Abstract
The Caulobacter crescentus sensor kinase DivJ is required for an early cell division step and localizes at the base of the newly formed stalk during the G1-to-S-phase transition when the protein is synthesized. To identify sequences within DivJ that are required for polar localization, we examined the ability of mutagenized DivJ sequences to direct localization of the green fluorescent protein. The effects of overlapping C-terminal deletions of DivJ established that the N-terminal 326 residues, which do not contain the kinase catalytic domain, are sufficient for polar localization of the fusion protein. Internal deletions mapped a shorter sequence between residues 251 and 312 of the cytoplasmic linker that are required for efficient localization of this sensor kinase. PleC kinase mutants, which are blocked in the swarmer-to-stalked-cell transition and form flagellated, nonmotile cells, also fail to localize DivJ. To dissect the cellular factors involved in establishing subcellular polarity, we have examined DivJ localization in a pleC mutant suppressed by the sokA301 allele of ctrA and in a pleD mutant, both of which display a supermotile, stalkless phenotype. The observation that these Mot+ strains localize DivJ to a single cell pole indicate that localization may be closely coupled to the gain of motility and that normal stalk formation is not required. We have also observed, however, that filamentous parC mutant cells, which are defective in DNA segregation and the completion of cell separation, are motile and still fail to localize DivJ to the new cell pole. These results suggest that formation of new sites for DivJ localization depends on events associated with the completion of cell separation as well as the gain of motility. Analysis of PleC and PleD mutants also provides insights into the function of the His-Asp proteins in cell cycle regulation. Thus, the ability of the sokA301 allele of ctrA to bypass the nonmotile phenotype of the pleC null mutation provides evidence that the PleC kinase controls cell motility by initiating a signal transduction pathway regulating activity of the global response regulator CtrA. Analysis of the pleD mutant cell cycle demonstrates that disruption of the swarmer-to-stalked-cell developmental sequence does not affect the asymmetric organization of the Caulobacter cell cycle.
Phosphorelay is a central mechanism of signal transduction in prokaryotic cells. The phosphotransfer proteins originally described consist of two components, a sensor kinase, or histidine protein kinase, and a response regulator (34). Many response regulators are transcription factors whose activities are modulated in response to phosphorylation. These His-Asp proteins are also organized in multicomponent phosphorelays containing a histidine kinase (H1), a response regulator (D1), a histidine phosphotransferase (H2), and a second response regulator (D2) in which phosphate transfer to the conserved His and Asp residues occurs in the sequence H1 → D1 → H2 → D2 (3).
Histidine kinase-mediated signal transduction is important in the control of cellular responses to changes in growth conditions, stress, osmolarity, cell density, and other environmental conditions (15). In the dimorphic bacterium Caulobacter crescentus, His-Asp proteins also respond to internal cell cycle cues and are essential for the regulation of cell cycle progression and polar morphogenesis. Because asymmetric cell division generates two different cell types in C. crescentus, the mother stalked cell and the new swarmer cell, these signal transduction pathways must regulate developmental events spatially as well as temporally (28, 40). The stalked and swarmer cells inherit distinctive developmental programs. During the stalked-cell cycle, the structures required to build a new swarmer cell, including the flagellum and bacteriophage receptor sites, are assembled at the stalk-distal pole. Immediately before cell division, the cell gains motility with the start of flagellum rotation. After cell separation is completed, the old stalked cell continues to divide asymmetrically while the new swarmer cell undergoes pilus assembly at the base of the flagellum, loss of motility, flagellum ejection, and stalk formation during its differentiation into a stalked cell (see below and Fig. 1A).
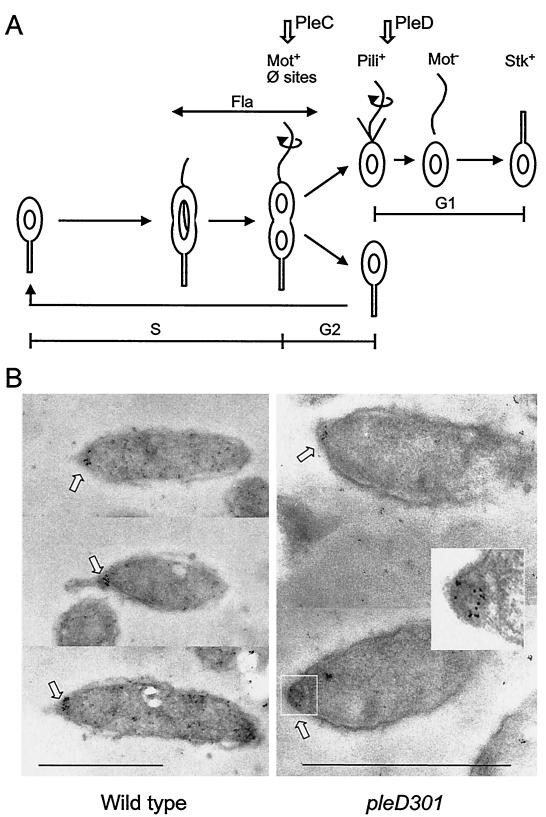
FIG. 1.
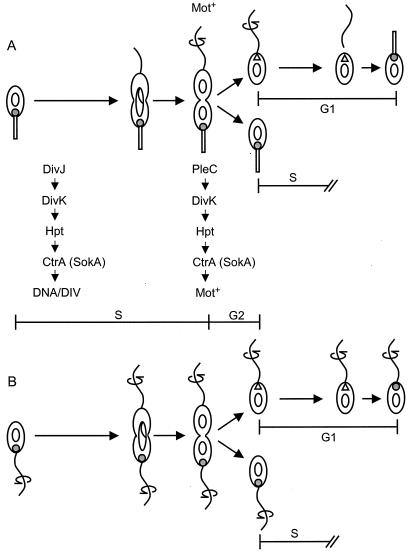
C. crescentus cell cycle and polar localization of the DivJ kinase. (A) Sequence of developmental events in the cell cycle. The vertical arrows at the top of figure indicate the points at which the PleC and PleD functions are required in the sequence of morphological events. (B) Photomicrographs showing immunogold localization of the DivJ protein in thin sections of cells of parent strain CB15 (15-nm-diameter gold particles) and pleD mutant strain PC5375 (5-nm-diameter gold particles). Insert panel shows an enlargement with enhanced contrast of the pleD mutant cell pole in the lower right panel. The bars represent 1 μm.
C. crescentus contains 61 histidine kinases and 43 response regulators (29). Four of these kinases, DivJ, PleC, CckA, and DivL, and the response regulators DivK, CtrA, and PleD have been implicated in aspects of cell cycle regulation. The sensor kinases DivJ, CckA, and DivL have been shown to control the cell division cycle by regulating activity of the global transcription regulator CtrA (reviewed in references 17 and 30). At least in the case of DivJ, the activity of CtrA is controlled by a multicomponent phosphorelay pathway mediated by the essential response regulator DivK (52).
The DivJ, PleC, and CckA kinases and the essential response regulator DivK are differentially localized to the cell poles of C. crescentus cells during cell cycle progression (18, 19, 50). PleC is required for the gain of cell motility (44), and its localization to the flagellated pole of the predivisional cell coincides with the initiation of flagellar rotation and the gain of motility late in the cell cycle (Fig. 1A) (50). PleC is then segregated with the swarmer cell at division and remains at the flagellated pole of the new swarmer cell until the end of the G1 phase. At the G1-to-S-phase transition, transcription (31) and translation (50) of the divJ gene are initiated and the DivJ kinase is localized to the new stalked-cell pole concomitant with the loss of PleC from this cell pole (Fig. 1A) (50). DivJ function is required for an early cell division step in the stalked-cell cycle (14, 31).
In the studies reported here, we have analyzed both the sequences within DivJ and the cellular factors that are required for the polar localization of the DivJ kinase in C. crescentus. The effects of overlapping C-terminal deletions of DivJ identify an N-terminal sequence which does not contain the kinase catalytic domain and which is sufficient for polar localization of the fusion protein. We also present evidence that a short cytoplasmic linker between the transmembrane region and catalytic domain is required for efficient polar localization. An examination of mutants defective in polar morphogenesis and cell division progression indicate that developmental events associated with gain of motility and cell separation are required to establish the correct subcellular address for polar localization.
