Abstract
Pathogenic Yersinia spp. secrete Yops (Yersinia outer proteins) via the type III pathway. The expression of yop genes is regulated in response to environmental cues, which results in a cascade of type III secretion reactions. yscM1 and yscM2 negatively regulate the expression of Yersinia enterocolitica yop genes. It is demonstrated that yopD and lcrH are required for yscM1 and yscM2 function and that all four genes act synergistically at the same regulatory step. Further, SycH binding to the protein products of yscM1 and yscM2 can activate yop gene expression even without promoting type III transport of YscM1 and YscM2. Reverse transcription-PCR analysis of yopQ mRNA as well as yopQ and yopE gene fusion experiments with the npt (neomycin phosphotransferase) reporter suggest that yscM1 and yscM2 regulate expression at a posttranscriptional step. The 178-nucleotide 5′ untranslated region (UTR) of yopQ mRNA was sufficient to confer yscM1 and yscM2-mediated regulation on the fused reporter, as was the 28-nucleotide UTR of yopE. The sequence 5′-AUAAA-3′ is located in the 5′ yop UTRs, and mutations that alter the sequence motif either reduced or abolished yscM1- and yscM2-mediated regulation. A model is proposed whereby YopD, LcrH, YscM1, YscM2, and SycH regulate yop expression in response to specific environmental cues and by a mechanism that may involve binding of some of these factors to a specific target sequence within the UTR of yop mRNAs.
Many gram-negative bacterial pathogens employ a virulence strategy known as type III secretion to establish infections in animal and plant hosts (24, 33). Three pathogenic Yersinia species, Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, share a common tropism for lymphatic tissues in their mammalian hosts (13, 45). Yersiniae use the type III pathway for the secretion of virulence factors and for the targeting of Yops (Yersinia outer proteins) into the cytoplasm of host macrophages (36, 38, 50, 51). These mechanisms manipulate the host's innate immune response, thereby preventing bacterial phagocytosis and killing (17). Y. enterocolitica transports 14 proteins via the type III pathway: LcrV, YopB, YopD, YopE, YopH, YopM, YopN, YopO, YopP, YopQ, YopR, YopT, YscM1, and YscM2 (1, 19, 25, 26, 32, 35, 40, 41, 44, 49, 52). The genes encoding secretion substrates and type III machinery components are located on a 70-kb virulence plasmid (13).
Expression of yop genes and secretion of encoded polypeptides are tightly regulated as Yersinia responds to specific environmental cues with type III transport reactions (9, 10, 37). Bacterial host entry is accompanied by a temperature shift to 37°C, activating the transcription of type III genes via the regulatory factor LcrF (VirF) (12, 29, 30, 54, 57). Y. enterocolitica assembles the type III secretion apparatus when cued by an extracellular amino acid signal (37, 39). A third signal, host serum proteins, triggers the secretion of YopB, YopD, YopR, and LcrV into the extracellular milieu (36-38). Bacterial contact with macrophages or tissue culture cells results in the insertion of needle complexes, a surface extension of type III machines, into the eukaryotic plasma membrane (31). This mechanism is thought to generate the fourth signal, i.e., a drop in the environmental calcium concentration, also referred to as low-calcium response (Lcr) or the calcium signal, from 1.2 mM (extracellular host fluids) to about 100 nM (intracellular fluid) (37). The calcium signal activates type III transport of YopE, YopH, YopM, YopN, YopO, YopP, YopT, and YscM1 into the eukaryotic cytosol (also named type III targeting or translocation) (5, 7, 27, 35, 36, 38, 41, 42, 50, 51).
Only when Y. enterocolitica is activated by a calcium signal is YopQ polypeptide synthesized and secreted, a phenotype which greatly facilitates genetic analysis of yop gene expression (2). Several genes are responsible for the regulation of yopQ expression (2). Mutations in class I genes (yopN, tyeA, sycN, yscB, and lcrG) bypass the requirement for the calcium signal, causing expression and secretion of YopQ in the presence of calcium at 37°C (2, 9, 11, 37). Mutations in class II genes, yopD and lcrH, lead to a phenotype that allows for the synthesis but not for the secretion of YopQ in the presence of calcium at 37°C (2, 37). yopD encodes a secretion substrate for the type III pathway (26), while lcrH (sycD) encodes a regulatory protein in the bacterial cytoplasm (4). LcrH (SycD) binds to YopB and YopD and has been implicated in promoting the type III secretion of the two polypeptides (53). Mutations in class III genes, i.e., any of the 21 ysc genes that encode the type III machinery, abolish yopQ expression in the presence and absence of calcium (2).
The lcrQ gene specifies a 12-kDa type III secretion substrate and is needed to inhibit yop gene expression when the type III pathway of Y. pseudotuberculosis is not induced (43, 48). Overexpression of lcrQ abolishes yop expression even under conditions that induce type III secretion (43, 48). Unlike Y. pseudotuberculosis and Y. pestis, each of which harbors a single lcrQ gene, the Y. enterocolitica virulence plasmid encodes two lcrQ homologs, yscM1 and yscM2 (52). Knockout mutations in yscM1 and yscM2, but not single deletions, cause mutant Yersinia strains to overexpress yop genes (52). Overexpression of each gene alone (yscM1 or yscM2) is sufficient to repress the synthesis of Yops (7, 52). Thus, yscM1 and yscM2 must display overlapping and at least partially redundant functions (52). yscM1 and yscM2 are expressed in the presence of Ca2+ (+Ca2+ conditions); however, their protein products are secreted only when calcium has been depleted from the culture medium (7, 43). A regulatory function of yscM1 and yscM2 for the expression and secretion of YopQ has hitherto not been examined.
Previous work on the regulation of yop expression suggested a mechanism whereby the protein product of lcrQ blocks the transcription of yop genes (7, 43, 52). Once the type III machinery is assembled and functional, LcrQ (YscM1 and YscM2) is depleted from the bacterial cytoplasm and transcription of yop genes is activated (43, 52). This model resembles the transcriptional control of flagellar biosynthesis, a type III pathway that is regulated by a sigma factor (sigma-28 or FliA) for RNA polymerase and by flgM (34). The flgM gene product functions as an anti-sigma factor and controls transcription by sequestration of FliA from RNA polymerase. As Salmonella enterica serovar Typhimurium completes the flagellar hook and basal body, FlgM is depleted from the cytoplasm by secretion, thereby allowing FliA-mediated transcription of flagellin genes (34). During tissue culture infection, Y. pseudotuberculosis LcrQ and Y. enterocolitica YscM1 are injected into the cytosol of eukaryotic cells (7). YscM1 and YscM2 both bind to the bacterial chaperone SycH, which is required for the effective type III transport of YopH and YscM1 (7). A sycH mutant is defective in both YopH and YscM1 targeting, and the overall expression of yop genes during infection is decreased (7). These and other observations suggest that SycH plays an important role in regulating yop expression (7, 56).
