Abstract
Site-directed mutagenesis was used to investigate a region of the PheP protein corresponding to the postulated consensus amphipathic region (CAR) in the GabP protein. Whereas some critical residues are conserved in both proteins, there are major differences between the two proteins which may reflect different functions for this region. Replacement of R317, Y313, or P341 by a number of other amino acids destroyed the PheP function. An R317E-E234R double mutant exhibited low levels of PheP transport activity, indicating that there is a possible interaction between these two residues in the wild-type protein. E234 is highly conserved in members of the superfamily of amino acid-polyamine-organocation transporters and also is critical for PheP function in the wild-type protein. Second-site suppressors were isolated for mutants with mutations in E234, Y313, R317, and P341. Most suppressor mutations were found to cluster towards the extracellular face of spans III, IX, and X. Some mutations, such as changes at M116, were able to suppress each of the primary changes at positions E234, Y313, R317, and P341 but were unable to restore function to a number of other primary mutants. The possible implications of these results for the tertiary structure of the protein are discussed.
The GabP and PheP proteins are members of the amino acid transport (AAT) family within the superfamily of amino acid-polyamine-organocation transporters (15). Approximately 30% of the residues are identical in these two proteins. Both GabP and PheP are polytopic integral membrane proteins containing 12 membrane-spanning regions, as determined by hydropathy plots and studies of fusion proteins (5, 7). GabP permease is primarily a transporter for γ-aminobutyric acid, and PheP is primarily a transporter for the aromatic amino acids phenylalanine and tyrosine (4, 8).
Hu and King (2) have identified a sensitive polar surface (SPS) extending from the latter half of transmembrane span VIII into the following cytoplasmic loop of GabP, which is part of a consensus amphipathic region (CAR) and is postulated to play a major role in substrate translocation (2). In the case of PheP, the cytoplasmic loop between transmembrane spans VIII and IX has been shown to be very sensitive to single amino acid insertions and to contain two highly conserved amino acid residues, R317 and P341, each of which is critical for protein function (6, 9). Since Hu and King have suggested that the role proposed for the CAR in GabP may also apply to all other members of the AAT family, we carried out a similar analysis of the putative SPS in PheP to test the generality of the results obtained with GabP (2). This analysis revealed three residues in this region of PheP that are critical for protein function and a fourth, in an earlier cytoplasmic loop, with which one of these three residues may interact. Starting with primary mutants with mutations at one of these three positions and also with mutants with mutations at the previously studied residue P341, we selected and analyzed a collection of second-site suppressors. The possible implications for the tertiary structure of the protein are discussed below.
MATERIALS AND METHODS
Bacterial strains and media.
The following Escherichia coli strains were used: JM101 [Δ(lac pro) supE thi (F′ traD36 proAB lacIq ZΔM15)] for work involving bacterial phage M13mp18; XL1-Red [endA1 gyrA96 thi-1 hsdSR17 supE44 relA1 lac mutD5 mutS mutT Tn10 (Tetr); Stratagene], which was used to enhance the occurrence of random mutations; JP4538 (pheA pheP aroP tyrP), a host strain for the isolation of second-site suppressor mutants; and JP6488 (aroP pheP), which was used for transport assays. The minimal media used were the half-strength medium 56 described by Monod et al. (5) supplemented with 0.2% glucose and required growth factors. Luria agar and Lennox broth were used as complete media. Kanamycin was used at a final concentration of 25 μg/ml.
Site-directed mutagenesis of the pheP gene.
Oligonucleotide-directed site-specific mutagenesis (13) was used to introduce various amino acid substitutions in the pheP gene. When numerous amino acid substitutions of the same residue were desired, oligonucleotides containing a mixture of nucleotides at each of the three bases of the target codon were used. A construct (mpMU3137 [6]) containing a modified pheP gene (with ATG instead of GTG as the start codon) was used as a template for mutagenesis. All the mutations were verified by DNA sequencing of the entire pheP gene, and the 2.3-kb EcoRI-SalI fragment containing the pheP gene was cloned into the corresponding sites on low-copy-number plasmid pLG339 (10); the uptake of l-[14C]phenylalanine was measured in the background of strain JP6488.
Random mutagenesis and screening for second-site suppressor mutants.