MATERIALS AND METHODS
Strains and culture conditions.
C. crescentus strains were all derived from strain CB15 (ATCC 19089). A complete list of strains appears in Table 1. The divJ null mutation (divJ356::Ω) was constructed by deleting approximately two-thirds of the gene, including the H and N domains, and inserting the Ω interposon (Spcr) (37). The pleC ctrA double-mutant strain PC3257 was constructed by generalized transduction of the ctrA mutant strain PC3247, containing the ctrA allele sokA301 (52), with a pleC376::aacC1 allele (6). Synchronous cultures were grown in M2 minimal salt medium containing 0.2% glucose (20). All other experimental cultures were grown in peptone-yeast extract (PYE) medium (36). Unless specified otherwise, cultures were grown at 30°C. Spectinomycin, gentamicin, and tetracycline antibiotics were added where appropriate at 50, 10, and 2 μg ml−1, respectively. Viability in this study was defined as the number of CFU per unit of optical density at 650 nm (OD650) on PYE plates at 30°C relative to that of the parent strain CB15.
TABLE 1.
Strains and plasmids used in this study
| Strain | Genotype or description | Reference, source, and/or constructiona |
|---|---|---|
| CB15 | Parent strain | ATCC 19089 |
| CB15F | Synchronizable CB15 strain | 11 |
| PC3017 | divJ356::Ω (Spcr) | This study |
| PC3247 | sokA301 (ctrA allele) | 52 |
| PC3257 | pleC376::aacC1 sokA301(Ts) | This study |
| PC4695 | pleD302::kan | 14 |
| PC5225 | pleC301::Tn5 | 44 |
| PC5375 | pleD301F | 44 |
| PC9808 | CB15/pTR100 | CB15 × pTR100 |
| PC9809 | divJ::Ω/pTR100 | PC3017 × pTR100 |
| PC9810 | divJ::Ω/pdivJ-gfp (complete DivJ ORFd) | PC3017 × pMZ2b |
| PC9811 | divJ::Ω/pdivJ1-612-gfp (DivJ1-204) | PC3017 × pMZ5 |
| PC9812 | divJ::Ω/pdivJ1-978-gfp (DivJ1-326) | PC3017 × pMZ68 |
| PC9813 | divJ::Ω/pdivJ1-735-gfp (DivJ1-245) | PC3017 × pMZ71 |
| PC9814 | divJ::Ω/pdivJ1-1671-gfp (DivJ1-557) | PC3017 × pMZ73 |
| PC9816 | pleC301::Tn5/pdivJ-gfp | PC5225 × pMZ2 |
| PC9817 | ftsA (divE309)(Ts)/pdivJ-gfp | PC3236c × pMZ2 |
| PC9818 | pleD302::kan pdivJ-gfp | PC4695 × pMZ2 |
| PC9819 | ftsI (divA305)(Ts)/pdivJ-gfp | PC7167 (32) × pMZ2 |
| PC9820 | ftsW (divB306)(Ts)/pdivJ-gfp | PC7101 (32) × pMZ2 |
| PC9822 | parC (divF310)(Ts)/pdivJ-gfp | PC8861 (48) × pMZ2 |
| PC9835 | divJ::Ω/pdivJ1-933-gfp (DivJ1-311) | PC3017 × pSAS10b |
| PC9836 | divJ::Ω/pdivJ1-864-gfp (DivJ1-288) | PC3017 × pSAS11 |
| PC9837 | divJ::Ω/pdivJ1-813-gfp (DivJ1-271) | PC3017 × pSAS12 |
| PC9838 | divJ::Ω/pdivJ1-771-gfp (DivJ1-257) | PC3017 × pSAS13 |
| PC9839 | pleD301F pdivJ-gfp | PC5375 × pMZ2 |
| PC9854 | divJ::Ω/pdivJΔ811-870-gfp (DivJΔ271-290) | PC3017 × pSAS21 |
| PC9857 | divJ::Ω/pdivJΔ751-810-gfp (DivJΔ251-270) | PC3017 × pSAS24 |
| PC9858 | divJ::Ω/pdivJΔ691-750-gfp (DivJΔ231-250) | PC3017 × pSAS25 |
| PC9860 | divJ::Ω/pdivJΔ631-690-gfp (DivJΔ211-230) | PC3017 × pSAS27 |
| PC9861 | pleC376::aacC1 sokA301/pdivJ-gfp | PC3257 × pMZ2 |
| PC9867 | divJ::Ω/pdivJΔ871-936-gfp (DivJΔ291-312) | PC3017 × pSAS31 |
| PC9879 | divJ::Ω/pdivJΔ811-936-gfp (DivJΔ271-312) | PC3017 × pSAS37 |
| PC9880 | divJ::Ω/pdivJΔ751-936-gfp (DivJΔ251-312) | PC3017 × pSAS38 |
| PC9881 | divJ::Ω/pdivJΔ751-870-gfp (DivJΔ251-290) | PC3017 × pSAS39 |
| PC9882 | divJ::Ω/pdivJΔ586-645-gfp (DivJΔ196-215) | PC3017 × pSAS40 |
All plasmids were moved by conjugation into recipient strains with the E. coli mobilizing strain pRK2013 (33).
The pMZ and pSAS series of plasmids are derivatives of pTR100 (49). The divJ::gfp fusions cloned in these series of plasmids include 115 bp of DNA upstream of the divJ start codon (see Materials and Methods).
Derivative of PC1049 (32).
ORF, open reading frame.
Cell synchrony and segregation experiments.
Cultures were synchronized (11) by harvesting swarmer cells from exponential cultures via Ludox density gradient centrifugation. Swarmer cells were resuspended in M2 medium at an OD650 of 0.3 and incubated at 30°C. For Western blot analysis, 1-ml aliquots of culture were removed every 10 min and the cells were pelleted by centrifugation, resuspended in 100 μl of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, immediately frozen on dry ice, and stored at −80°C. Progression through the cell cycle was monitored by phase-contrast microscopy. When cell division was >90% complete, the swarmer cells were separated a second time from the stalked cells by Ludox density gradient centrifugation. Purified swarmer cells and stalked cells were pelleted by centrifugation, resuspended in 1× sample buffer, frozen, and stored as before. Samples from the each synchrony were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis with affinity-purified polyclonal anti-DivJ serum at a dilution of 1:10. Equal amounts of cell culture were loaded on the gel for each time point. For samples from the second segregation, equal numbers of cells (expressed as OD) were loaded in the lanes. For flow cytometry experiments, the same synchrony procedures were followed, except for sample removal, which was performed as described below, and for the initial resuspension of purified swarmer cells in M2 medium, which was carried out at an OD650 of 0.1.
Flow cytometry.
Cells were prepared for analysis as follows. At the indicated time points, 90 μl of cells was mixed with 210 μl of 100% ethanol in Eppendorf tubes and stored at 4°C. To stain cells, fixed samples were pelleted, followed by resuspension in a solution of 1 μM SYTOX green nucleic acid stain (Molecular Probes, Eugene, Oreg.) in sterile Tris-buffered saline (20 mM Tris-HCl [pH 7.5], 150 mM NaCl) containing 0.2 mg of RNase ml−1. Samples were then incubated at 4°C overnight. The following day, samples were analyzed with the FACSort flow cytometer (BD Biosciences).
Plasmid constructions.