A requirement of class II genes for LcrQ function, yopD in Y. pestis and lcrH in Y. pseudotuberculosis, has been reported previously (48, 55). Y. enterocolitica YopD and LcrH have been suggested elsewhere to regulate expression of yopQ by a posttranscriptional mechanism that involves binding of YopD and LcrH to a −45 to +45 nucleotide sequence of yopQ mRNA (2). Δ(yopD), Δ(lcrH), and Δ(yscM1 yscM2), but not Δ(sycH), mutations allow mutant Yersinia to bypass the requirement for an amino acid signal to catalyze type III secretion during tissue culture infection (37). In this report we sought to determine whether Y. enterocolitica yscM1 and yscM2 operate at the same step of regulation as do yopD and lcrH. It is shown that yopD and lcrH are required for yscM1 and yscM2 function and that all four genes act synergistically at the same regulatory step. Further, SycH binding to the protein products of yscM1 and yscM2 can activate yop gene expression. Reverse transcription-PCR (RT-PCR) analysis of yopQ mRNA suggests that yscM1 and yscM2 have a minimal effect on transcriptional inhibition. Experiments that fused yopQ and yopE gene sequences to the npt (neomycin phosphotransferase) reporter suggest that yscM1 and yscM2 regulate expression at a posttranscriptional step. The 178-nucleotide 5′ untranslated region (UTR) of yopQ mRNA is sufficient to confer yscM1- and yscM2-mediated regulation on the fused reporter, as is the 28-nucleotide UTR of yopE. Mutations in nucleotide sequence 5′-AUAAA-3′, which is found in many 5′ yop UTRs, either reduced or abolished yscM1- and yscM2-mediated regulation, suggesting that the conserved sequence element targets transcripts for YopD-, LcrH-, YscM1-, YscM2-, and SycH-mediated regulation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this study are summarized in Table 1. Y. enterocolitica strains EC3 Δ(yopD lcrH), EC4 Δ(yopD yscM1 yscM2), EC6 Δ(yopD lcrH yscM1 yscM2), and EC7 Δ(lcrH yscM1 yscM2) were constructed by allelic exchange by using the suicide plasmids pVLΔyopD, which carries a frameshift-stop codon mutant allele of yopD, and pCT129, which carries a deletion mutant allele of lcrH (7, 15, 36, 38, 39). Strain EC3 was generated through mating of Escherichia coli S17-1 pVLΔyopD with Y. enterocolitica CT133. Selection of merodiploids was performed on Luria-Bertani agar supplemented with nalidixic acid (35 μg/ml) and chloramphenicol (20 μg/ml) and incubation at 26°C. Double crossovers were selected by incubation on Luria-Bertani agar supplemented with nalidixic acid (35 μg/ml) and 5 mM sucrose. Mutants were screened by inducing type III secretion in tryptic soy broth (TSB) supplemented with 5 mM EGTA at 37°C. Protein was precipitated with trichloroacetic acid (TCA) and detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. Yersinia strains EC4, EC6, and EC7 were generated by mating E. coli S17-1 carrying pVLΔyopD or pCT129 with Y. enterocolitica EC2 or EC4, respectively.
TABLE 1.
Bacterial strains and plasmids
| Designation | Property | Reference |
|---|---|---|
| Y. enterocolitica strains | ||
| W22703 | O:9 serotype; pYVe227 Nalr; wild-type isolate | 14 |
| CT133 | Δ(lcrH) Nalr | 2 |
| EC2 | Δ(yscM1 yscM2) Nalr | 7 |
| EC3 | Δ(yopD lcrH) Nalr | This study |
| EC4 | Δ(yopD yscM1 yscM2) Nalr | This study |
| EC5 | Δ(yscM2) Nalr | 7 |
| EC6 | Δ(yopD lcrH yscM1 yscM2) Nalr | This study |
| EC7 | Δ(lcrH yscM1 yscM2) Nalr | This study |
| MC3 | Δ(yopQ) Nalr | 3 |
| VTL1 | Δ(yopN) Nalr | 36 |
| VTL2 | Δ(yopD) Nalr | 38 |
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 28 |
| S17-1 | λpir+ | 16 |
| Plasmids | ||
| pCT129 | pLC28 derivative, lcrH deletion construct | 2 |
| pDA183 | yopQ pro-QUTR-npt Cmr | 3 |
| pDA184 | yopQ pro-QUTR-yopQ (1-15aa)-npt Cmr | 3 |
| pDA208 | yopQ pro-QUTR-yopQ (1-10aa)-npt Cmr | 3 |
| pDA209 | yopQ pro-QUTR-yopQ-nptUTR-npt Cmr | 3 |
| pDA218 | yopQ pro-QUTR-yopQ Cmr | 3 |
| pDA219 | tac promoter fused to yopQ; Cmr | 3 |
| pDA243 | yopQ pro-QUTR-yopQ-npt Cmr | 3 |
| pDA259 | tac promoter fused to gst; Cmr | 7 |
| pDA325 | tac promoter fused to lcrH; Cmr | 2 |
| pDA330 | yopQ pro-nptUTR-npt Cmr | 2 |
| pEC52 | yopQ pro-QUTR(−178-−148)-nptUTR-npt Cmr | This study |
| pEC53 | yopQ pro-QUTR(−178-−120)-nptUTR-npt Cmr | This study |
| pEC54 | yopQ pro-QUTR(−178-−88)-nptUTR-npt Cmr | This study |
| pEC55 | yopQ pro-QUTR(−178-−58)-nptUTR-npt Cmr | This study |
| pEC56 | yopQ pro-QUTR(−178-−28)-nptUTR-npt Cmr | This study |
| pEC57 | yopQ pro-QUTR(−178-−12)-nptUTR-npt Cmr | This study |
| pEC58 | yopQ pro-QUTR(−178-−1)-nptUTR-npt Cmr | This study |
| pEC70 | yopQ pro-QUTR(−150-−1)-nptUTR-npt Cmr | This study |
| pEC71 | yopQ pro-QUTR(−120-−1)-nptUTR-npt Cmr | This study |
| pEC72 | yopQ pro-QUTR(−90-−1)-nptUTR-npt Cmr | This study |
| pEC73 | yopQ pro-QUTR(−60-−1)-nptUTR-npt Cmr | This study |
| pEC74 | yopQ pro-QUTR(−30-−1)-nptUTR-npt Cmr | This study |
| pEC80 | yopQ pro-yopEUTR-npt Cmr | This study |
| pEC83 | npt pro-nptUTR-npt Cmr | This study |
| pEC84 | npt pro-QUTR-npt Cmr | This study |
| pEC102 | yopE pro-nptUTR-npt Cmr | This study |
| pEC112 | yopQ pro-nptUTR(yopEUTR hyb)-npt Cmr | This study |
| pEC138 | yopQ pro-yopEUTR(A−11C)-npt Cmr | This study |
| pEC139 | yopQ pro-yopEUTR(A−10C)-npt Cmr | This study |
| pEC140 | yopQ pro-yopEUTR(T−9G)-npt Cmr | This study |
| pEC141 | yopQ pro-yopEUTR(A−8C)-npt Cmr | This study |
| pEC142 | yopQ pro-yopEUTR(A−7C)-npt Cmr | This study |
| pEC143 | yopQ pro-yopEUTR(A−6C)-npt Cmr | This study |
| pEC144 | yopQ pro-yopEUTR(T−5G)-npt Cmr | This study |
| pEC145 | yopQ pro-yopEUTR Δ(−11-−5)-npt Cmr | This study |
| pEC148 | yopQ pro-nptUTR(yopQUTR hyb)-npt Cmr | This study |
| pEC260 | tac promoter fused to gst; Tetr | 7 |
| pEC345 | tac promoter fused to yscM1; Cmr | 7 |
| pEC346 | tac promoter fused to gst-yscM1; Cmr | 7 |
