Abstract
Mutations in yjfQ allowed us to identify this gene as the regulator of the operon yjfS-X (ula operon), reported to be involved in l-ascorbate metabolism. Inactivation of this gene renders constitutive the expression of the ula operon, indicating that YjfQ acts as a repressor. We also demonstrate that this repressor regulates the nearby yjfR gene, which in this way constitutes a regulon with the ula operon.
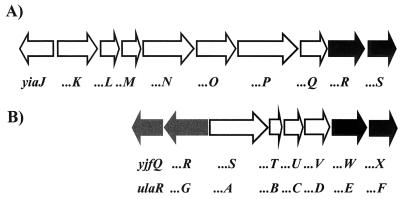
Analysis of the Escherichia coli genome revealed two different paralog proteins encoded in operon yiaK-S (centisome 80.7) and operon ula (yjfS-X, centisome 95.3) (10) (Fig. 1). The yiaK-S operon has been partially characterized at both the genetic and biochemical levels (5, 6, 12), although no specific role has been established so far for this system. Sequence similarity studies have indicated that this operon is involved in the metabolism of an unknown carbohydrate (14). Previous work by Sanchez et al. (12) identified the yiaP gene product purified from mutant cells selected for their ability to grow on the rare pentose l-lyxose as a kinase able to phosphorylate l-xylulose. However, the yiaK-S operon seems only fortuitously used for the metabolism of the intermediate l-xylulose formed from l-lyxose since, of the nine genes in the yiaK-S cluster, only three participate in the metabolism of this ketose. These three genes, yiaP, yiaR, and yiaS, encode the above-mentioned kinase (12), a 3-epimerase, and a 4-epimerase, respectively (6), and account for the transformation of l-xylulose into d-xylulose-5-phosphate, which is subsequently metabolized by the pentose phosphate pathway.
FIG. 1.
Genetic map of the region encompassing the yiaK-S operon (A) and the yjfR-X regulon (B). Arrows indicate the extent and direction of transcription of the genes. The genes encoding the sets of paralog proteins YiaR/YjfW and YiaS/YjfX are indicated in black, and the genes identified in this study as belonging to the ula regulon are indicated in grey.
The natural origin of l-xylulose may result from the action of the yiaK-S-encoded proteins on the unknown substrate. Expression of the yiaK-S cluster has only been detected in mutant strain JA134, selected for its ability to grow on l-lyxose (5). In this strain, the regulator gene is inactivated by a genome rearrangement mediated by IS1 transposition, which leads to the constitutive expression of the yiaK-S operon (1).
The yjfS-X (ula) operon is responsible for the anaerobic metabolism of l-ascorbate (16). In the corresponding proposed pathway, l-xylulose 5-phosphate is generated as an intermediary that is also transformed into d-xylulose-5-phosphate by the YjfW (UlaE), encoding a 3-epimerase, and YjfX (UlaF), encoding a 4-epimerase. These two enzyme proteins are paralogs to the YiaR (3-epimerase activity) and YiaS (4-epimerase activity) encoded in the yiaK-S operon.
Here, we report that mutations suppressing the inability to metabolize l-xylulose-5-phosphate in a YiaR mutant by means of the constitutive expression of the paralog YjfW (UlaE) displaying the same 3-epimerase activity mapped in yjfQ, a gene thus shown to encode the repressor of the yjfS-X (ula) operon. We also show that this operon is part of a regulon that includes the divergently transcribed yjfR gene.
Identification of YjfQ as a repressor of yjfR-X.
A spontaneous l-lyxose-positive strain was obtained from strain JA195 (strain JA134 yiaR::cat), which cannot metabolize this pentose, by culturing these cells in minimal medium (3) containing 0.2% casein acid hydrolysate and 40 mM l-lyxose. Upon exhaustion of the casein acid hydrolysate, the culture mass entered a second phase of growth, which was interpreted as growth on the sugar pentose. The ability to utilize l-lyxose was checked in solid medium (3), and in this way clone JA211 was isolated.