Escherichia coli strain DH5α [F− endA1 hsdR17(rK− mK+) supE44 thi-1 λ− recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 φ80dlacZΔM15; Bethesda Research Laboratories] was used for cloning recombinant DNA. The plasmid used to promote transfer in conjugations was pRK2013 (33). The various C-terminal deletion fusions of divJ to gfp were constructed as follows. Each divJ fragment was amplified by PCR with primers that generated an XbaI site 115 bp upstream of the divJ start codon and a BamHI site at the 3′ end of the divJ fragment. Similarly, PCR was used to place a BamHI site immediately 5′ of the gfp start codon (green fluorescent protein [GFP] Mut2) (7) and a HindIII site 3 bp after the stop codon of the gfp open reading frame. The gfp PCR product was then digested and ligated between the BamHI and HindIII sites of pBluescript II (Stratagene, La Jolla, Calif.), creating pSAS20. Subsequently, XbaI- and BamHI-digested divJ PCR fragments were ligated between the XbaI and BamHI sites of pSAS20 to generate divJ::gfp fusions with a glycine-serine linker resulting from the artificial BamHI site. The various divJ::gfp fusions were moved as XbaI-HindIII fragments from the pBluescript derivative vectors into the shuttle plasmid pTR100 (49) between the XbaI and HindIII polylinker sites present in that vector. Internal deletions in divJ were made by overlapping PCRs of the individual divJ fragments flanking the deleted sequence (54). The resulting internal divJ deletions contained flanking 5′ XbaI and 3′ BamHI sites that were used to make the subsequent fusions of internally deleted divJ to gfp as described above. All fusions and deletion constructs were verified by sequencing and Western blot analysis. PCR primer sequences used in the constructions are available upon request. All PCRs were carried out with Pfu polymerase (Stratagene).
Fluorescence microscopy.
Cells were prepared for light microscopy as follows. Twenty-five microliters of cell culture was adsorbed to polylysine-coated coverslips for 3 min. The coverslips were then placed onto a 25-μl pool of 0.1 mg of FM4-64 (Molecular Probes) ml−1 for 3 min. FM4-64 was used to visualize cell membranes and stalks (35). The coverslips were then sealed to microscope slides, and cells were visualized by using a Zeiss LSM 510 confocal microscope. Images were acquired with Zeiss LSM software. Adobe Photoshop version 5.0 software was used to prepare the images shown in the figures. The frequencies of DivJ-GFP localization to the stalk poles shown in Fig. 2 and 3 and Table 2 were determined by analysis of ≥200 exponential-phase cells for each strain. Only cells with clearly identifiable stalks were scored for the GFP signal. In all of the strains studied here, cells without a GFP signal localized at the cell pole displayed very faint whole-cell fluorescence.
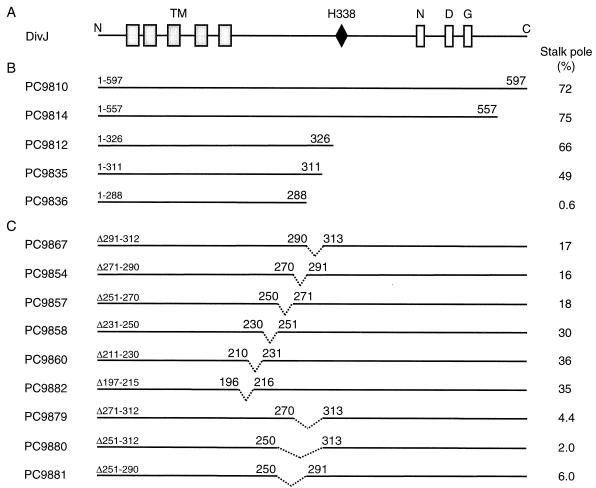
FIG. 2.
Organization of DivJ protein sequence and location of C-terminal deletions. (A) DivJ sequence and conserved histidine kinase sequence motifs. TM, transmembrane regions. (B) C-terminal deletions in DivJ. (C) Internal deletions in the cytoplasmic domain of DivJ. The efficiency of localization as measured by the percentage of stalked cells observed with stalk pole-localized DivJ-GFP is indicated in panels B and C.
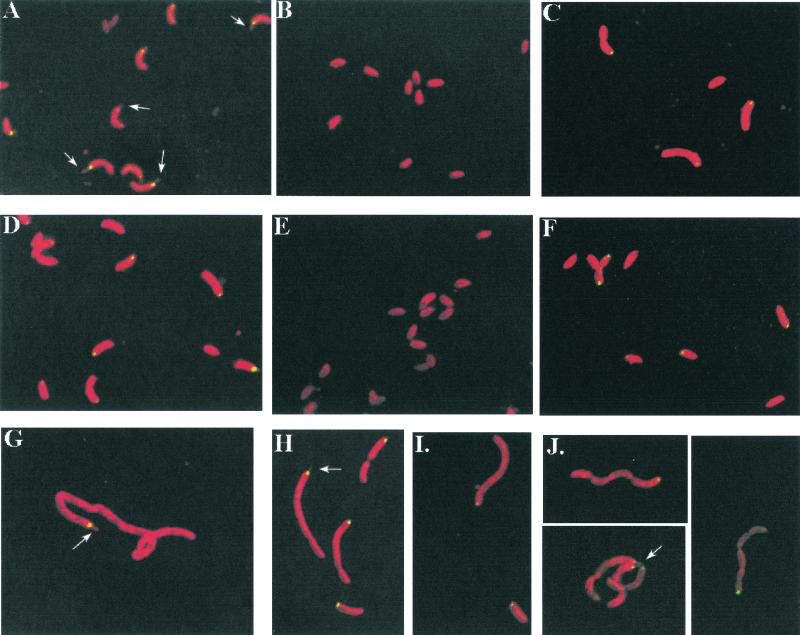
FIG. 3.
Localization of DivJ-GFP fusion proteins in various C. crescentus strains. (A) Strain PC9810, full-length DivJ-GFP in a divJ::Ω background; (B) PC9816, DivJ-GFP in a pleC mutant; (C) PC9861, full-length DivJ-GFP in a pleC sokA double mutant; (D) PC9818, full-length DivJ-GFP in a pleD divJ::Ω background; (E) PC9839, full-length DivJ-GFP in a synchronizable pleD divJ::Ω background, high-density fraction; (F) PC9839, low-density fraction; (G) PC9817, full-length DivJ-GFP in a ftsA(Ts) background; (H) PC9819, full-length DivJ-GFP in a ftsI(Ts) background; (I) PC9820, full-length DivJ-GFP in a ftsW(Ts) background; (J) PC9822, full-length DivJ-GFP in a parC(Ts) background. Cells in panels A through D and G through J were photographed during exponential-phase growth. Cells in panels G through J were shifted to 35°C for ∼7 h prior to photography. Cells in panels E and F were isolated from fractions resulting from centrifugation in a Ludox solution, washed, and then resuspended in M2 medium prior to photographing. Arrows point to stalk structures. The stalk pole DivJ-GFP localization observed in the low-density fraction isolated from a Ludox solution was lower than that seen in exponential-phase cultures (compare with Fig. 2B). This difference is probably due to the fact that the low-density fraction contains a mixture of stalked cells and predivisional cells, some of which had divided to give swarmer cell progeny by the time of collection and observation.
TABLE 2.
Effects of deletion mutations in divJ on DivJ-GFP localization, growth, and viabilitya
| Strain | Relevant genotype | DivJ-GFP foci at stalk poles (%)b | Doubling time (min) | Relative viability |
|---|---|---|---|---|
| CB15 | Parent strain | NA | 105 | 1.0 |
| PC9808 | CB15/pTR100 | NA | 118 | 0.71 |
| PC3017 | divJ::Ω | NA | 123 | 0.56 |
| PC9809 | divJ::Ω/pTR100 | NA | 169 | 0.42 |
| PC9810 | divJ::Ω/pdivJ-gfp | 72 | 124 | 0.68 |
| PC9814 | divJ::Ω/pdivJ1-1668-gfp (DivJ1-557) | 75 | 116 | 0.60 |
| PC9812 | divJ::Ω/pdivJ1-975-gfp (DivJ1-326) | 66 | 152 | 0.40 |
| PC9813 | divJ::Ω/pdivJ1-735-gfp (DivJ1-245) | <1 | 168 | 0.40 |
| PC9879 | divJ::Ω/pdivJΔ811-936 (DivJΔ271-312) | 4 | 124 | 0.62 |
| PC9880 | divJ::Ω/pdivJΔ751-936 (DivJΔ251-312) | 2 | 123 | 0.62 |
Strains were grown and plated on PYE medium with antibiotics as appropriate. Results presented are the averages from three to five experiments. For DivJ-GFP scoring, over 200 cells were scored per strain. See Materials and Methods for details.
NA, not applicable.
DivJ antiserum.