| pEC347 | tac promoter fused to gst-yscM1; Tetr | 7 |
| pEC348 | tac promoter fused to yscM2; Cmr | 7 |
| pEC349 | tac promoter fused to gst-yscM2; Cmr | 7 |
| pEC350 | tac promoter fused to gst-yscM2; Tetr | 7 |
| pKR12 | tac promoter fused to yopD; lcrH Cmr | This study |
| pVLΔyopD | pLC28 derivative; yopD deletion construct | 38 |
Plasmid pEC52 was constructed through a three-way ligation of pDA330 vector backbone liberated with EcoRI and BamHI to a yopQ promoter sequence containing additional bases (−178 to −148 positions) of the 5′ yopQ UTR generated with the primers YopQ1 (3) and YopQUTR7, 5′-AACATATGACTCCGTGACGTTGCTCATT-3′, yielding a fragment with 5′ EcoRI and 3′ NdeI ends and a fragment containing the 5′ UTR of neomycin phosphotransferase (npt) linked 5′ to the npt open reading frame (ORF) generated with the primers Npt1Nde, 5′-AACATATGATCAAGAGACAGGATGAGGAT-3′, and Npt3 (3), yielding a fragment with 5′ NdeI and 3′ BamHI ends. pEC53 to pEC57 were generated through ligation of the pEC52 vector backbone liberated with EcoRI and NdeI combined with 5′ EcoRI-3′ NdeI yopQ promoter fragments containing 3′ truncations of the 5′ yopQ UTR generated through combination of the primer YopQ1 with the following primers: YopQUTR6 (pEC53), 5′-AACATATGATGCATCGGAATATTTCAAG-3′; YopQUTR5 (pEC54), 5′-AACATATGAAAATAGAATATCTACTCTCAATG-3′; YopQUTR4 (pEC55), 5′-AACATATGTGTATAATTAAACTCACTCCGTA-3′; YopQUTR3 (pEC56), 5′-AACATATGACTACACTATTTAATAACCGTCA-3′; and YopQUTR2 (pEC57), 5′-AACATATGAAAATTTACTTTATAAACTACACTAT-3′. pEC58 was generated from the primer YopQUTR1, 5′-AAGGTACCAGTGACTACTCCAAAATTTACTT-3′, yielding a fragment with a 3′ KpnI site. This fragment was ligated with a fragment generated from the primers Npt1Kpn and Npt3. Constructs containing 5′ truncations of the 5′ yopQ UTR were constructed through a three-way ligation of pDA330 vector backbone liberated with EcoRI and BamHI to a yopQ promoter fragment generated with the primers YopQ1 and YopQUTR8N, 5′-AACATATGTTATTTATTTTAAAGCTACTGATGT-3′, yielding a 5′ EcoRI-3′ NdeI yopQ promoter fragment. Second fragments with 5′ NdeI and 3′ BamHI ends were generated by using pEC58 as a template for PCRs that utilized a common (3′) primer, Npt3, in combination with the primers YopQUTR9 (5′-AACATATGGTGGAGAATACTTGAAATATTCC-3′), YopQUTR10 (5′-AACATATGGGGGAGCTCATTGAGAGTAG-3′), YopQUTR11 (5′-AACATATGTTTATAAATTACGGAGTGAGTTTA-3′), YopQUTR12 (5′-AACATATGCACTCGTAGTGACGGTTATTA-3′), and YopQUTR13 (5′-AACATATGGTTTATAAAGTAAATTTTGGAGTAG-3′) to yield pEC70 to pEC74, respectively. pEC80 was constructed through a three-way ligation of pDA183 vector backbone liberated with EcoRI and NdeI to a yopQ promoter sequence generated with the primers YopQ1 and YopQUTR8K, yielding a fragment with 5′ EcoRI and 3′ KpnI ends, and a set of oligonucleotides, E1 (5′-CTTGTTTTAATAGCCAAGGGAATAAATAGTCCA-3′) and E2 (5′-TATGGACTATTTATTCCCTTGGCTATTAAAACAAGGTAC-3′), that contain the yopE UTR between 5′ KpnI and 3′ NdeI restriction sites. pEC138 to pEC145 were constructed through ligation of oligonucleotide pairs into pEC80 vector backbone liberated with KpnI and NdeI. All sets of oligonucleotides were identical to E1 and E2 with the exception of the transversion substitutions at positions indicated in Table 1. pEC83 was generated through the ligation of pDA183 vector backbone liberated with EcoRI and NdeI combined with a 5′ EcoRI-3′ NdeI npt promoter fragment containing the npt UTR generated from NptProm5′Eco, 5′-AAGAATTCGCGCAAGGGCTGCTAAAG-3′, and Npt3. pEC84 was generated through three-way ligation of pDA183 vector backbone liberated with EcoRI and NdeI to npt promoter sequence generated from primers NptProm5′Eco and NptProm-UTR3′Bgl, 5′-AAAGATCTTGATCCCCTGCGCCATCAG-3′, yielding a 5′ EcoRI and 3′ BglII fragment, and yopQ UTR generated with primers YopQUTR15, 5′-AAAGATCTTCATATAAACAATGAGCAACGTC-3′, and YopQUTR16, 5′-AACATATGAGTGACTACTCCAAAATTTACTT-3′, yielding a 5′ BglII and 3′ NdeI fragment. pEC102 was generated through ligation of pDA330 vector backbone liberated with EcoRI and KpnI to a yopE promoter sequence generated from primers YopE1 and YopEproREVKpn, 5′-AAGGTACCAGGTTATCTTAGTGGGAAAATAG-3′, yielding a 5′ EcoRI and 3′ KpnI fragment. pEC112 and pEC148 were constructed through ligation of a pDA330 backbone liberated with KpnI and BamHI combined with 5′ KpnI-3′ BamHI 5′ npt UTR mutations and an npt ORF generated through combination of Npt3 with the following primers: pEC112, Npt(QUTR)hyb, 5′-AAGGTACCTGACTGACTGATTATAAAACAGGATGAGGATCGTTTCG-3′, and pEC148, Npt(EUTR)hyb2, 5′-AAGGTACCTGACTGACTGATCAAGAGACAGGATGAGGAATAAATCGCATGATTGAACAAGATGG-3′. pKR12 was generated by three-way ligation of pEC345 (6) vector backbone liberated with NdeI and BamHI to a yopD coding sequence generated through primers YopDstart, 5′-AACATATGACATATAAATATCAAGACAGACAG-3′, and YopDstop, 5′-AAGGATCCGTCAGACAACACCAAAAGCGG-3′, yielding a 5′ NdeI and 3′ BamHI fragment with a 5′ BglII and 3′ BamHI fragment generated from primers LcrHSDBgl, 5′-AAAGATCTAGGTAATTATGCAACAAGAGAC-3′, and LcrHstop, 5′-AAGGATCCTCATGGGTTATCACCGCACT-3′, containing the lcrH gene with its native ribosome binding sequence. Ligations were transformed into E. coli DH5α and screened by restriction analysis. Plasmid constructs were sequenced with fluorescently labeled dideoxy chain termination PCRs (University of Chicago CRC DNA Sequencing Facility). All DNA cloning manipulations were performed with pCR2.1 (Invitrogen) in E. coli DH5α. Plasmid constructs were electroporated into yersiniae, and transformants were selected on TSB agar supplemented with nalidixic acid (35 μg/ml) and chloramphenicol (20 μg/ml) and grown at 26°C (8). Yersinia strains containing the expression vectors pEC260, pEC347, and pEC350 were also cultured with a supplement of tetracycline (5 μg/ml).