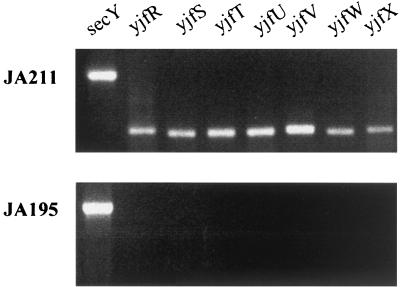
The presence in the genome of yjfW (ulaE), which encodes a paralog of YiaR, led us to test whether this protein accounted for the observed complementation of the deficient YiaR function of strain JA195. To this end, the expression of the yjfW (ulaE) gene in this mutant was analyzed by reverse transcription-PCR (11) with primers designed to detect an internal 300-bp fragment of the gene (Table 1). The RNA was obtained with the SV total RNA isolation system of Promega (Madison, Wis.). An internal fragment of 632 bp (see primers in Table 1) of secY, a gene encoding a membrane protein that is constitutively expressed, was used as a control. Electrophoretic analysis of the reverse transcription-PCR products revealed constitutive expression of the yjfW (ulaE) gene in the isolated mutant but not in the parental strain JA195 (Fig. 2). It is thus likely that the mutation affected the function of the regulator of the yjfS-X (ula) operon, which would act as a repressor.
TABLE 1.
PCR and reverse transcription-PCR primers
| Gene or fragment | Forward primer | Reverse primer |
|---|---|---|
| yifW fragment | TATCAGGAAGCCAATAACG | GAAGACGCCAGGTTTGGTG |
| secY fragment | GGCCTGGTGATTAACCCG | CCGAATTCCTGGTACAAATGCTCCGGACTTCTTC |
| yifR and yifS promoter region | GCGAATTCAATTGCGCCGTTAATGC | TGCTCAGGATCCAGGATTCACGGG |
| yifQ gene region | AAGACAACTTCGAGTACC | ACGCCTGAGGCGTTAGATG |
| yifQ promoter region | GCGAATTCAAGACAACTTCGAGTACC | CGGGATCCGCCCAATTGTGCGAGC |
| yifR fragment | CCGTACTGCACTGATCACC | GATATCGTGGTGGAACGGG |
| yifS fragment | ACATCATGTTCCAGCAGGC | GGAGACGATGTTGTCGTGG |
| yifT fragment | TACGTATTCTGGCTGTGTG | CACATCCTGCGGGAAATG |
| yifU fragment | ATAAATCCATCCGCCTGC | CGCCGCCATGGTGATGAGG |
| yifV fragment | AAAGCGCTCTACCCGCAC | GCTTCGCCCCACGCCACGC |
| yifX fragment | AAAGCGGCCGATATGGTGG | CCAGCGTTTCGATAATCAC |
| Tn10 | TAAGGTGGATACACATCTTGTC | CCAAAATCATTAGGGGATTCATCAG |
FIG. 2.
Expression of the yjfR-X region detected via reverse transcription-PCR with primers designed to pick up 300-bp internal fragments of the genes listed above each lane. Levels of mRNA were normalized to that of the secY transcript (632-bp internal fragment) in all samples.
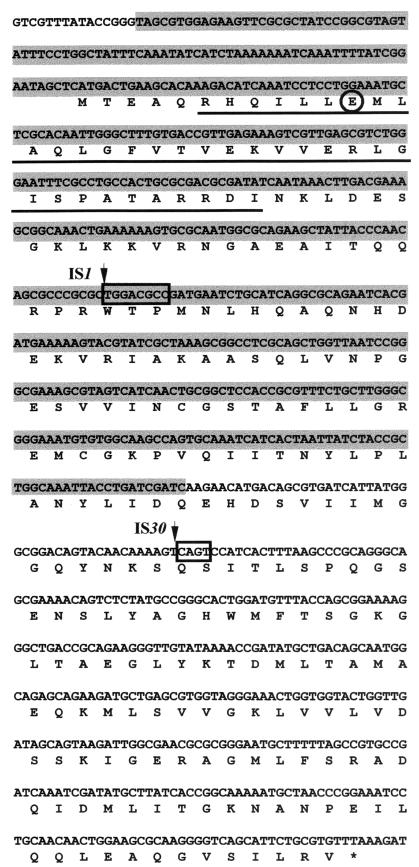
Genome analysis of the yjfW (ulaE)-proximal region allowed us to identify gene yjfQ, which had been classified as a putative regulator of unknown function (2). The corresponding amino acid sequence (P39299) was used as the query sequence for a BLAST P search, and a high degree of similarity was found between this sequence and regulator proteins of the DeoR repressor family. Accordingly, a helix-turn-helix motif was found in the amino-terminal end, matching the signature sequence of the DeoR family proposed by Reizer et al. (10),RX3[LIVM]X3[LIVM]X20T[LIVMA]R[RKAN]D[LIVMF](Fig. 3).