High-titer polyclonal antiserum against DivJ was raised in rabbits and affinity purified on an antigen column by the methods of Harlow and Lane (12). Affinity-purified serum recognized a single band on a Western blot of total Caulobacter protein.
Immunoelectron microscopy.
Cells from exponential or synchronized cultures of C. crescentus strain CB15 were harvested by centrifugation and resuspended in a 50:50 mixture of M2 medium and McLean-Nakane PLP (paraformaldehyde lysine periodate) fixative (25) at room temperature. After 30 min, cells were pelleted and resuspended in fresh fixative for a total fixation time of 2 h. Cells were then rinsed two to three times with ice-cold M2 medium and preimbedded in 2% low-melting-temperature agarose. Samples were dehydrated by a graded ethanol series, infiltrated with a 50:50 mix of ethanol and Lowicryl K4M for several hours, and then infiltrated with 100% resin overnight. Resin was polymerized by UV irradiation for 36 to 48 h at −35°C. Sections (95 nm) were cut from the blocks and mounted on 200-mesh Formvar- and carbon-coated nickel grids. Immunolabeling of the sections was carried out by the droplet method in a humidified chamber. Sections were incubated for 1 h at room temperature in blocking buffer (0.1 M phosphate [pH 7 to 7.4], 1% ovalbumin [grade 5; Sigma], 2 to 5% normal goat serum, 0.05% Tween 20). Sections were then incubated with primary antibody (affinity purified) diluted 1:10 in blocking buffer for 3 to 4 h. After washing with blocking buffer, sections were incubated with secondary antibody (15-nm-diameter colloidal gold conjugate; E. Y. Laboratories, San Mateo, Calif.) diluted 1:10 in blocking buffer. After labeling, sections were washed sequentially with blocking buffer, 0.1 M phosphate buffer (pH 7.2), and 0.1 M cacodylate buffer (pH 7.2). Specimens were fixed in 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 10 min and then rinsed in cacodylate buffer (0.1 M, pH 7.2) followed by distilled water. Samples were then treated with 2% potassium ferrocyanide-reduced osmium tetroxide. After rinsing with distilled water, samples were stained with 1% uranyl acetate (aqueous) for 1 to 2 min and finally rinsed with distilled water. Specimens were examined in a JEOL JEM100C transmission electron microscope at an accelerating voltage of 80 kV.
A total of 111 sections with cells containing particle clusters of three or more gold particles were recorded and photographed. In these samples, 84.6% of the gold clusters were unambiguously associated with the stalked-cell pole, 13.5% were associated with a cell pole but the nature of the pole was ambiguous, 0.9% were unambiguously associated with the stalk-distal pole, and 0.9% were associated with the cell membrane at a nonpolar position. Only 5% of the clusters in the cells were not membrane associated, and thus they were not counted. Clusters that were associated with cellular debris in the plastic (Fig. 1B, top) were not counted. Labeled sections were also prepared from synchronized cultures at different points in the cell cycle. When these were embedded, sectioned, and labeled in the same manner as that for the exponential cultures described above, labeled cells were found only in the samples that contained stalked cells. Labeling was not detected in samples containing only swarmer cells.
The same procedure was employed for the examination of DivJ localization in exponential cells of pleD mutant PC5375, which were prepared as described above, except that 5-nm-diameter-colloidal-gold-conjugated secondary antibody was used. Approximately 300 thin sections were examined for clusters of three or more gold particles (see Fig. 1B).
RESULTS
Identification of DivJ sequences required for polar localization.
Experiments using the DivJ kinase fused to the GFP reporter protein have demonstrated that DivJ, which is expressed during the swarmer-to-stalked-cell transition (31, 50), localizes to the base of the stalk at a time coincident with stalk formation in C. crescentus (50). We examined DivJ localization in thin sections of synchronous C. crescentus cells of strain CB15 by immunogold electron microscopy to visualize DivJ in wild-type strain CB15. These results (Fig. 1B and Materials and Methods) confirmed that DivJ is localized to the base of the stalk in stalked cells (50). The relatively high resolution provided by electron microscopy, compared to that by fluorescence microscopy, also demonstrates the extremely tight clustering of DivJ protein at the base of the stalk.
DivJ is a 597-residue protein with an N-terminal sequence containing five predicted transmembrane regions (45) connected by a cytoplasmic linker to a C-terminal catalytic domain, which contains the conserved site of phosphorylation, His-338, and the N, D, and G boxes (Fig. 2A). To identify sequences that are required for DivJ localization, we constructed overlapping C-terminal deletions of the protein (Fig. 2B). The effects of these deletions fused to GFP were examined in divJ null mutant PC3017 for polar localization (Fig. 2) as well as for cell growth and viability (Table 2). Strain PC3017 grew more slowly and displayed a reduced viability compared to that of parent strain CB15 (Table 2). The full-length divJ::gfp fusion strain PC9810 (DivJ1-597) localized DivJ efficiently to the base of the stalk (Fig. 2B and 3). We consistently observed that ca. 70% of the stalked cells in exponential cultures contained a single focus of fluorescence at the base of the stalk (Fig. 2B). The divJ null mutant containing the plasmid vector alone (PC9809) exhibited an increased doubling time and low viability relative to those of an isogenic wild-type strain (PC9808). The full-length DivJ-GFP fusion in strain PC9810 complemented the growth rate and viability defects of the divJ null mutant PC9809 (Table 2), indicating that the DivJ fusion protein is active.
The extent of polar localization of DivJ-GFP in strains PC9814 (DivJ1-557) and PC9812 (DivJ1-326), which carry C-terminal DivJ deletions of 40 and 271 residues, respectively, was similar to that in strain PC9810 (full-length DivJ) (Fig. 2B). These results indicate that the N-terminal 326 amino acids are sufficient for polar localization of the protein. This result is of particular interest because the DivJ1-326 deletion removes the canonical histidine and conserved sequence motifs of the DivJ catalytic domain (Fig. 2B). As expected, this DivJ′-GFP construct did not complement the growth phenotype of the divJ null mutation (Table 2). However, the normal stalked-cell pole localization of the DivJ1-326 protein (Fig. 2B) indicates that the kinase catalytic domain histidine is not required for subcellular localization.
The fraction of cells with localized DivJ was only moderately reduced in the DivJ deletion strain PC9835 (DivJ1-311) (Fig. 2B). Strain PC9836 (DivJ1-288), in which 307 residues of the DivJ sequence were truncated, displayed negligible polar localization (Fig. 2B) and growth and viability defects very similar to those seen in PC9812 (Table 2 and data not shown). Longer C-terminal deletions of DivJ (see strains PC9837, PC9838, PC9813, and PC9811 in Table 1) also displayed extremely low levels of localized DivJ (data not shown) and presumably remove the same sequence(s) required for polar localization. Thus, this set of C-terminal deletions appears to map a sequence required for polar localization to the N-terminal 326 residues that is separable from the catalytic domain of the DivJ kinase. Western blot analysis showed that the DivJ′-GFP fusion protein levels of strains with deletions in DivJ were not reduced compared to those found in strain PC9810 containing full-length DivJ-GFP protein (data not shown).
Internal deletions in the DivJ kinase linker region map a sequence required for DivJ stalk pole localization.
The difference in polar localization between strain PC9812 (DivJ1-326) and strain PC9836 (DivJ1-288) suggested that amino acids 289 to 325 might be required for localization of DivJ (Fig. 2B). To examine this possibility, we constructed a set of internal deletions in DivJ, each of which removed ca. 20 amino acids, and evaluated their effects on the pattern of DivJ localization. Deletion of DivJ residues 291 to 312 (PC9867), 271 to 290 (PC9854), or 251 to 270 (PC9857) reduced DivJ localization to ca. 17%, while the more N-terminal deletions of residues 231 to 250 (PC9858), 211 to 230 (PC9860), or 196 to 215 (PC9882) reduced localization to between 30 and 36% (Fig. 2B).