Yersinia secretion.
Yersiniae were grown to stationary phase overnight in TSB supplemented with nalidixic acid (35 μg/ml) (and chloramphenicol [20 μg/ml] where needed) and diluted 1:50 into 30 ml of fresh TSB medium supplemented with 5 mM CaCl2 or EGTA (±Ca2+) and antibiotic in 125-ml glass Erlenmeyer flasks. Cultures were incubated on a rotary shaker for 2 h at 26°C and induced at 37°C for 3 h. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 1 mM immediately upon temperature shift. Cultures were centrifuged at 15,000 × g for 15 min, and the supernatant was separated from the cell pellet. Proteins in both fractions were precipitated on ice with 10% TCA, centrifuged at 15,000 × g for 15 min at 4°C, washed in acetone, and suspended in sample buffer. All samples were analyzed by SDS-15% PAGE. Following electrotransfer to a polyvinylidene difluoride membrane, proteins were examined by immunoblotting with purified rabbit polyclonal antisera raised against Yersinia antigens and glutathione S-transferase (Schneewind laboratory), anti-NptII (U.S. Biological), and anti-Cat (Sigma). Horseradish peroxidase-conjugated anti-rabbit immunoglobulin G was used to generate chemiluminescent signals that were visualized on a Fluorchem 8800 Imaging System (Alpha Innotech).
Expression experiments.
Yersiniae transformed with both reporter and expression constructs were grown to stationary phase overnight in TSB supplemented with chloramphenicol (20 μg/ml) and tetracycline (5 μg/ml) and diluted 1:50 into 4 ml of fresh TSB supplemented with 5 mM EGTA (−Ca2+) and antibiotics in 10-ml borosilicate glass tubes. Cultures were incubated on a wheel for 2.5 h at 26°C and induced at 37°C for 1.5 h. Expression constructs were induced at time of temperature shift with IPTG, added to 1 mM. Readings of optical density at 600 nm (OD600) were taken for standardization, and 1-ml aliquots of cultures were precipitated on ice with 10% TCA. Samples were centrifuged at 15,000 × g for 15 min at 4°C, washed with acetone, solubilized in sample buffer, and loaded on SDS-polyacrylamide gels based on correction for OD600. Samples were analyzed by immunoblotting. Timed promoter experiments were performed in 125-ml glass Erlenmeyer flasks containing 50 ml of TSB supplemented with 5 mM CaCl2 with a 1:50 dilution of stationary-phase culture and appropriate antibiotics. Cultures were immediately incubated at 37°C for the time course. Two 1-ml aliquots were collected at each time point for measurement of OD600 and TCA precipitation. Precipitated proteins were processed for analysis as described above, and all quantification of immunoreactive signals was performed with Fluorchem software (Alpha Innotech).
RT-PCR analysis.
Yersinia cultures were prepared as explained under “Yersinia secretion” above except that the culture volumes were increased to 500 ml. Cultures were centrifuged at 15,000 × g for 15 min, and the supernatant was separated from the cell pellet. The cell pellet was resuspended in 30 ml of lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl). Aliquots containing proteins from each fraction were precipitated on ice with 10% TCA, centrifuged at 15,000 × g for 15 min at 4°C, washed in acetone, and suspended in sample buffer. Protein fractions were analyzed by SDS-PAGE and immunoblotting. The pellet fraction was next subjected to lysis with a French pressure cell (16,000 lb/in2). One-hundred-microliter aliquots were used for total RNA isolation with the SV Total RNA isolation kit (Promega) according to the manufacturer's protocol. Eluted RNA samples were digested with 2 U of DNase (Promega) for 1 h at 37°C. Samples were extracted with phenol-chloroform, ethanol precipitated, and suspended in 90 μl of nuclease-free H2O. OD260/280 measurements were taken to calculate the concentration of each sample and for standardization. RT reactions (25-μl reaction mixtures) were performed with 0.6 μg of total RNA in the presence or absence of reverse transcriptase (Promega) and primer YopQcod2 (100 pmol), 5′-AAGGATCCTCATCCCATAATACATTTTTGAT-3′, according to protocol. Each reaction mixture was diluted to 100 μl in H2O, and 20 μl of the sample was used as template for PCRs (100-μl reaction mixtures) with primers 5′ Q2Bgl5 (20 pmol), 5′-AAAGATCTTGGAGTAGTCACTATGTTTATT-3′, and 3′ YopQcod2 (20 pmol) (40 cycles, 1-min extension at 72°C) (Qiagen). Virulence plasmid DNA (pYVe227) was used as a positive-control template for PCR. The 549-bp yopQ ORF was analyzed by 2% agarose gel electrophoresis and compared to a molecular size standard, a 1-kb DNA ladder (Invitrogen). The gel was stained with ethidium bromide and visualized on a UV light table.
RESULTS
yscM1, yscM2, yopD, and lcrH act synergistically to regulate yopQ expression.
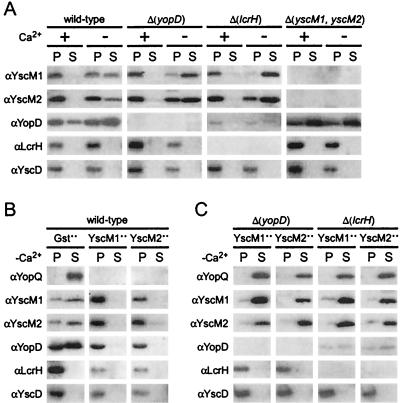
Y. enterocolitica W22703 (wild type) was grown at 37°C in TSB supplemented with either 5 mM calcium chloride (+Ca2+) or 5 mM EGTA (−Ca2+). Cultures were centrifuged, and the extracellular medium was separated (supernatant) from the bacterial sediment (pellet). Protein in both fractions was precipitated with TCA and analyzed by SDS-PAGE and immunoblotting (Fig. 1A). Wild-type yersiniae secreted YopD both in the presence and in the absence of calcium. Although YopE was synthesized in the presence of calcium, yersiniae did not secrete the polypeptide unless calcium was chelated from the growth medium. YopQ synthesis and secretion occurred only in the absence of calcium.