Random mutations were introduced by propagating plasmid DNA carrying the pheP gene with one of the primary mutations in strain XL1-Red for up to four cycles. Plasmid DNA was isolated and transformed into strain JP4538 (pheA pheP aroP tyrP), and transformants were screened for growth on minimal media containing 10−5 M phenylalanine. This medium can only support the growth of cells which have some capacity to transport phenylalanine (i.e., wild-type revertants or suppressed mutants).
Cloning and sequence analysis of second-site suppressor mutations.
The 2.3-kb EcoRI-SalI fragment containing the pheP gene was cloned into the corresponding sites on M13mp18. Automated DNA sequencing, performed with a model 377 DNA sequencer and ABI BigDye terminators (Perkin-Elmer Corporation), was performed for the entire pheP gene to identify any second-site mutation(s) and to confirm the presence of the primary mutation and the absence of any other changes.
Transport assays.
Active transport was measured in E. coli JP6488 (aroP pheP) transformed with plasmids expressing wild-type or mutant pheP genes, as previously described (6). Mid-log-phase cells were washed in half-strength buffer 56 containing 0.2% glucose and 80 μg of chloramphenicol per ml and resuspended in the same buffer to an optical density at 600 nm of approximately 0.45. Cells were preincubated at 30°C for 5 min, and l-[14C]phenylalanine (final concentration, 10 μM) was added. Aliquots (150 μl) were removed at the appropriate times and filtered through cellulose acetate filters, which were then washed twice with half-strength buffer 56. Intracellular radioactivity was determined by liquid scintillation counting.
RESULTS
Topology of span VIII of PheP and GabP.
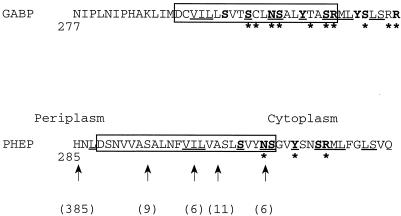
Figure 1 shows the amino acid sequences encompassing transmembrane span VIII and the CAR in both GabP and PheP.
FIG. 1.
Comparison of the sequences of GabP and PheP in the region involving putative span VIII and part of the following cytoplasmic loop. The polar residues proposed to provide a sensitive polar surface for the CAR of GabP are indicated by boldface type, as are the equivalent residues in PheP. The membrane spans identified by hydropathy plots are enclosed in boxes. The positions of pheP′-phoA-′pheP sandwich fusions are indicated by arrows, and the alkaline phosphatase activity values are given in parentheses. Identical amino acids in the two proteins are underlined, and asterisks indicate positions where amino acid substitutions reduced the level of transport activity to less than 25% of the wild-type level. The GabP data are from reference 2, and the PheP data are from this study.
The difference in the postulated positions of span VIII in the two proteins reflects major differences in hydropathy plots. While the results of previous alkaline phosphatase fusion studies support the general position of span VIII, they do not provide evidence that permits the location of either span to be determined more precisely (3, 7). In this study, a number of PheP′-PhoA-′PheP sandwich fusions were constructed with the phoA gene inserted at S294, I300, and A303 of PheP, and the alkaline phosphatase activities were 9, 6, and 11 U, respectively. The previously reported value for the activity of the periplasmic fusion at H285 was 385 U, and the previously reported value for the activity of the cytoplasmic fusion at N309 was 6 U (7). Consequently, it seems that at least for PheP, the arrangement of span VIII described previously (7) is correct.
Mutational analysis of span VIII of PheP.
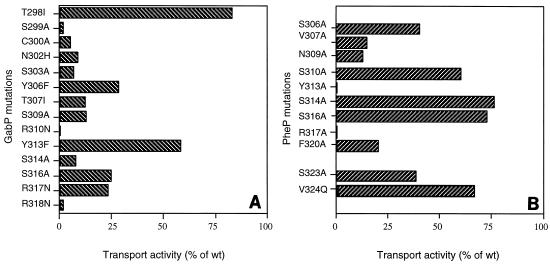
The residues of the putative SPS for the two proteins are shown in Fig. 1, and the data show that the central core (namely, SxxNSxxYxxSR) is the same for both proteins. In the case of GabP, replacement of residues of the SPS by nonpolar amino acids or by noncharged amino acids for each of the three arginine residues has been shown to result in a major reduction in transport activity (2). To test the role of the corresponding amino acid residues in the function of PheP, site-directed mutagenesis was used to replace each of the amino acids of the putative SPS of PheP with alanine, and the EcoRI-SalI fragment containing the mutant pheP gene was cloned into the corresponding sites on pLG339 (10). Uptake of [14C]phenylalanine (10 μM) was measured in transformants of E. coli JP6488 (aroP pheP) with plasmid pLG339 carrying the mutated pheP gene. The results of these assays are summarized in Fig. 2, which includes the results for substitutions of some other residues in this region and also the previously published results for GabP.