The lower levels of localization in the first set of three deletions suggested that residues critical for the correct polar localization of DivJ may lie between residues 250 and 312. To confirm this conclusion and to map the extent of the localization sequence in more detail, we constructed larger internal deletions of 40 to 60 amino acids spanning residues 271 to 312 (PC9879), 251 to 312 (PC9880), and 251 to 290 (PC9881). Cells containing any one of these larger DivJ deletions showed very little polar localization (Fig. 2B). These data confirm the conclusion from the examination of the smaller deletions that a sequence between residues 251 and 312 may be critical for polar localization. They also show that deletion of a larger sequence in this region has a more deleterious effect on the localization of DivJ. It should be noted that the internal deletions examined here could alter the conformation of the mutant DivJ sequence in such a way as to interfere with its polar localization (see Discussion).
Role of CtrA in the regulation of PleC-dependent motility and DivJ localization.
Mutations in pleC block developmental steps late in the cell cycle, leading to the gain of motility (see arrow in Fig. 1A) and the swarmer-to-stalked-cell transition in the next cycle (Fig. 1A) (44). The observation that DivJ is not localized at the pole in pleC cells has suggested that polar localization may depend on stalk formation (50). To dissect the cellular factors that are required for polar localization of the DivJ protein, we have examined DivJ localization in two strains that are motile (Mot+) but fail to undergo normal stalk formation (Stk−). The first of these was a pleC strain in which the Mot− phenotype is suppressed by the sokA301 allele of ctrA.
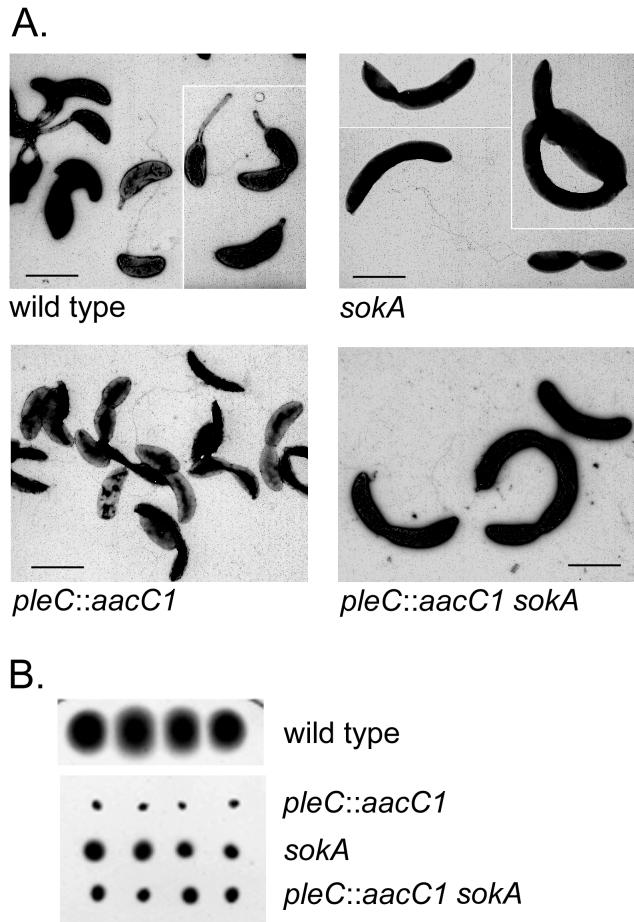
The DivJ kinase regulates cell division via a phosphorelay (DivJ → DivK → Hpt → CtrA) that controls the activity of CtrA, which is an essential response regulator in C. crescentus (39). Evidence for this pathway was originally obtained by the isolation of sokA (suppressor of divK) suppressor mutations in ctrA that bypass cell division defects of divJ and divK mutations (52). The complex epistatic relationship between the divJ and pleC mutations in the control of both motility and cell division (6, 42) suggests that PleC may also regulate CtrA activity. To test this possibility, we examined the ability of the sokA301 allele of ctrA to bypass the Mot− phenotype of a pleC disruption mutation. As shown in Fig. 4B, the sokA301 mutation suppresses the Mot− swarm phenotype of the pleC::aacC1 null mutation. Light microscopy confirmed that the pleC::aacC1 sokA301 double mutant is motile and that, as judged by the disproportionately large number of Mot+ cells, it displays a supermotile phenotype similar to that of the sokA301 strain (52). A partial cell division defect of the temperature-sensitive sokA strain at 30°C (53) explains the slightly larger cell size of sokA301 strains (Fig. 4A). This larger cell size may account for the small swarm size of the pleC::aacC1 sokA301 strain in motility agar compared to that of the wild-type strain (Fig. 4B) (6).
FIG. 4.
CtrA is a target of the PleC kinase in the regulation of cell motility. (A) Electron micrographs of negatively stained cells (52) from the strains indicated. Bar, 1 μm. (B) Suppression of the Mot− phenotype of pleC by the sokA301 mutation as assayed in motility agar.
These motility results and electron microscopy demonstrate that (i) the sokA suppressor mutation bypasses the Mot− defect of the pleC null mutation but that the suppressed supermotile cells do not form stalks (Fig. 4A) and (ii) the pleC::aacC1 sokA301 cells display the same supermotile phenotype as the sokA301 cells (Fig. 4B), indicating that the sokA allele is epistatic to the pleC motility phenotype. We also conclude from these results that the PleC kinase regulates motility directly or indirectly by controlling the activity of the global transcription regulator CtrA (see Fig. 7 and Discussion).
FIG. 7.
Model for the regulation of DivJ localization and asymmetric DNA initiation in wild-type C. crescentus and pleD mutant cells. (A) Strain CB15 cell cycle. DivJ regulates a phosphorelay pathway controlling CtrA activity early in the stalked-cell cycle, as shown (52). Localization of DivJ at the stalked-cell pole is dependent on events occurring late in the cell cycle, presumably at the new cell pole (▵). Subsequent developmental events at this pole during the G1-to-S-phase transition make the pole competent as a target of DivJ localization (see Discussion). Motility in the predivisional cell requires PleC (44), which is located in the swarmer-cell compartment (50), and initiates a signal transduction pathway (PleC → DivK → Hpt → CtrA) controlling CtrA activity and the gain of motility, as shown in Fig. 4. (B) Asymmetric control of DNA replication in the pleD mutant cell cycle. The localization of DivJ uniquely to one cell pole of the old, S-phase progeny cell at division (Fig. 6) indicates that DivJ localization is regulated by asymmetrically distributed factors, as described above for CB15.
Examination of a DivJ fusion protein in the pleC::Tn5 sokA301 strain PC9861, which had very few detectable stalks, showed that 26% of the cells contained DivJ-GFP localized at the pole and that the GFP signal was localized to only one pole in single and predivisional cells (Fig. 3C). This result suggests that normal stalk formation is not required for polar localization of the DivJ kinase and that the failure of DivJ to localize in pleC mutants may result instead from the inability of these cells to complete the developmental program leading to gain of motility (Fig. 1A).
Localization of DivJ in pleD mutants.
The second Mot+ Stk− strain that we examined for DivJ localization was a pleD mutant. pleD mutants are motile but fail to turn off motility, eject the flagellum, or undergo normal stalk formation (1a, 14, 44). As a result, pleD mutants form two motile flagellated cells at division and thus appear to be blocked after the PleC-dependent stage of polar morphogenesis (see arrow in Fig. 1A) (13). We initially examined these cells for DivJ localization by immunogold electron microscopy. Unlike the pleC::Tn5 mutant, in which no clusters of gold particle were detected (data not shown), clusters of gold particles were located in longitudinally sectioned pleD mutant cells. Although only ca. 5% of the sections contained gold clusters, 90% of the clusters were associated at a cell pole that usually had a nub or protrusion, which we interpreted to be a site of incipient stalk formation (Fig. 1B). It was not possible to conclude from these data whether the gold particles were localized to only one of the two cell poles (see below).