FIG. 1.
Regulation of yop expression in Y. enterocolitica. Y. enterocolitica strains were cultured in TSB supplemented with 5 mM CaCl2 (+Ca2+) or 5 mM EGTA (−Ca2+) for 2 h at 26°C and then induced for type III secretion at 37°C for 3 h. Cultures were centrifuged to separate the bacterial pellet (P) from the culture supernatant (S). Proteins in both fractions were precipitated with TCA and analyzed by SDS-PAGE and immunoblotting. (A) Protein of Y. enterocolitica strain W22703 (wild type) was analyzed by immunoblotting for the synthesis and secretion of YopQ, YopE, and YopD. (B and C) Synthesis and secretion of YopQ in various Y. enterocolitica strains were analyzed by immunoblotting with antisera raised against purified YopQ. Plasmid-encoded genes were overexpressed (++) by adding 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon.
Knockout mutations in class II genes (yopD and lcrH) allowed yopQ expression in the presence of calcium; however, the mutant yersiniae did not secrete YopQ polypeptide under these conditions. Knockout mutations in yscM1 and yscM2 alone resulted in an intermediate phenotype, as only small amounts of YopQ were detected in bacteria grown in the presence of calcium. A strain carrying deletions in both genes, the Δ(yscM1 yscM2) strain, displayed a class II phenotype with yopQ expression but no secretion in the presence of calcium. The knockout mutation of sycH did not affect the calcium regulation of yopQ expression or YopQ secretion (data not shown). In contrast, overexpression of sycH from the IPTG-inducible tac promoter resulted in a class II phenotype (Fig. 1B).
We tested whether the phenotype of yopQ expression without YopQ secretion in the presence of calcium was altered in mutant strains lacking multiple class II genes. The combined deletions of Δ(yopD) and Δ(lcrH); Δ(yopD) and Δ(yscM1 yscM2); Δ(lcrH) and Δ(yscM1 yscM2); or Δ(yopD), Δ(lcrH), and Δ(yscM1 yscM2) allowed for the expression of yopQ in the presence of calcium. To our surprise, some secretion of YopQ could be observed when the mutant yersiniae were grown in the presence of calcium. This phenotype was distinct, as overexpression of yopQ from an inducible promoter in the Δ(yopQ) strain did not result in secretion of YopQ in the presence of calcium (Fig. 1C). It should also be noted that this phenotype was distinct from the secretion of YopQ by class I mutant strains, as Δ(yopN) mutants quantitatively transport YopQ into the extracellular medium in the presence of calcium (Fig. 1B).
yopD and lcrH are required for yscM1 and yscM2 function.
Three of the four class II gene products are transported by the Yersinia type III pathway. We tested whether knockout mutations in yopD, lcrH, yscM1, and yscM2 affected the expression or the type III transport of class II gene products. Wild-type yersiniae secreted YscM1 and YscM2 in the absence but not in the presence of environmental calcium ions (Fig. 2A). As expected, YopD was secreted both in the presence and in the absence of calcium, whereas LcrH was not secreted (37). Knockout mutations of yopD or lcrH reduced neither Yersinia secretion nor the concentration of YscM1 and YscM2. Consistent with previous reports, the deletion of lcrH (sycD) caused a significant reduction in the bacterial concentration of YopD (53). Δ(yscM1 yscM2) mutant yersiniae synthesized greater amounts of YopD and LcrH than did the wild-type strain (Fig. 2A). It seems that the phenotype of Δ(yopD) and Δ(yscM1 yscM2) mutant yersiniae cannot be explained as the altered stability or the increased secretion of class II gene products. It is conceivable, however, that the regulatory role of the lcrH gene product involves stabilizing YopD in the cytoplasm of yersiniae.
FIG. 2.
yopD and lcrH are required for the function of YscM1 and YscM2. Y. enterocolitica strains were analyzed for type III secretion as described in the legend to Fig. 1. (A) Y. enterocolitica strains W22703 (wild type), VTL2 [Δ(yopD)], CT133 [Δ(lcrH)], and EC2 [Δ(yscM1 yscM2)] were analyzed by immunoblotting with antisera raised against purified YscM1, YscM2, YopD, LcrH, and YscD. The type III machinery component YscD is located within bacteria. (B) Y. enterocolitica W22703 (wild type) harboring pDA259 (encoding Gst), pEC345 (encoding YscM1), or pEC348 (encoding YscM2) were cultured in −Ca2+. Plasmid-borne genes were overexpressed (++) by adding 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon. (C) Yersinia strains VTL2 [Δ(yopD)] and CT133 [Δ(lcrH)] were transformed with pEC345 and pEC348 and analyzed by immunoblotting.
Previous work showed that the overexpression of LcrQ (YscM1 or YscM2) blocked the expression of yop genes as well as the type III transport of yop gene products (7, 43). We wondered whether knockout mutations in yopD and lcrH affected the ability of YscM1 and YscM2 to prevent type III secretion. Overexpression of yscM1 or yscM2 in wild-type yersiniae abolished the secretion of YscM1, YscM2, and YopD (Fig. 2B). A block in YopQ secretion could not be assessed, as overexpression of yscM1 or yscM2 prevented the expression of yopQ. Overexpression of yscM1 or yscM2 in Δ(yopD) and Δ(lcrH) mutant yersiniae affected neither the expression nor the secretion of YscM1, YscM2, and YopQ (Fig. 3C). Thus, yopD and lcrH are absolutely required for YscM1- and YscM2-mediated repression of yop gene expression.
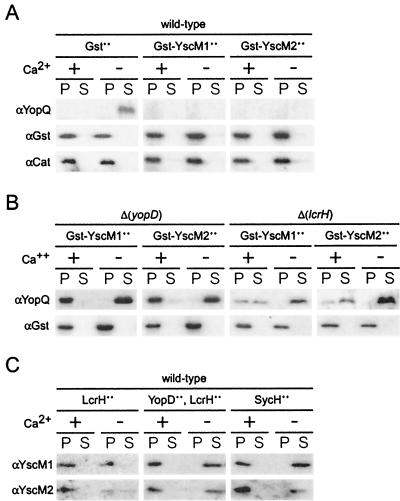
FIG. 3.
Nonsecretable YscM1 and YscM2 also require yopD and lcrH for function. Y. enterocolitica strains were analyzed for type III secretion as described in the legend to Fig. 1. Y. enterocolitica strains W22703 (wild type) (A and C) and VTL2 [Δ(yopD)] and CT133 [Δ(lcrH)] (B) were analyzed by immunoblotting with antisera raised against purified YopQ, Gst, YscM1, YscM2, and Cat. Plasmids pEC260 (encoding Gst), pEC347 (encoding Gst-YscM1), pEC350 (encoding Gst-YscM2), pDA325 (encoding LcrH), pKR12 (encoding LcrH and YopD), and pEC441 (encoding SycH) were transformed into Yersinia strains, and expression was induced by the addition of 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon.
