA and ΔAΔB was detected.
We determined the transcription of the sgmA promoter in mutants ΔA, ΔB, and ΔAΔB (Fig. 6D). S1 nuclease mapping showed that almost no transcription occurred in mutants ΔA and ΔAΔB, indicating that the binding of AdpA to the A site is essential for the transcription of sgmA. This finding is in agreement with the above-described observation that no transcription of sgmA occurred in the adpA disruptant. On the other hand, sgmA was transcribed at days 1 and 2 in mutant ΔB to a similar or only a slightly reduced extent compared with sgmA transcription in the wild type. However, no enhancement of sgmA transcription at day 3 was observed in this mutant. The B site is therefore required for the marked transcriptional activation at day 3.
The mechanism by which the binding of AdpA to both the A and the B sites markedly enhances sgmA promoter activity remains to be elucidated. For adsA, encoding an extracytoplasmic function sigma, AdpA binds a single site, at nucleotide positions +7 to +46, with respect to its transcriptional start point, for transcriptional activation. The A site essential for the switching on of sgmA transcription is comparable to this binding site, in that both are essential for transcriptional activation by AdpA. The AdpA-binding position with respect to the −35 and −10 sequences seems to be unimportant for AdpA to activate transcription. AdpA is believed to recruit RNA polymerase to the promoters to be activated. When sgmA and adsA were compared, the transcription of the former was markedly and abruptly increased at day 3. Since the B site for sgmA is necessary for this marked enhancement, we speculate that an unknown repressor able to bind at or near the B site exists until day 2. This putative repressor would disappear or lose its activity at day 3, thus allowing AdpA to bind the B site, resulting in the full activation of sgmA transcription at day 3. This hypothesis is in agreement with the observation that the marked enhancement at day 3 also occurs in the wild-type strain, where AdpA is overexpressed (Fig. 4). Further examinations are necessary to test this hypothesis.
Phenotypes of sgmA disruptants.
To determine a possible role for sgmA in secondary metabolism and morphological development, we generated sgmA disruptants by replacing most of sgmA with the kanamycin resistance gene (Fig. 7A). The correct gene disruption was confirmed by Southern hybridization with appropriate probes (Fig. 7B). We independently isolated two sgmA disruptants. The disruptants produced streptomycin and a yellow pigment and formed aerial mycelium and spores on various media containing different carbon sources. However, careful examination of the time course of morphological development showed that the timing of aerial mycelium formation by the two independently isolated sgmA disruptants was delayed by about half a day compared to that of the wild type (Fig. 7C). sgmA on a low-copy-number plasmid (plasmid pSGML) in the disruptants caused the host to form aerial mycelium at about 90 h, while mutant ΔsgmA, harboring only vector pKU209, formed aerial mycelium at about 100 h. It seems that pKU209 causes a slight delay in aerial mycelium formation. These findings show that sgmA plays some role in aerial mycelium formation, although it is not essential for growth, secondary metabolism, or morphological development.
Consistent with the idea that sgmA is involved in morphological development, the aerial hypha formation of mutants ΔA and ΔAΔB was also delayed by 10 to 12 h. However, mutant ΔB, in which sgmA is transcribed but with no marked enhancement at day 3, formed aerial hyphae in the same time course as the wild type. This means that the enhancement of sgmA transcription at day 3 is not required for normal development but that the transcription induced by AdpA at days 1 and 2 is necessary. Overexpression of sgmA did not apparently affect the timing of aerial mycelium formation; the wild-type strain harboring pIJ702-sgmA developed aerial hyphae at the same time as the wild-type strain harboring pIJ702-d.
We also examined the timing of streptomycin production by using a bioassay with Bacillus subtilis as an indicator. No apparent difference between the wild type and mutant ΔsgmA could be detected. The sgmA disruptants produced a yellow pigment, whose production was under the control of A-factor, in the same time course as the wild type. This result implies that sgmA disruption has no effects, if any, on secondary metabolism.
Possible role of sgmA in aerial mycelium formation.
Most cells in substrate mycelium die during aerial mycelium formation, because the aerial mycelium reuses material first assimilated into the substrate mycelium (20, 31). A detailed morphological study on the colony development of Streptomyces antibioticus (5, 21) has shown that, in substrate mycelium, the nucleoid structure is disorganized, followed by progressive degradation of cytoplasmic contents and distortion of the hyphal shape. In addition, hyphal death occurs at a specific region and time; this process is known as programmed cell death. It is therefore conceivable that many hydrolytic enzymes, such as proteases, nucleases, and lipases, required for the degradation of cytoplasmic contents are produced at a specific time at the beginning of aerial mycelium formation. Since A-factor switches on aerial mycelium formation, it is also reasonable to assume that A-factor switches on the expression of these hydrolytic enzyme genes to ensure ordered development. As proposed for nucleases and proteases (23), we assume that the extracellular location of the hydrolytic enzymes would permit them to gain access to the cellular contents after the lysis of substrate hyphae. Although SgmA is not a major protease in the extracellular fraction of S. griseus (30), SgmA is presumably a protease of the hydrolytic enzymes for supplying aerial hyphae with amino acids. It is also likely that SgmA plays a role in processing some of the hydrolytic enzymes.