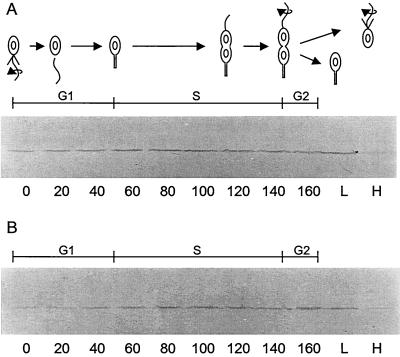
As another approach to studying DivJ localization in the pleD strain, we used Western blot analysis of synchronous cells to examine DivJ synthesis during the cell cycle and segregation at division. The high-density cell fraction of pleD301 strain PC5375 isolated from a Ludox gradient (Materials and Methods) has been shown to contain a synchronous population of motile swarmer cells (44). In our experiments, these cells initially contained a very low level of the DivJ kinase on Western blots and the levels increased as the cells progressed through the cell cycle (Fig. 5B). This pattern of DivJ accumulation is similar to that in the parent CB15F strain (Fig. 5A) (50) and reflects the periodic transcription and translation of divJ that occurs during the G1-to-S-phase transition in wild-type cells (31, 50). When the high- and low-density cell fractions were isolated at the end of the this synchronous cell division cycle, most of the DivJ protein fractionated with the low-density cells of the parent CB15 strain (Fig. 5A) and the pleD mutant strain (Fig. 5B). These results suggest that DivJ localizes to only one pole of predivisional pleD cells and then segregates at division to the lower-density population of daughter cells.
FIG. 5.
Levels of DivJ detected on Western blots of synchronously dividing cells. (A) Wild-type strain CB15F; (B) pleD301F strain PC5375. Cell division, which occurred at 160 min in each culture, was monitored by phase-contrast microscopy. H and L in panel A and panel B correspond to cells isolated from the high- and low-density fractions, respectively, after centrifugation on Ludox gradients (see Materials and Methods).
The asymmetric localization of DivJ in the pleD mutant is supported by two additional observations. First, exponential cultures of cells expressing the full-length DivJ-GFP fusion protein displayed fluorescence at only one pole of single and dividing cells (Fig. 3D). Second, we examined high- and low-density cells of pleD mutant strain PC9839 isolated from a synchronously dividing strain culture for DivJ localization. Cells in both density fractions were motile, and, as expected from the results shown in Fig. 5B, only the low-density cells showed significant polar localization of DivJ-GFP (49% of cells in the low-density fraction [Fig. 3F] versus <1% of cells in the high-density fraction [Fig. 3E]). These results and those presented above on the pleC sokA301 mutant (Fig. 2 and 3B and C) support the conclusion that normal stalk formation is not required for DivJ localization.
Cell division in pleD cells is asymmetric.
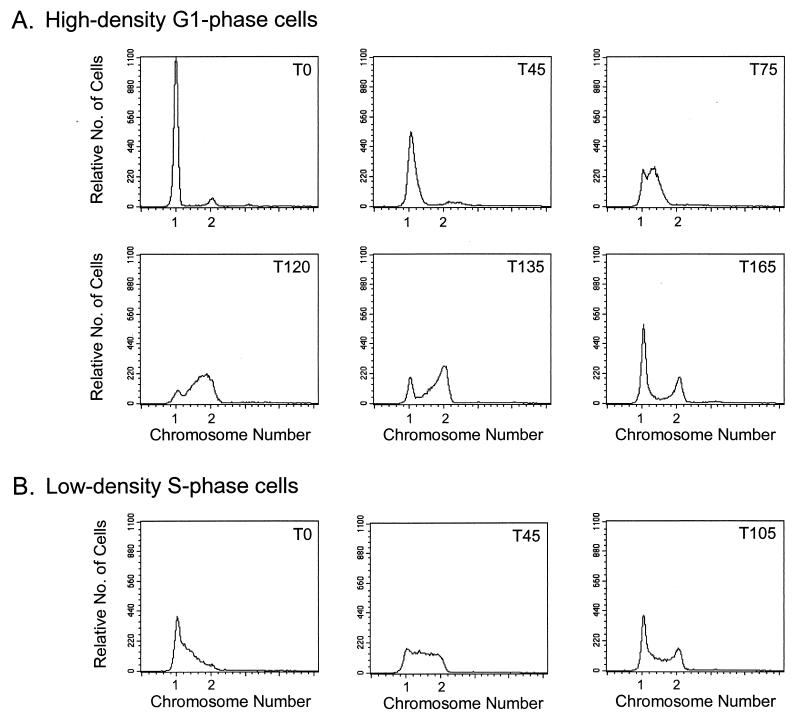
Despite the symmetrical morphology of predivisional pleD cells, the differential polar localization of DivJ within these cells indicates that the two daughter cells are developmentally distinct (Fig. 3B and 5B). To determine if pleD cells are at different stages in the DNA synthetic and cell division cycles, we performed fluorescence-activated cell sorter analysis on high- and low-density cells isolated from a synchronously dividing pleD culture. The high-density cells, in which little or no DivJ-GFP fusion protein could be initially detected, underwent a G1 period of 45 to 60 min that was followed by initiation of DNA synthesis between time points T45 and T75 (in minutes) (Fig. 6A). Consistent with this identification of the high-density cells as G1-phase cells, DivJ localization increased from <1% at 0 min to 8% at 20 min to 17% at 40 min and to 30% at 60 min as the cells were incubated at 30°C. (Note that DNA initiation and stalk formation occur at ca. 60 min in wild-type cells.)
FIG. 6.
Cell cycle analysis of the synchronous pleD mutant strain PC9839. DNA content in synchronous cells was determined by flow cytometry at various times after collection from Ludox gradients and resuspension in fresh medium. (A) Cells from the high-density fraction display a G1 arrest in DNA replication (T0 and T45). The G1 period was followed by entry into S phase (T75) and then G2 phase (T120 and 135). By T165, many of the cells had divided as indicated by the large 1n peak in the corresponding panel. (B) Cells from the high-density fraction were allowed to grow to the midpoint of division, as approximated by phase-contrast microscopy, and then fractionated a second time. Cells collected from the low-density cell fraction (T0) were predominantly in S phase, as were those at T45. S phase was followed by entrance into G2 phase, which occurred between T75 (data not shown) and T105. By T105, a large portion of the cells had divided, as indicated by the large 1n peak in the corresponding panel. T, time (minutes) after resuspension. See Materials and Methods for details of cell fractionation and analysis.
By contrast, a substantial number of the lower-density, S-phase cells had initiated DNA replication at the time of collection (time point T0 in Fig. 6B). These cells, in which DivJ was localized to one pole (Fig. 5B), displayed an S-G2-phase cell cycle typical of wild-type stalked cells of the same density (see legend to Fig. 6B). Consistent with the conclusion that the high-density progeny represent G1-phase cells (Fig. 6A) and low-density cells represent S-phase cells (Fig. 6B), we observed that the two cell populations divided at 165 and 105 min, respectively. Taken together, these results indicate that progeny pleD cells are developmentally distinct and follow asymmetric patterns of DNA initiation and cell division. As depicted in Fig. 7, this pattern of cell cycle regulation is characteristic of wild-type C. crescentus cells, despite the inability of the pleD mutant to complete the development transition required for swarmer-to-stalked-cell differentiation (9).
Requirement of a cell division checkpoint for pole localization of DivJ.
Polar morphogenesis in C. crescentus is coupled to successive stages of cell cycle progression. Among these regulated polar events are flagellum biosynthesis, which requires DNA replication; gain of motility and stalk formation, which require completion of a cell division progression (DIVp); and pilus formation, which requires completion of cell separation (reviewed in reference 30). This dependence of polar morphogenesis on cell cycle progression provides a potentially useful way to determine if normal passage through early, middle, and late cell division cycle checkpoints is necessary for correct polar localization of DivJ. We have examined strains with temperature-sensitive mutations in three genes (ftsA, ftsI, and ftsW) required for cell division progression (32). Early exponential cultures of these strains containing the divJ::gfp fusion were shifted from 30 to 35°C and examined after 7 h at the nonpermissive temperature. When cell cycle progression is blocked, only the newly divided swarmer cells undergo stalk formation. In stalked cells, subsequent development at the stalk-distal cell pole is arrested after flagellum biosynthesis, and consequently, the cells do not gain motility or form a second stalk (16). We visualized a focus of fluorescence at only one pole in filamentous cells of the ftsA (Fig. 3G), ftsW (Fig. 3H), and ftsI (Fig. 3I) strains. DivJ-GFP was localized at the base of the stalk in all filamentous cells in which a stalk was visualized. The failure to observe multiple sites of localization suggests that the stalk-distal pole is not competent for DivJ localization and that completion of a late step in cell division is necessary for polar localization of this sensor kinase.