FIG. 3.
Nucleotide sequence and amino acid translation of the yjfQ gene, encoding the repressor of the ula operon. The IS1 and IS30 insertion sites are marked by arrows, and the target sequences duplicated in the transposition processes are boxed. The amino acid position changed in the point mutant analyzed is circled. The nucleotide sequence deleted in strain JA211 is shaded, and the amino acid sequence at the N-terminal end corresponding to the helix-turn-helix motif is underlined.
To determine whether the mutation responsible for the growth of strain JA211 on l-lyxose is located in yjfQ, this gene was amplified by PCR with primers against the yjfQ region (Table 1). The resulting PCR product was smaller than that obtained from the parental strain JA195 (data not shown). DNA sequencing of the corresponding PCR product in an automated ABI 377 DNA sequencer with a fluorescent dye termination method revealed a 486-bp deletion in the yjfQ gene (Fig. 3). We conclude that yjfQ encodes the repressor of the yjfS-X (ula) operon, which becomes constitutive upon inactivation, and propose that the yjfQ gene should be referred to as ulaR, according to its regulatory function for the ula regulon.
Sixteen additional independent spontaneous l-lyxose-positive mutants were isolated from strain JA195 as described for strain JA211. Analysis by PCR of the yjfQ region and comparison of the electrophoretic mobilities of the PCR products showed changes in the yjfQ gene size in nine of the isolated mutants. Six displayed no size difference, and one gave no PCR product, probably due to a large undetermined insertion or deletion mutation. These results suggest that, at least in the mutants displaying changes in size, the suppressing mutations were also located in the yjfQ gene. The location of the suppressing mutations was confirmed by sequencing the PCR products. In this way, we were able to identify an IS1 insertion in two of the mutants and an IS30 insertion in the other seven mutants. In all cases, duplicated base pairs of the target sequences were also identified (Fig. 3). Sequencing of one of the mutants yielding no difference in gene size indicated that the codon encoding E12 was replaced by a stop codon.
It has to be underlined that isolation of several l-lyxose-positive spontaneous mutants from a strain unable to metabolize this pentose due to a point mutation (A126T) in yiaR (strain JA192) (6), following the procedure indicated at the beginning of this section, in all cases selected revertants of this gene rather than suppressing mutations in unlinked genes.
To analyze yjfQ (ulaR) gene expression, a 246-bp fragment encompassing the corresponding 5′ promoter region was prepared by PCR with the primers shown in Table 1 and cloned in pRS551 (13). Single-copy fusions on the E. coli chromosome were obtained by the method of Elliot (4) and transferred to different genetic backgrounds by P1 transduction (7). Expression of the fused promoter Φ(yjfQ-lacZ) in aerobic cultures on casein acid hydrolysate of either strain JA195 or strain JA211 yielded similar levels of β-galactosidase activity (ca. 300 Miller units). The low and constitutive expression of this gene is consistent with its regulatory function, and the similar level of expression in the wild-type and repressor mutant strains eliminates the possibility of any autogenous effect on the regulation of its expression.
Defining the ula regulon formed by yjfS-X and yjfR operons.
The control of the repressor yjfQ (ulaR) on the structural gene yjfW (ulaE) was extended to the other genes in the cluster. Transcription of the rest of the genes in strain JA211 and the parental strain JA195 was analyzed by reverse transcription-PCR with primers directed against yjfS (ulaA), yjfT (ulaB), yjfU (ulaC), yjfV (ulaD), and yjfX (ulaF) (Fig. 2). The constitutive expression of all these genes in strain JA211 pointed to a coordinated expression by this repressor. Likewise, the divergently transcribed gene yjfR (proposed name ulaG) was also studied and shown to be controlled by theYjfQ (UlaR) repressor. Furthermore, the expression of the yjfS-X gene cluster from a single promoter was evidenced by the polarity effects caused by a Tn10 insertion in the yjfS (ulaA) gene on the downstream transcribed genes.
The mutant was obtained by infection of strain JA211 with phage lambda NK1098 (15) and selected by the loss of ability to grow on l-lyxose. The insertion was mapped by inverse PCR by the method of Ochman et al. (8) with primers designed from the Tn10 sequence (Table 1). These observations indicated that all genes in the yjfS-X cluster were expressed as a single transcriptional unit. All genes in the regulon, except the divergently transcribed ulaG, have been assigned a function in l-ascorbate metabolism. The gene ulaG displayed no homology to any other protein of known function. However, since in the metabolic pathway for l-ascorbate suggested by Yew and Gerlt (16) a hydrolase or a tautomerase is possibly required, it is likely that the ulaG gene product could account for one of these two functions.