FIG. 2.
Transport activities of mutants with mutations in span VIII of GabP (A) (data from reference 2) and PheP (B). The values are the steady-state levels of [14C]phenylalanine uptake compared to the wild-type (wt) level, expressed as percentages; the wild-type level (10.1 nmol/mg [dry weight]) was defined as 100%. The region from position 298 to position 317 of GabP corresponds to the region from position 305 to position 324 of PheP.
As Fig. 2 shows, in the case of PheP, substitutions at N309, Y313, R317, and F320 each resulted in a reduction in transport activity to a level that was less than 25% of the wild-type level. The corresponding positions in GabP are N302, Y306, R310, and Y313. Of these, changes at positions 302 and 310 had dramatic effects on transport, whereas the changes resulting from substitutions at Y306 and Y313 were less marked. Conversely, whereas substitutions in GabP at positions 299, 300, 303, 307, 309, and 314 each reduced transport activity to a level that was less than 25% of the wild-type level, the corresponding changes at positions 306, 307, 310, 314, and 316 in PheP had significantly less dramatic effects on the levels of transport activity.
Further analysis of R317 and Y313.
The two substitutions in PheP that had the greatest effect involved the amino acids Y313 and R317. In order to test the specific requirement for each of these amino acids at these positions, site-directed mutagenesis was performed as previously described, and the purified mutants were analyzed to determine their transport activities. The results of these assays are shown in Table 1, which shows that each of these amino acids is highly specific for its position and that the presence of these amino acids is critical for functional activity of the PheP protein. Even if Y313 was replaced by phenylalanine or if R317 was replaced by lysine, there was no residual transport activity. In the 36 members of the AAT family, arginine is always present at positions corresponding to position 317. On the other hand, although aromatic residues are often found at positions corresponding to position 313, the overall distribution is 23 tyrosine residues, 9 phenylalanine residues, two serine residues, one glycine residue, and one threonine residue.
TABLE 1.
Phenylalanine transport activities of mutants with substitutions at E234, Y313, R317, and P341
| Residue | Phenylalanine transport activity of mutanta:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | A | V | I | L | F | P | S | T | W | N | Q | C | K | R | H | D | E | |
| E234 | 0 | 0 | 0 | 15 | ||||||||||||||
| Y313 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | |||||||||
| R317 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||
| P341b | 0 | 5 | 3 | 17 | 0 | 0 | 0 | 0 | ||||||||||
Steady-state level of [14C]phenylalanine uptake, expressed as a percentage of the uptake by wild-type permease (10.1 nmol/mg [dry weight]), which was defined as 100%.
The data for P341 have been published previously (6).
Search for an amino acid capable of interacting with R317.
Critical charged residues are sometimes found to interact with an oppositely charged residue appropriately positioned in the protein (11). In order to search for a possible complementing residue for R317, we started with mutant R317E and used this template to introduce additional mutations, either E118R, E226R, or E234R. A negative charge at each of these positions is highly conserved throughout the AAT family. At the position equivalent to position 118 in PheP, there is a glutamate or aspartic acid in 31 of the 36 members. At positions corresponding to position 226, glutamate is present in 32 cases, and the other four proteins have an aspartic acid. At positions corresponding to position 234, glutamate is present in all 36 members of the family.
Plasmids carrying the pheP gene with double mutations were introduced into strain JP4538 (pheA pheP aroP tyrP) and selected on minimal media supplemented with high levels (millimolar levels) of phenylalanine. After purification, the transformants were tested for the ability to grow on minimal media supplemented with 10−5 M phenylalanine and were also assayed for transport activity. Of the three double mutants, only the E234R-R317E mutant showed any ability to grow on the selective medium and only this mutant showed any transport activity. Although the level of activity was very low (2% of the wild-type level), it was reproducible, as was the slow but positive growth on the medium containing a low level (10−5 M) of phenylalanine. Under the same conditions the R317E and E234R single mutants and the other double mutants failed to show any sign of growth.