We also examined the temperature-sensitive parC mutant PC9822 for localization of DivJ at the nonpermissive temperature in an experiment similar to those described above. This strain, which is defective in the completion of chromosome segregation and cell separation (48), progresses far enough in the cell cycle to synthesize flagella, gain motility, and form a new, second stalk in some cells (16). Because the mutant cells are blocked in the completion of cell division, however, they do not assemble pili, which would normally occur at the flagellated pole after cell separation (43). Only one focus of DivJ localization was visualized in parC filaments (Fig. 3J), indicating that new sites of DivJ localization were not formed after the temperature shift. Assays for DivJ by Western blot analysis demonstrated that accumulation of the fusion protein was not reduced in the division mutants at the nonpermissive temperature compared to that in the CB15 parent strain (data not shown). We conclude from these results that, in addition to the gain of motility, a very late cell cycle event(s) associated with the completion of cell separation is required for DivJ localization (see Discussion).
DISCUSSION
The C. crescentus histidine kinase DivJ is regulated temporally and spatially in the cell cycle (50). We show here that the N-terminal 326 residues of DivJ are both necessary and sufficient for normal spatial regulation of DivJ. The N-terminal 326 residues do not contain the catalytic domains of DivJ, and therefore, the localization and the catalytic functions of DivJ are mediated by separate regions of the protein. Our results also suggest that localization of DivJ is dependent on a cytoplasmic linker region that is located between residues 250 and 312. Additionally, we show that polar localization of DivJ is dependent on cellular events associated with the gain of motility and a late step in cell cycle progression but not on normal stalk formation. The observation that the ctrA allele sokA301 restores motility and DivJ polar localization in pleC mutants also suggests that the PleC kinase regulates motility directly or indirectly by controlling the activity of the global transcription regulator CtrA (Fig. 4 and 7).
Sequence features of DivJ localization sequences.
The C-terminal deletion analysis of the DivJ protein revealed that the highly conserved H, N, D, and G boxes found in the C terminus of histidine kinases were required for normal growth, as expected (Table 2). This same analysis showed that these conserved catalytic sequence motifs are not required for localization of DivJ at the stalk pole. Thus, the DivJ1-326-GFP fusion localized to the stalked-cell pole as frequently as the full-length DivJ-GFP fusion (Fig. 2B). However, a longer C-terminal deletion that leaves 311 N-terminal residues showed a mild impairment (ca. 32% decrease) in DivJ stalked-cell pole localization compared to the full-length DivJ fusion, while a C-terminal deletion leaving 288 N-terminal residues eliminated DivJ localization to the stalked-cell pole (Fig. 2). As considered below, sequences between residues 288 and 326 of the linker region appear to contain information important for the polar localization of DivJ.
Interestingly, none of the ∼20-amino-acid sequences deleted between residues 313 and 250 of DivJ (PC9867, PC9854, and PC9857 [Fig. 2]) diminished stalked-cell pole localization to the extent observed in the C-terminal deletion represented by DivJ1-288. Larger deletions of the residue 251 to 312 sequence, however, drastically reduced the polar localization of DivJ (Fig. 2B). Computer analysis of the DivJ secondary structure predicts a short helix followed by a coil and a long helix between residues 251 and 326 (residues 253 to 265, 266 to 295, and 296 to 326, respectively) (2). Separate computer analyses designed to detect coiled-coil motifs identified putative coiled-coil motifs between residues 296 and 342 (51) and residues 311 and 342 (41). Thus, it is possible that localization of DivJ is sensitive to the presence or absence of these predicted structural motifs in the linker region in a hierarchical fashion. In this way, removing part of the long helix could diminish DivJ localization only partially (DivJ1-311). However, removing a portion of the coil plus the entire long helix (DivJ1-288) or removing the short helix plus the entire coil and half of the long helix (DivJΔ251-312) could eliminate DivJ localization completely. Alternatively, we cannot rule out the possibility that the various mutations have more global effects on the overall structure of the protein. Western blot analysis does indicate, however, that none of the DivJ deletions examined in these studies affect DivJ′-GFP protein stability (data not shown).
The transmembrane region of DivJ (Fig. 2B, DivJ1-288), is not sufficient for polar localization of the protein kinase. A more detailed mutational analysis of the N-terminal DivJ sequence should reveal whether the linker region alone, or the linker region in combination with the membrane region of the protein, is sufficient for DivJ localization. In this regard, it is interesting to note that DivJ differs from many of the membrane-bound proteins studied in E. coli and Bacillus subtilis, where the localization determinant appears to be contained entirely in the transmembrane region (4, 5, 8, 10, 21, 23, 46, 55). In the absence of a transmembrane region, these mutant proteins are either nonfunctional or negligibly functional. Regarding function, the availability of DivJ mutants that do not localize but retain their catalytic function allows us to address the question of how important DivJ localization is to its intracellular function. We have observed that some of the mutant proteins that lack the localization signal, e.g., DivJΔ271-312 (PC9879), seem to complement the growth defect of the divJ null mutation (Table 2). We also note that BLAST analysis (2) has identified a putative signal transduction protein that shares significant homology to the DivJ251-326 region in both Methylobacterium extorquens and Xylella fastidiosa. The spatial localization of these proteins is unknown.
Developmental regulation of DivJ localization.
Although the DivJ protein sequence studied here is required for DivJ localization, it is only one component of the machinery involved in protein localization to the stalked-cell pole. Morphogenetic events potentially required for DivJ localization include motility, flagellum ejection, and stalk biogenesis. In a previous study, it was shown that DivJ fails to localize in a pleC mutant (50). Because pleC mutants do not gain motility and fail to develop stalks, these data suggested that progress through one or both of these developmental events is requisite for DivJ stalked-cell pole localization. In this work, we have dissected these possibilities by examining developmental events, beginning with gain of motility in the late-predivisional cell and culminating in the loss of motility and stalk formation by the new swarmer cell (Fig. 1A). Analysis of pleC sokA301 and pleD strains demonstrated that neither flagellum ejection nor normal stalk formation is needed for DivJ localization (Fig. 2 and 4). Rather, these experiments suggest that DivJ localization is closely associated with the gain of motility.
We also investigated the role of cell division progression in directing subcellular localization by examining the ftsA, ftsI, and ftsW mutants, which at the nonpermissive temperature form filamentous cells blocked in different stages of cell division. Fluorescence microscopy indicated that DivJ localization occurs at the older, stalked-cell pole of these filamentous cells. The failure of these fts division mutants to develop multiple DivJ-GFP foci suggests that a late cell division step is required for the elaboration of new sites for localization of this histidine kinase. A similar pattern of DivJ localization was observed in parC mutants, which are defective in chromosome partitioning (48). Since these filamentous cells are motile and sometimes form a new second stalk (16), these results indicate that, while gain of motility seems to be required for polar localization of DivJ, it may not be sufficient. We suggest that new polar sites for DivJ localization must also depend in some way on the completion of chromosome segregation and cell separation. As depicted in Fig. 7A, we suggest that this cell cycle-dependent event(s) occurs at the time of cell separation and that subsequent development during the G1-to-S-phase transition makes the new pole competent for the localization of DivJ (see legend to Fig. 7A). This formulation is consistent with the previously described observation (50) that DivJ ectopically expressed in swarmer cells is not sufficient for polar localization of the kinase in these cells.
Other developmental events whose timing implicates them in DivJ localization are localization of the DivK (19) and FtsZ (38) proteins as well as the initiation of DNA replication (Fig. 1A). The response regulator DivK is a soluble protein that is phosphorylated by DivJ (13, 50), and DivK localization depends directly or indirectly on the presence of DivJ (19). Therefore, DivK seems unlikely to be required for DivJ polar localization. Prior to its assembly at midcell, C. crescentus FtsZ has been shown to localize at the stalk pole (38). This event makes FtsZ an intriguing candidate as a recruiter of DivJ based on the observation in E. coli that FtsZ is required for the recruitment of FtsA, which in turn is needed to recruit additional division proteins such as FtsN and FtsW (1, 24, 26, 47). Similarly, in B. subtilis, FtsZ is required to recruit the DivIB and DivIC proteins to the division septum (22). Initiation of DNA replication could also play a role in the localization of DivJ since it has been shown to be required for the polar localization of ParB in Caulobacter (27).
pleD divides asymmetrically.