As Δ(yopD) and Δ(lcrH) mutant yersiniae secreted YscM1 and YscM2, the class II phenotype of these variants may be caused by the depletion of YscM1 and YscM2 from the bacterial cytoplasm. If so, overexpression of Gst-YscM1 or Gst-YscM2, two nonsecretable hybrids that regulate the expression of yop genes, should complement the class II phenotype of Δ(yopD) and Δ(lcrH) mutant strains (7). As expected, Gst-YscM1 and Gst-YscM2 blocked the synthesis of YopQ in wild-type yersiniae (Fig. 3A). Overexpression of Gst-YscM1 and Gst-YscM2 in Δ(yopD) mutant yersiniae did not complement the class II phenotype, as YopQ synthesis in the presence of calcium and YopQ secretion in the absence of calcium still occurred (Fig. 3B). This result was consistent with similar studies of LcrQ in Δ(yopD) Y. pestis (55). A similar result was observed when Gst-YscM1 and Gst-YscM2 were analyzed in Δ(lcrH) mutants (Fig. 3B); however, overexpression of Gst-YscM1 and Gst-YscM2 caused the secretion of YopQ in the presence of calcium, a phenotype distinct from the non-gst-fused forms of yscM1 and yscM2. In sum, yopD, lcrH, and yscM1 or yscM2 is each required for the control of yop gene expression at a unique step. Further, yscM1 or yscM2 appears to fulfill overlapping but nonredundant functions and can be inactivated by the binding of the chaperone SycH to YscM1 and YscM2.
Although overexpression of sycH (pEC441) increased the secretion of YscM1 and YscM2 in the absence of calcium, it did not induce the secretion of YscM1 and YscM2 in the presence of calcium (7). Overexpression of sycH did induce the expression of yopQ in the presence of calcium (Fig. 1B), suggesting that the binding of SycH to YscM1 and YscM2 may be sufficient to inactivate the regulatory properties of class II gene products (Fig. 3C). This mechanism of induction is not essential for the regulation of yopQ in the presence of calcium, as Δ(sycH) mutants express yopQ in a manner that is indistinguishable from that of wild-type strains. As a control, overexpression of both yopD and lcrH did not affect the secretion of YscM1 and YscM2 (Fig. 3C). Surprisingly, the overexpression of LcrH alone reduced the secretion of YscM1 and YscM2 in the absence of calcium (Fig. 3C). These results are consistent with the assignment of a regulatory role to lcrH, independent of YopB and YopD stabilization, that coordinates the regulatory circuit with the type III machinery (21-23).
YscM1 and YscM2 regulate yopQ expression at a posttranscriptional step.
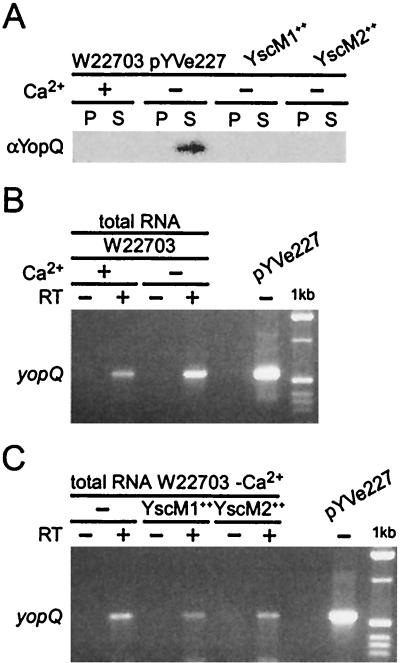
Wild-type Y. enterocolitica W22703 does not express yopQ when cultures are grown at elevated temperatures in the presence of calcium. Previous data suggested that LcrQ (YscM1-YscM2) overexpression blocks the expression of various yop genes at a transcriptional step (43, 52). We tested whether the yopQ mRNA transcript was present in cells grown under these conditions. Y. enterocolitica W22703 (wild type) was grown at 37°C in TSB supplemented with either 5 mM calcium chloride (+Ca2+) or 5 mM EGTA (−Ca2+). Y. enterocolitica W22703 (wild type) harboring low-copy-number plasmids expressing yscM1 (pEC345) or yscM2 (pEC348) under control of the IPTG-inducible tac promoter was grown at 37°C in TSB in the absence of calcium. Cultures were centrifuged, and the extracellular medium was separated (supernatant) from the bacterial sediment (pellet). Protein in both fractions was precipitated with TCA and analyzed by SDS-PAGE and immunoblotting. YopQ was synthesized and secreted in the wild-type strain only in the absence of calcium. Overexpression of yscM1 or yscM2 completely blocked the synthesis of yopQ in the absence of calcium (Fig. 4A). Total RNA was isolated from the pellet fraction of the cultures, and quantitative RT-PCR analysis was performed on each of the strains. Amplification of the yopQ coding sequence demonstrated the presence of yopQ mRNA transcript in Y. enterocolitica W22703 in both the presence and absence of calcium (Fig. 4B). Overexpression of YscM1 or YscM2 had only a slight effect on the concentration of yopQ mRNA transcript (Fig. 4C). pYVe227 virulence plasmid DNA containing yopQ was used as a control for PCR amplifications. Together these results suggest that YscM1 and YscM2 block the expression of yopQ at a posttranscriptional step.
FIG. 4.
Transcription of yopQ mRNA is not blocked by YscM1 and YscM2 overexpression. (A) Y. enterocolitica W22703 (wild type) and strains harboring plasmids pEC345 (encoding YscM1) and pEC348 (encoding YscM2) were analyzed for type III secretion as described in the legend to Fig. 1. Each fraction was analyzed by immunoblotting with antisera raised against purified YopQ. (B and C) Total RNA was isolated from the bacterial sediment (pellet), and RT-PCR analysis of yopQ mRNA was performed (+) or reverse transcriptase was omitted from the reaction mixture (−) prior to PCR amplification. The 549-bp yopQ ORF was analyzed by 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized on a UV light table. The size of the amplicon was compared to a molecular size standard (1 kb). Virulence plasmid DNA (pYVe227) was used as a positive control for the PCR amplification step.