We also tested possible interactions between E234 and other positively charged residues in cytoplasmic loops by constructing a number of other double mutants (namely, the E234R-K406E, E234R-R393E, and E234R-R26E double mutants). None of these double mutants was able to grow on the selective media (data not shown), strengthening the significance of the results obtained with the E234R-R317E double mutant.
Selection of second-site suppressor mutations.
Data obtained in this study revealed that R317, Y313, and E234 have a critical role in the transport activity of the PheP protein. Previous studies (6) showed that P341 has a similar critical role (Table 1). Amino acid residues Y313, R317, and P341 are all located within or immediately adjacent to the cytoplasmic loop between transmembrane spans VIII and IX, and E234 was implicated in possible interactions with R317. We decided, therefore, to use null mutants with mutations at each of these positions to select suppressed strains which regained transport activity as a consequence of a second substitution elsewhere in the protein and to characterize such mutations in the hope that they might shed light on the important roles of these four residues.
Potential second-site mutations were randomly introduced into various pheP mutant templates by introducing plasmids carrying pheP with the primary mutations into the mutator strain XL1-Red. Plasmid DNA was isolated after one to four growth cycles in XL1-Red and transformed into a transport-negative strain requiring phenylalanine for growth (JP4538 [pheA pheP aroP tyrP]). Colonies were screened for growth on minimal media containing 10−5 M phenylalanine. Plasmid DNA was extracted from the colonies that grew and was sequenced to determine whether the original mutation was present. Plasmids retaining the original mutation were retransformed into the transport-negative host to test for functional activity, and the entire pheP gene was sequenced to identify the second-site suppressing mutation(s). Approximately 52% of the colonies that grew were primary-site revertants and were not studied further. The results obtained for mutants with mutations at each one of the four chosen positions are described below.
Second-site suppressors of mutations changing arginine at position 317.
Six primary mutants were used: the R317G, R317C, R317E, R317W, R317H, and R317K mutants. Eighteen independent second-site suppressors were characterized, and they restored transport activity to levels that were between 5 and 49% of the wild-type steady-state levels of accumulation. The residues altered in these suppressed strains are indicated in Fig. 3, and the levels of suppression for individual mutants are shown in Table 2.
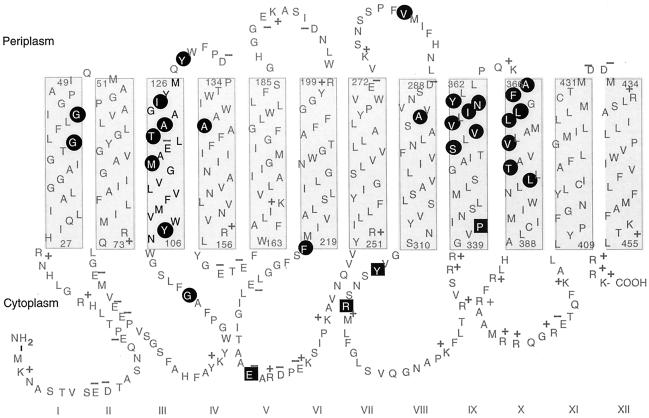
FIG. 3.
Topological model of PheP, showing the locations of all second-site suppressor mutations (highlighted in circles) identified by using primary mutants (highlighted in squares) of E234, Y313, R317, and P341.
TABLE 2.
Second-site suppressors of R317X, P341X, Y313E, and E234X of PheP
| Mutation | Location of suppressor mutationa | Activity (% of wild type)b |
|---|---|---|
| R317X mutations | ||
| R317C M116V | Span 3 | 49 |
| R317G M116V | Span 3 | 30 |
| R317G M116T | Span 3 | 30 |
| R317E M116T | Span 3 | 17 |
| R317C A121T | Span 3 | 9 |
| R317W I124S | Span 3 | 10 |
| R317K A140V | Span 4 | 7 |
| R317C F220L | CL3 | 6 |
| R317H F220L | CL3 | 18 |
| R317G V281A | PL4 | 5 |
| R317C S353L | Span 9 | 5 |
| R317C V356M | Span 9 | 7 |
| R317G N359S | Span 9 | 11 |
| R317G Y360H | Span 9 | 5 |
| R317G F367V | Span 10 | 9 |
| R317E F367S | Span 10 | 6 |
| R317W L370P | Span 10 | 19 |
| R317G L379P | Span 10 | 23 |
| P341X mutations | ||
| P341R G40S | Span 1 | 4 |
| P341R M116T | Span 3 | 15 |
| P341K T120I | Span 3 | 6 |
| P341Q A121T | Span 3 | 16 |
| P341Q Y128C | PL2 | 5 |
| P341R V355M | Span 9 | 8 |
| P341Q F367S | Span 10 | 15 |
| P341R F367Y | Span 10 | 16 |
| P341Q T377M | Span 10 | 5 |
| Y313E M116I | Span 3 | 4.2 |
| E234X mutations | ||
| E234G L369P | Span 10 | 20 |
| E234G I358T | Span 9 | 3.5 |
| E234G Y128C | PL2 | 3 |
| E234I V355A | Span 9 | 2 |
| E234I V356A | Span 9 | 0.5 |
| E234I V356M | Span 9 | 1 |
| E234I N359S | Span 9 | 2.5 |
PL, periplasmic loop; CL, cytoplasmic loop.