We have used three separate approaches to show that pleD progeny cells, which are morphologically indistinguishable at division, divide asymmetrically and display different patterns of chromosome replication. Each approach relied on the fact that the two cell types of synchronizable C. crescentus strains can be fractionated by density. Using a synchronizable pleD strain, we isolated high- and low-density cell fractions, resuspended them in growth medium, and then examined cells from each population over time. The cells were analyzed for DNA content, DivJ protein levels, and polar DivJ-GFP localization. The parental strain CB15 divides asymmetrically with the high-density fraction corresponding to swarmer cells that are silent for chromosome replication and undergo a G1 arrest some 45 to 60 min before initiating DNA synthesis. Conversely, the low-density cell fraction contains stalked cells, which actively replicate DNA (S-phase cells). Using flow cytometry, we have shown that the high- and low-density cell fractions of the synchronizable pleD mutant strain exhibit this same pattern of asymmetric DNA replication (Fig. 7). In addition, Western blot analysis showed that, as is the case for the parent strain CB15, the DivJ protein in the pleD mutant differentially segregates to the low-density, S-phase cell fraction (Fig. 5). Examination of DivJ-GFP in the two density fractions by fluorescence microscopy confirmed these results and showed that DivJ was localized to one cell pole (Fig. 3C). As summarized in Fig. 7B, we propose that pleD mutant cells divide asymmetrically based on their asymmetric pattern of chromosome replication and differential localization of the DivJ kinase at cell division. We also conclude that the developmental events accompanying swarmer-to-stalked-cell differentiation in C. crescentus, including stalk biogenesis, are not essential for the asymmetric regulation of DNA initiation observed in C. crescentus progeny cells after division (Fig. 7B).
Acknowledgments
Work from this laboratory was supported by Public Health Service grant GM58794 from the National Institutes of Health.
We are grateful to Michael Zinda for plasmid constructs used in the initial phase of this study and to Joseph Goodhouse for invaluable assistance with electron microscopy and confocal microscopy. We also thank Andrew Beavis for assistance in flow cytometry and Ray Webster for assistance in preparing figures.
REFERENCES
- 1.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 1a.Aldridge, P., and U. Jenal.1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleby, J. L., J. S. Parkinson, and R. B. Bourret. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86:845-848. [DOI] [PubMed] [Google Scholar]
- 4.Arigoni, F., A.-M Guerout-Fleury, I. Barak, and P. Stragier. 1999. The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Mol. Microbiol. 31:1407-1415. [DOI] [PubMed] [Google Scholar]
- 5.Bath, J., L. J. Wu, J. Errington, and J. C. Wang. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995-997. [DOI] [PubMed] [Google Scholar]
- 6.Burton, G., G. B. Hecht, and A. Newton. 1997. Roles of the histidine protein kinase PleC in Caulobacter crescentus motility and chemotaxis. J. Bacteriol. 179:5849-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 8.Dai, K., Y. Xu, and J. Lutkenhaus. 1996. Topological characterization of the essential Escherichia coli cell division protein FtsN. J. Bacteriol. 178:1328-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degnen, S., and A. Newton. 1972. Chromosome replication during development in Caulobacter crescentus. J. Mol. Biol. 64:671-680. [DOI] [PubMed] [Google Scholar]
- 10.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Hecht, G., T. Lane, N. Ohta, J. Sommer, and A. Newton. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecht, G., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer to stalked cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. ASM Press, Washington, D.C.
- 16.Huguenel, E. D., and A. Newton. 1982. Localization of surface structures during procaryotic differentiation: role of cell division in Caulobacter crescentus. Differentiation 21:71-78. [DOI] [PubMed] [Google Scholar]
- 17.Hung, D., H. McAdams, and L. Shapiro. 2000. Regulation of the Caulobacter cell cycle, p. 361-378. In Y. V. Brun and L. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 18.Jacobs, C., I. Domian, J. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111-120. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs, C., D. Hung, and L. Shapiro. 2001. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 98:4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, R., and B. Ely. 1977. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katis, V. L., and R. G. Wake. 1999. Membrane-bound division proteins DivIB and DivIC of Bacillus subtilis function solely through their external domains in both vegetative and sporulation division. J. Bacteriol. 181:2710-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katis, V. L., R. G. Wake, and E. J. Harry. 2000. Septal localization of the membrane-bound division proteins of Bacillus subtilis DivIB and DivIC is codependent only at high temperatures and requires FtsZ. J. Bacteriol. 182:3607-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Londono-Vallejo, J.-A., Frehel, Claude, Stragier, Patrick. 1997. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24:29-39. [DOI] [PubMed] [Google Scholar]
- 24.Ma, X., Q. Sun, R. Wang, G. Singh, E. L. Jonietz, and W. Margolin. 1997. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J. Bacteriol. 179:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLean, W., and P. F. Nakane. 1974. Periodate-lysine-paraformaldehyde fixative. A new fixative for immunoelectron microscopy. J. Histochem. Cytochem. 22:1077-1083. [DOI] [PubMed] [Google Scholar]
- 26.Mercer, K. L. N., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohl, D. A., and J. W. Gober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 28.Newton, A., and N. Ohta. Role of multiple sensor kinases in cell cycle progression and differentiation in Caulobacter crescentus. In M. Inouye (ed.), Histidine kinases in signal transduction, in press. Academic Press, New York, N.Y.
- 29.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potcka, W. C. Nelson, A. Newton, S. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta, N., T. Grebe, and A. Newton. 2000. Signal transduction and cell cycle checkpoints in developmental regulation of Caulobacter, p. 341-359. In Y. Brun and L. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 31.Ohta, N., T. Lane, E. G. Ninfa, J. M. Sommer, and A. Newton. 1992. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc. Natl. Acad. Sci. USA 89:10297-10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohta, N., A. J. Ninfa, A. D. Allaire, L. Kulick, and A. Newton. 1997. Identification, characterization and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J. Bacteriol. 179:2169-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta, N., E. Swanson, B. Ely, and A. Newton. 1984. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J. Bacteriol. 158:897-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 35.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. Perez, Y. L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA marker. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 38.Quardokus, E. M., N. Din, and Y. V. Brun. 2001. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 39:949-959. [DOI] [PubMed] [Google Scholar]
- 39.Quon, K., G. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro, L., and R. Losick. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89-98. [DOI] [PubMed] [Google Scholar]
- 41.Singh, M., B. Berger, P. S. Kim, J. M. Berger, and A. G. Cochran. 1998. Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc. Natl. Acad. Sci. USA 95:2738-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer, J. M., and A. Newton. 1991. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics 129:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer, J. M., and A. Newton. 1988. Sequential regulation of developmental events during polar morphogenesis in Caulobacter crescentus: assembly of pili on swarmer cells requires cell separation. J. Bacteriol. 170:409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer, J. M., and A. Newton. 1989. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J. Bacteriol. 171:392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 46.Wang, L., and J. Lutkenhaus. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29:731-740. [DOI] [PubMed] [Google Scholar]
- 47.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward, D., and A. Newton. 1997. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 26:897-910. [DOI] [PubMed] [Google Scholar]
- 49.Weinstein, M., R. C. Roberts, and D. R. Helinski. 1992. A region of the broad-host-range plasmid RK2 causes stable in planta inheritance of plasmids in Rhizobium meliloti cells isolated from alfalfa root nodules. J. Bacteriol. 174:7486-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wheeler, R. T., and L. Shapiro. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell. 4:683-694. [DOI] [PubMed] [Google Scholar]
- 51.Wolf, E., P. S. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, J., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. USA 95:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, J., N. Ohta, J.-L. Zhao, and A. Newton. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. USA 96:13068-13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yon, J., and M. Fried. 1989. Precise gene fusion by PCR. Nucleic Acids Res. 17:4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, X. C., A. H. Tran, Q. Sun, and W. Margolin. 1998. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J. Bacteriol. 180:1296-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]