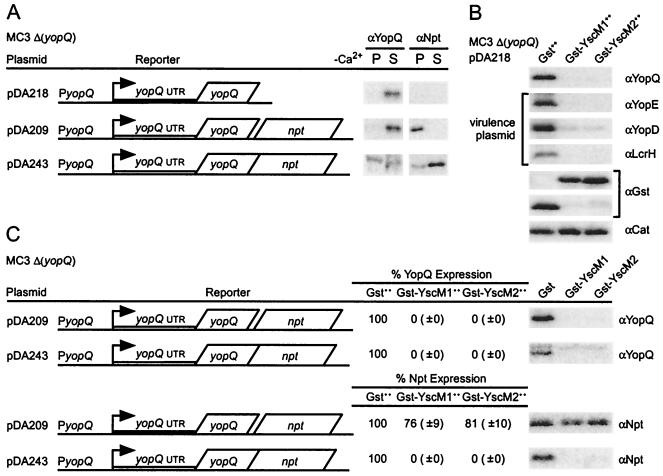
To analyze the regulatory properties of YscM1 and YscM2, plasmid-carried gene fusions were employed that tethered yopQ to neomycin phosphotransferase (npt), a gene that is not a member of the Yersinia virulence regulon (virulon) or type III pathway and that is therefore not regulated by yopD, lcrH, yscM1, and yscM2 (2). Y. enterocolitica MC3 harbors a knockout mutation of yopQ [Δ(yopQ)]. Plasmid pDA218 carries the wild-type yopQ gene. When pDA218 is transformed into strain MC3, the plasmid restores yopQ expression and YopQ secretion (2). pDA209 carries a fusion of yopQ to a promoterless npt gene. Upon induction of the type III pathway, a single transcript specifying two polypeptides, YopQ and Npt, is generated. Finally, pDA243 carries a translational fusion between yopQ and npt. The resulting hybrid, YopQ-Npt, is transported by the type III pathway and synthesized in a manner that is subject to regulation by the Yersinia virulon (Fig. 5A) (3). Overexpression of gst-yscM1 and gst-yscM2, but not of gst alone, abolished the expression of yopQ encoded by pDA218 (Fig. 5B). This result indicates that plasmid-expressed yopQ is regulated in a manner that is indistinguishable from that of virulence plasmid-carried yopQ. Overexpression of gst-yscM1 and gst-yscM2 also abolished the expression of yopQ carried by pDA209, whereas the expression of npt, located at the 3′ end of the pDA209 transcript, was not significantly reduced (Fig. 5C). As a control, the translational hybrid yopQ-npt was regulated in a manner resembling that of wild-type yopQ. These results suggest that, similarly to LcrH and YopD (2), YscM1 and YscM2 also regulate yopQ expression at a posttranscriptional step, as the expression of fused npt is coregulated in translational but not in transcriptional hybrids.
FIG. 5.
Gst-YscM1 and Gst-YscM2 block the expression of plasmid-encoded YopQ at a posttranscriptional step. (A) Y. enterocolitica MC3 [Δ(yopQ)] harboring plasmid pDA218, pDA209, or pDA243 was cultured in −Ca2+ and analyzed for synthesis and secretion of YopQ as described in the legend to Fig. 1 with YopQ- and Npt-specific antisera. (B) Y. enterocolitica MC3 [Δ(yopQ)] harboring pDA218 was transformed with pEC260 (encoding Gst), pEC347 (encoding Gst-YscM1), or pEC350 (encoding Gst-YscM2). The expression of pEC260-, pEC347-, or pEC350-carried genes was induced by the addition of 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon. Protein precipitated from total cultures was analyzed by immunoblotting for the synthesis of YopQ, Gst, or Cat (plasmid encoded) and YopE, YopD, or LcrH (virulence plasmid encoded) with specific antisera. (C) Y. enterocolitica MC3 [Δ(yopQ)] harboring plasmid pDA209 or pDA243 was cultured in −Ca2+ and analyzed for synthesis of YopQ and Npt. Y. enterocolitica MC3 [Δ(yopQ)] harboring pDA209 or pDA243 was transformed with pEC260 (encoding Gst), pEC347 (encoding Gst-YscM1), or pEC350 (encoding Gst-YscM2). The expression of pEC260-, pEC347-, or pEC350-carried genes was induced as described for panel B. Quantification of immunoreactive signals is reported as the percent amount of the Npt signal compared to the signal for bacteria overexpressing Gst.
YscM1- and YscM2-mediated regulation requires the 5′ UTR of yop mRNA.
We sought to identify yopQ nucleotide sequences that were sufficient to confer yopD-, lcrH-, yscM1-, and yscM2-mediated regulation on npt. yopQ was truncated at its 3′ end, and DNA fragments were fused to npt in a manner that allowed translation of yopQ-npt hybrids. Plasmids were transformed into Y. enterocolitica MC3, and class II-mediated yopQ-npt regulation was measured by overexpressing Gst-YscM1 or Gst-YscM2. yopQ-npt expression was calibrated by measuring the concentration of YopQ-Npt in Yersinia cultures expressing Gst alone (100%). Deletion of yopQ codons 16 to 193 diminished the repressive effect of class II genes by a factor of about 5 to 10, as 11 and 17% of YopQ1-15-Npt were detected in extracts with overexpressed Gst-YscM1 and Gst-YscM2, respectively (Fig. 6). These values must be compared with 0% of YopQ1-193-Npt in Yersinia harboring Gst-YscM1 or Gst-YscM2 (Fig. 5C). Further 3′ truncations of yopQ coding sequence did not affect class II regulation, as yopQ1-10-npt and yopQ1-npt displayed similar concentrations of YopQ-Npt in Yersinia expressing Gst-YscM1 and Gst-YscM2. It should be noted that the yopQ1-10-npt hybrid (pDA208) contains sufficient information for the polypeptide to be secreted by the type III pathway, whereas the yopQ1-npt hybrid (pDA183) does not (data not shown) (3, 46). The 178-nucleotide 5′ yopQ UTR (3) appears sufficient to confer class II-mediated repression when appended to transcripts that are not part of the yop virulon (pDA183 in Fig. 6). As a control, plasmid-carried npt encompassing the yopQ promoter and 5′ npt UTR (pDA330) (2) was not subject to Gst-YscM1- and Gst-YscM2-mediated regulation (Fig. 6). Deletion of yopD or lcrH abolished the regulation of 5′ yopQ UTR fusions to npt, indicating that 5′ yopQ UTR fusions repressed reporter synthesis via class II genes.
FIG. 6.
The 5′ UTR of yopQ mRNA is the target of Gst-YscM1- and Gst-YscM2-mediated repression. Y. enterocolitica strains MC3 [Δ(yopQ)], VTL2 [Δ(yopD)], and CT133 [Δ(lcrH)] harboring plasmid pDA183, pDA184, pDA208, or pDA330 were cultured in −Ca2+ and analyzed for synthesis of YopE and Npt. Y. enterocolitica strains were transformed with pEC260 (encoding Gst), pEC347 (encoding Gst-YscM1), or pEC350 (encoding Gst-YscM2). The expression of pEC260-, pEC347-, or pEC350-carried genes was induced by the addition of 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon. Quantification of immunoreactive signals is reported as the percent amount of the Npt signal compared to the signal for bacteria overexpressing Gst.