Steady-state level of [14C]phenylalanine uptake, expressed as a percentage of the uptake by wild-type permease.
In span III, changes along one face at M116, A121, and I124 all resulted in significant suppression, and M116V and M116T occurred a number of times. Another cluster of changes involving residues along one face of the periplasmic end of span IX (i.e., S353, V356, N359, andY360) also resulted in strong suppression. In addition, two leucine-to-proline changes at positions 370 and 379 of span X resulted in strong levels of suppression. Also, two changes in phenylalanine residues, one located in the cytoplasmic loop containing E234 (F220) and the other located just at the top of span X (F367), restored significant activity.
Second-site suppressors of mutations changing proline at position 341.
The following three primary mutants were used: the P341Q, P341R, and P341K mutants. Nine independent mutants were characterized. The relative positions of the second-site changes are shown in Fig. 3, and the details of their suppressed levels are shown in Table 2. Again, changes at residues T120, A121, M116, and Y128, which cluster along one face of the upper region of span III, were observed, and there were three independent isolates involving changes in F367 at the top of span X (previously observed with R317 mutants). Additional mutations involved V355 close to the span IX cluster, G40S, and T377M.
Second-site suppression of mutations changing tyrosine at position 313.
The primary mutant used was the Y313E mutant. Only one suppressor was isolated. This suppressor had a change in a residue in span III that has been observed previously (namely, M116I).
Second-site suppressors of mutations changing glutamate at position 234.
Two of the four mutants obtained by site-directed mutagenesis (Table 1) (the E234I and E234G mutants) were used. Seven independent mutants were characterized; the positions of the substitutions are shown in Fig. 3, and the levels of activity are shown in Table 2.
Again, three of the mutations affecting span IX (namely, V355A, V356A, and N359S) had been found previously, whereas I358T was isolated for the first time. In span X the introduction of a proline residue at position 369 caused strong suppression. One change in the upper part of span III involving a previously identified residue (namely, Y128C) was also observed.
Further investigations of changes at M116.
Because changes at M116, particularly M116T and M116V, restored significant transport activity to mutants with primary changes at P341, R317, or Y313, we constructed a number of additional mutants to determine the specificity of M116 suppressors and also to test the effects of other substitutions on an otherwise wild-type protein. The results are shown in Table 3.
TABLE 3.
Suppressor effects of M116 mutations of PheP
| Primary mutation | Secondary mutation | Activity (% of wild type)a |
|---|---|---|
| E234G | M116T | 74 |
| E234G | M116V | 103 |
| E234D | M116V | 145 |
| E234R | M116V | 0 |
| Y313E | M116T | 20 |
| Y313E | M116V | 37 |
| R317C | M116V | 49 |
| R317G | M116V | 30 |
| R317G | M116T | 30 |
| R317E | M116T | 17 |
| E226A | M116V | 0 |
| E226G | M116V | 0 |
| E118C | M116V | 0 |
| E161Q | M116V | 0 |
| K168E | M116V | 0 |
| E234G | wtb | 0 |
| E234D | wt | 15 |
| E234R | wt | 0 |
| Y313E | wt | 0 |
| E118C | wt | 0 |
| E161Q | wt | 7 |
| E226G | wt | 13 |
| E226A | wt | 0 |
| K168E | wt | 18 |
| M116T | wt | 14 |
| M116V | wt | 14 |
| M116E | wt | 92 |
| M116K | wt | 107 |
| M116G | wt | 88 |
Steady-state level of [14C]phenylalanine uptake, expressed as a percentage of the uptake by the wild-type permease (10.1 nmol/mg [dry weight]), which was defined as 100%.
wt, wild type.