Our unpublished results have revealed that several protease genes, in addition to sgmA, are also activated by AdpA. These proteases are produced in a pre-pro form. Almost all of these proteases perhaps degrade proteins in substrate hyphae at a programmed time. This may be because aerial mycelium formation by mutant ΔsgmA is delayed by only half a day. The proteases that cleave the pre-pro and mature-pro junctions in the sgmA primary product are unclear, although SgmA itself may be responsible for these cleavages. Some of the proteases, including SgmA, produced in response to A-factor also may be involved in the cleavage of hydrolytic enzymes to yield their mature forms.
Acknowledgments
H. Yamazaki was supported by the Japan Society for the Promotion of Science. This work was supported by the Asahi Glass Foundation and by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-02-VI-2-4).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localisation of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. [DOI] [PubMed] [Google Scholar]
- 3.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 4.Chater, K. F., and R. Losick. 1997. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives, p. 149-182. In J. A. Shapiro and M. Dworkin (ed.), Bacteria as multicellular organisms. Oxford University Press, New York, N.Y.
- 5.Fernandez, M., and J. Sänchez. 2002. Nuclease activities and cell death processes associated with the development of surface cultures of Streptomyces antibioticus ETH7451. Microbiology 148:405-412. [DOI] [PubMed] [Google Scholar]
- 6.Hara, O., S. Horinouchi, T. Uozumi, and T. Beppu. 1983. Genetic analysis of A-factor synthesis in Streptomyces coelicolor A3(2) and Streptomyces griseus. J. Gen. Microbiol. 129:2939-2944. [DOI] [PubMed] [Google Scholar]
- 7.Hardisson, C., and M. B. Manzanal. 1976. Ultrastructural studies of sporulation in Streptomyces. J. Bacteriol. 127:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 9.Horinouchi, S. 1996. Streptomyces genes involved in aerial mycelium formation. FEMS Microbiol. Lett. 141:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Horinouchi, S., and T. Beppu. 1992. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 46:377-398. [DOI] [PubMed] [Google Scholar]
- 11.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 12.Horinouchi, S., H. Onaka, H. Yamazaki, S. Kameyama, and Y. Ohnishi. 2000. Isolation of DNA fragments bound by transcriptional factors, AdpA and ArpA, in the A-factor regulatory cascade. Actinomycetologica 14:37-42. [Google Scholar]
- 13.Ishikawa, J., and K. Hotta. 1999. Frame Plot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 14.Kakinuma, S., Y. Takada, H. Ikeda, H. Tanaka, and S. Omura. 1991. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester producers and identification of genes for kalafungin biosynthesis of the kalafungin producer. J. Antibiot. 44:995-1005. [DOI] [PubMed] [Google Scholar]
- 15.Katz, E., C. J. Thompson, and D. A. Hopwood. 1983. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J. Gen. Microbiol. 129:2703-2714. [DOI] [PubMed] [Google Scholar]
- 16.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima, S., T. Kumazaki, S. Ishii, and K. Miura. 1998. Primary structure of Streptomyces griseus metalloendopeptidase II. Biosci. Biotechnol. Biochem. 62:1392-1398. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 20.Méndez, C., A. F. Brana, M. B. Manzanal, and C. Hardisson. 1985. Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol. 31:446-450. [DOI] [PubMed] [Google Scholar]
- 21.Miguëlez, E., C. Hardisson, and M. B. Manzanal. 1999. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J. Cell Biol. 145:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakai, R., S. Horinouchi, and T. Beppu. 1988. Cloning and nucleotide sequence of a cellulase gene, casA, from an alkalophilic Streptomyces strain. Gene 65:229-238. [DOI] [PubMed] [Google Scholar]
- 23.Nicieza, R. G., J. Huergo, B. A. Connolly, and J. Sanchez. 1999. Purification, characterization, and role of nucleases and serine proteases in Streptomyces differentiation. J. Biol. Chem. 274:20366-20375. [DOI] [PubMed] [Google Scholar]
- 24.Oh, S. H., and K. F. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 26.O'Neill, G., S. H. Goh, R. A. Warren, D. G. Kilburn, and R. C. Miller, Jr. 1986. Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene 44:325-330. [DOI] [PubMed] [Google Scholar]
- 27.Robbins, P. W., R. B. Trimble, D. F. Wirth, C. Hering, F. Maley, G. F. Maley, R. Das, B. W. Gibson, N. Royal, and K. Biemann. 1984. Primary structure of the Streptomyces enzyme endo-β-N-acetylglucosaminidase H. J. Biol. Chem. 259:7577-7583. [PubMed] [Google Scholar]
- 28.Shinkawa, H., Y. Hatada, M. Okada, H. Kinashi, and O. Nimi. 1995. Nucleotide sequence of a principal sigma factor gene (hrdB) of Streptomyces griseus. J. Biochem. 118:494-499. [DOI] [PubMed] [Google Scholar]
- 29.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuyuki, H., K. Kajiwara, A. Fujita, T. Kumazaki, and S. Ishii. 1991. Purification and characterization of Streptomyces griseus metalloendopeptidases I and II. J. Biochem. 110:339-344. [DOI] [PubMed] [Google Scholar]
- 31.Wildermuth, H. 1970. Development and organization of the aerial mycelium in Streptomyces coelicolor. J. Gen. Microbiol. 60:43-50. [DOI] [PubMed] [Google Scholar]
- 32.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]