Promoter analysis of the divergently transcribed units was approached by constructing transcriptional fusions of the yjfR (ulaG) and yjfS (ulaA) genes. A fragment of 648 bp encompassing the corresponding intergenic region was prepared by PCR with the primers shown in Table 1 and cloned into the pRS550 and pRS551 vectors (13). Expression was analyzed in the genetic background of strain JA211 grown on casein acid hydrolysate under aerobic conditions. Analysis of φ(yjfR-lacZ) gave a β-galactosidase activity of 1,960 Miller units, while expression of φ(yjfS-lacZ) displayed a β-galactosidase activity of 3,990 Miller units. Negligible expression was detected when these fusions were analyzed in the genetic background of strain JA195, which encodes a functional YiaQ repressor when grown in casein acid hydrolysate.
Although it has been reported that the yjfS-X operon is involved in the anaerobic metabolism of l-ascorbate, our results show that in the absence of the YjfQ repressor, the yjfR-S regulon is constitutively expressed in aerobic conditions. Finally, the expression φ(yjfR-lacZ) and φ(yjfS-lacZ) was also analyzed in the genetic background of strain JA195 growing anaerobically on l-ascorbate. In these conditions, the level of β-galactosidase activity was 2,150 Miller units for φ(yjfR-lacZ) and 4,500 Miller units for φ(yjfS-lacZ). These results indicated that l-ascorbate or a derived metabolite abolished the repressor activity of UlaR in strain JA195.
Acknowledgments
This work was supported by grant BMC2001-3003 from the Dirección General de Investigación, Ministerio de Ciencia y Tecnología, Madrid, Spain, and by the “Comissionat per Universitats i Recerca de la Generalitat de Catalunya.” E.C. is a recipient of a scholarship from the Ministerio de Educación, Cultura y Deporte (Spain).
REFERENCES
- 1.Badia, J., E. Ibañez, M. Sabaté, L. Baldomà, and J. Aguilar. 1998. A rare 920-kilobase chromosomal inversion mediated by IS1 transposition causes constitutive expression of the yiaK-S operon for carbohydrate utilization in Escherichia coli. J. Biol. Chem. 273:8376-8381. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett, I. I. I., C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 3.Boronat, A., and J. Aguilar. 1979. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliot, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibañez, E., E. Campos, L. Baldomà, J. Aguilar, and J. Badia. 2000. Regulation of expression of the yiaKLMNOPQRS operon for carbohydrate utilization in Escherichia coli: involvement of the main transcriptional factors. J. Bacteriol. 182:4617-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibáñez, E., R. Giménez, T. Pedraza, L. Baldomà, J. Aguilar, and J. Badia. 2000. Role of genes yiaR and yiaS of Escherichia coli in the metabolism of endogenously formed l-xylulose. J. Bacteriol. 182:4625-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Ochman, H., M. M. Medhora, D. Garza, and D. L. Hartl. 1990. Amplification of flanking sequences by inverse PCR, p. 219-227. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 9.Reizer, J., T. M. Ramseier, A. Reizer, A. Charbit, and M. H. Saier. 1996. Novel phosphotransferase genes revealed by bacterial genome sequencing: a gene cluster encoding a putative N-acetylgalactosamine metabolic pathway in Escherichia coli. Microbiology 142:231-250. [DOI] [PubMed] [Google Scholar]
- 10.Reizer, J., A. Reizer, and M. H. Saier. 1997. Is the ribulose monophosphate pathway widely distributed in bacteria? Microbiology 143:2519-2520. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook, J., and D. W. Rusell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Sanchez, J. C., R. Gimenez, A. Schneider, W-D. Fessner, L. Baldomà, J. Aguilar, and J. Badia. 1994. Activation of a cryptic gene encoding a kinase for l-xylulose opens a new pathway for the utilization of l-lyxose by Escherichia coli. J. Biol. Chem. 269:29665-29669. [PubMed] [Google Scholar]
- 13.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 14.Sofia, H. J., V. Burland, D. L. Daniels, G. Plunkett, and F. R. Blattner. 1994. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 22:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Way, J. C., M. A. Davies, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 16.Yew, W.-S., and J. A. Gerlt. 2002. Utilization of l-ascorbate by Escherichia coli. K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 184:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]