As Table 3 shows, the primary mutation E234G was strongly suppressed by M116T and M116V. These two suppressors are also more efficient suppressors of Y313E than M116I is. The primary mutation E234D reduced the level of transport activity to 15% of the wild-type level, and addition of the M116V mutation restored activity to levels that were greater than the wild-type level. Of particular interest, however, is the observation that the primary mutation E234R was not suppressed by M116V, which effectively suppressed a number of other changes to E234. Evidence of the specificity of the M116 suppressors was provided by the observation that primary mutations in E118, E161, E226, and K168 were not suppressed by the very strong suppressor mutation M116V (Table 3).
Finally, when the M116V and M116T changes were made to the wild-type PheP protein, the level of transport activity was reduced to 14% of the wild-type level. On the other hand, changing M116 to glutamate, lysine, or glycine had only a minor effect on transport activity.
DISCUSSION
Comparison of GabP and PheP.
Although GabP and PheP both exhibit a CAR with a clearly identifiable SPS extending from the lower part of span VIII into the ensuing cytoplasmic loop, there are a number of differences between the two proteins that should be noted. (i) PheP does not contain the central cysteine residue essential for GabP activity. (ii) The hydropathy plots and the presence of a lysine immediately before span VIII in GabP but not in PheP suggest that there are slight differences in the positions of the transmembrane spans. (iii) Whereas replacing any one of nine residues of the SPS of GabP reduces the level of transport activity to less than 25% of the wild-type level, in the case of PheP only four residues have the same dramatic effect. (iv) The critical role of Y313 of PheP contrasts with the less significant role of Y306 of GabP. On the other hand, together, the three residues lying along one face of the helix in both proteins (asparagine, tyrosine, and arginine) clearly play a critical role. It would be interesting to know whether the proline in GabP corresponding to P341 of PheP is also critical for activity.
R317 and Y313.
Two lines of evidence suggest that there is close proximity in the tertiary structure of the PheP protein between residues E234 and R317. First, mutant PheP permease with both mutation E234R and mutation R317E exhibited some transport activity, whereas no transport activity was detected with mutants having either of the single mutations. Second, primary mutations at either of these positions can be partially overcome by a number of second-site mutations in span III. Hu and King have suggested that the CAR of GabP may function as a water-filled channel for the passage of substrate and that it may move up and down in the membrane (2). If there was movement of this region into the membrane either as a dynamic event or as a reentrant loop, as has been hypothesized for LacS (14) or GltP (12), a salt bridge between E234 and R317 could play a critical role. The failure of the suppressor M116V to suppress the primary mutation E234R, although it effectively suppressed a number of other substitutions at this position, would be explained if E234 was normally within the membrane as part of a salt bridge with R317. Replacement of E234 by arginine would not only destroy the salt bridge and the structure that it held together but would also leave an unneutralized positive charge within the membrane. Presumably, the changes at M116 can, in some way, compensate for the change in structure but cannot overcome the difficulties caused by the positively charged arginine. Salt bridges between two oppositely charged amino acids have been shown to be important in the case of membrane-embedded residues (11). If R317 and E234 are within the membrane, then Y313 and E226, two other critical residues of PheP, would be in the lipid environment. There is some ambiguity about the position of E226. Hydropathy plots originally indicated that E226 should lie in the middle of transmembrane span VI, but on the basis of low alkaline phosphatase activities with sandwich fusions at positions 204 and 209, the entry of span VI into the membrane was revised to position 199, with the result that E226 was positioned in the cytoplasmic loop following span VI. Other possibilities that might accommodate these results include an unusually long span VI that still starts at position 199 or a reentrant loop involving residues 199 to 214. More detailed studies will have to be undertaken to resolve this dilemma.
Second-site suppressors.
Perhaps the most compelling aspect of the second-site suppressor studies is the finding that there is a major overlap between suppressors for primary mutations in each of the four residues R317, P341, Y313, and E234. The failure of the suppressor mutation M116V to restore function to mutants with a number of other primary mutations in PheP (Table 3) confirms the overall specificity of these suppressors. Although there are some exceptions, the suppressor mutations form three distinct clusters, one in the top of span III, another in the top of span IX, and the third in the top of span X. Previous studies of AroP-PheP chimeras established that there is an important close interaction between span III and the more-distal span IX or X (1). It appears from the suppressor studies that Y313, R317, P341, and E234 all contribute to one important aspect of the tertiary structure of PheP and that an alteration in any one of them can perturb this structure. Such perturbations can be partially corrected by changes in the upper segments of spans III, IX, and X, which in turn may interact to constitute another major component of the tertiary structure of the protein. If the hypothesis of Hu and King that the CAR of GabP contributes to a water-filled channel is correct and can be extended to other proteins, we argue that in PheP, span III also makes a critical contribution and that the upper segments of spans IX and X can influence the channel, even though this effect may be indirect.
Acknowledgments
We are grateful to T. Betteridge for technical assistance. We thank Ji Yang and Judy Praszkier for critical reading of the manuscript.
This work was supported by the Australia Research Council Large Grants Scheme.
REFERENCES
- 1.Cosgriff, A., G. Brasier, J. Pi, C. Dogovski, J. P. Sarsero, and A. J. Pittard. 2000. A study of AroP-PheP chimeric proteins and identification of a residue involved in tryptophan transport. J. Bacteriol. 182:2207-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu, L. A., and S. C. King. 1998. Functional sensitivity of polar surfaces on transmembrane helix 8 and cytoplasmic loop 8-9 of the Escherichia coli GABA (4-aminobutyrate) transporter encoded by gabP: mutagenic analysis of a consensus amphipathic region found in transporters from bacteria to mammals. Biochem. J. 330:771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu, L. A., and S. C. King. 1998. Membrane topology of the Escherichia coli gamma-aminobutyrate transporter: implications on the topography and mechanism of prokaryotic transporters from the APC superfamily. Biochem J. 336:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metzer, E., and Y. S. Halpern. 1990. In vivo cloning and characterization of gabCTDP gene cluster of Escherichia coli K-12. J. Bacteriol. 172:3250-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monod, J., G. Cohen-Bazire, and M. Cohn. 1951. Sur la biosynthese de la β-galactosidase (lactase) chez Escherichia coli. La specificite de l'induction. Biochim. Biophys. Acta 7:585-599. [DOI] [PubMed] [Google Scholar]
- 6.Pi, J., C. Dogovski, and A. J. Pittard. 1998. Functional consequences of changing proline residues in the phenylalanine-specific permease of Escherichia coli. J. Bacteriol. 180:5515-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pi, J., and A. J. Pittard. 1996. Topology of the phenylalanine-specific permease of Escherichia coli. J. Bacteriol. 178:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pi, J., P. J. Wookey, and A. J. Pittard. 1991. Cloning and sequencing of the pheP gene, which encodes the phenylalanine-specific transport system of Escherichia coli. J. Bacteriol. 173:3622-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pi, J., P. J. Wookey, and A. J. Pittard. 1993. Site-directed mutagenesis reveals the importance of conserved charged residues for the transport activity of the PheP permease of Escherichia coli. J. Bacteriol. 175:7500-7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pouwels, P. H., B. E. Enger-Valk, and W. J. Brammar. 1985. Cloning vectors, p. I-A-ii-1. Elsevier Science Publishers, B. V., Amsterdam, The Netherlands.
- 11.Sahin-Toth, M., and H. R. Kaback. 1993. Properties of interacting aspartic acid and lysine residues in the lactose permease of Escherichia coli. Biochemistry 32:10027-10035. [DOI] [PubMed] [Google Scholar]
- 12.Slotboom, D. J., I. Sobczak, W. N. Konings, and J. S. Lolkema. 1999. A conserved serine-rich stretch in the glutamate transporter family forms a substrate-sensitive reentrant loop. Proc. Natl. Acad. Sci. USA 96:14282-14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandeyar, M. A., M. P. Weiner, C. J. Hutton, and C. A. Batt. 1988. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene 65:129-133. [DOI] [PubMed] [Google Scholar]
- 14.Veenhoff, L. M., E. R. Geertsma, J. Knol, and B. Poolman. 2000. Close approximation of putative alpha-helices II, IV, VII, X, and XI in the translocation pathway of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 275:23834-23840. [DOI] [PubMed] [Google Scholar]
- 15.Young, G. B., D. L. Jack, D. W. Smith, and M. H. Saier, Jr. 1999. The amino acid/auxin:proton symport permease family. Biochim. Biophys. Acta 1415:306-322. [DOI] [PubMed] [Google Scholar]