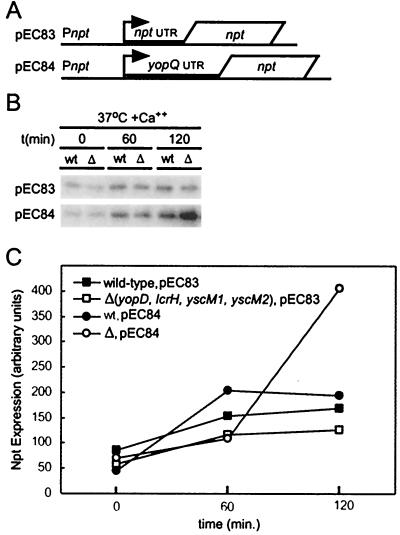
We wished to determine whether class II-mediated regulation at the 5′ yopQ UTR required a yop promoter. We observed that the npt promoter is constitutively expressed in yersiniae (E. D. Cambronne, unpublished data). The npt reporter gene of pEC83 and pEC84 was expressed from the npt promoter. pEC84-encoded transcripts, but not pEC83-encoded transcripts, harbor the 5′ yopQ UTR (Fig. 7A). The expression of pEC83- and pEC84-carried npt in Y. enterocolitica strains W22703 (wild type) and EC6 [Δ(yopD lcrH yscM1 yscM2)] was measured 0, 60, and 120 min after inoculating cultures containing 5 mM calcium chloride and incubation at 37°C (Fig. 7B). Expression of pEC84-carried npt, but not of pEC83-carried npt, was decreased in wild-type yersiniae compared to the class II mutant strain EC6. These data indicated that the yopQ promoter was not required for posttranscriptional repression of yop genes by YopD, LcrH, YscM1, and YscM2 (Fig. 7C).
FIG. 7.
The yopQ promoter is not required for class II-mediated posttranscriptional regulation of yopQ. Y. enterocolitica strains W22703 (wild type) and EC6 [Δ(yopD lcrH yscM1 yscM2)] were transformed with pEC83 or pEC84 (A). Both plasmids encode an npt reporter gene under the control of the npt promoter and either with or without 5′ yopQ UTR sequences. Y. enterocolitica strains were cultured in TSB supplemented with 5 mM CaCl2 (+Ca2+), and the type III virulon was induced at 37°C for 0, 60, and 120 min. Secretion was quenched by precipitating protein from total cultures followed by immunoblot analysis for the synthesis of Npt with specific antisera. Data generated in panel B were quantified and analyzed for panel C with chemiluminescent signals that were visualized on a Fluorchem 8800 Imaging System (Alpha Innotech). wt, wild type.
To test whether the 5′ UTR of other yop genes was also the target of class II gene-mediated regulation, the effect of Gst-YscM1 and Gst-YscM2 on the expression of yopE was analyzed (Fig. 8A). Gst-YscM1 and Gst-YscM2 abolished the expression of wild-type yopE. Plasmid pEC102 carried a fusion of the npt reporter gene to the yopE promoter. The expression of npt was reduced twofold by Gst-YscM1 and Gst-YscM2. However, fusion of the 28-nucleotide 5′ yopE UTR (20) to npt resulted in a greater-than-fivefold repression of npt in the presence of Gst-YscM1 or Gst-YscM2 (Fig. 8A). It therefore appears that the 5′ UTR of yopE may also be the target of class II-mediated gene regulation. A comparison of reported 5′ yop UTR nucleotide sequences revealed the presence of 5′-AUAAA-3′ in yopQ, yopE, and yopH (3, 6, 20) as well as in positions 5′ to the translational start sites of other yop genes (Fig. 8B). This sequence element was present four times in the 5′ yopQ UTR and once in the 5′ yopE UTR. Furthermore, the 5′ npt UTR (47) did not harbor this sequence element. Although the precise function of the 5′-AUAAA-3′ element is still unclear, the data are consistent with YopD, LcrH, YscM1, and YscM2 recognizing a feature of yop mRNAs to control gene expression at a posttranscriptional step (Fig. 8B).
FIG. 8.
The 5′ UTR of yopE mRNA is the target of Gst-YscM1- and Gst-YscM2-mediated repression. (A) Y. enterocolitica strain MC3 [Δ(yopQ)] harboring plasmid pEC80 or pEC102 was cultured in −Ca2+ and analyzed for synthesis of YopE and Npt. Y. enterocolitica strains were transformed with pEC260 (encoding Gst), pEC347 (encoding Gst-YscM1), or pEC350 (encoding Gst-YscM2). The expression of pEC260-, pEC347-, or pEC350-carried genes was induced by the addition of 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon. Quantification of immunoreactive signals is reported as the percent amount of the Npt signal compared to the signal for bacteria overexpressing Gst. (B) A conserved nucleotide sequence was identified in the 5′ UTR of yopQ, yopE, yopH, yopM, yopP, yopT, yscM1, yscM2, and sycH transcripts. Conserved sequences of individual genetic loci with nucleotide positions (relative to the translational AUG start codon) are indicated in the box with a consensus sequence listed at the bottom. Small boxes represent predicted ribosome binding sites.
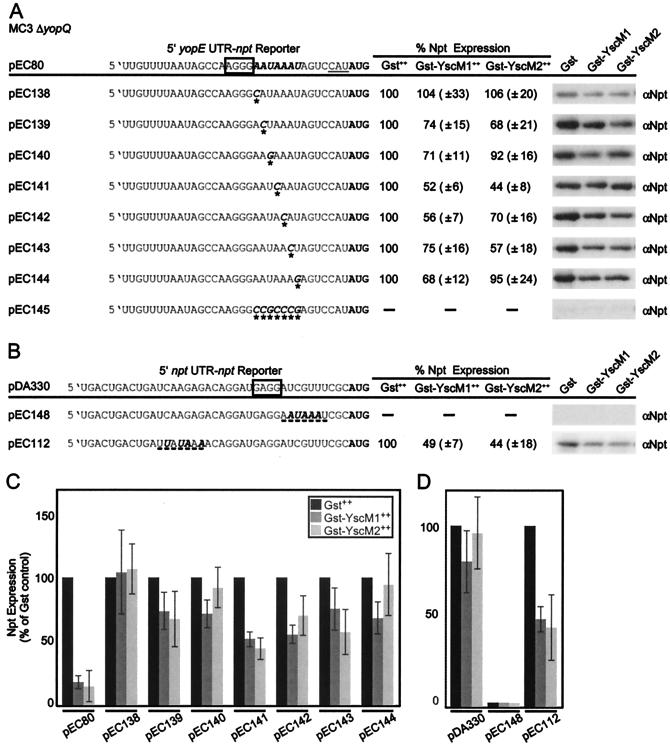
Because of the relatively short 28-nucleotide 5′ yopE UTR and single-copy 5′-AUAAA-3′, we chose pEC80 as a substrate for mutagenesis that introduced single nucleotide transversion substitutions in the predicted sequence motif, including positions both −1 and +1 relative to the 5′-AUAAA-3′ sequence (Fig. 9A). As the 5′ UTR of yop genes is AU rich, cytidyl was chosen to replace adenyl nucleotides and guanyl was chosen to replace uridyl nucleotides. Gst-YscM1 and Gst-YscM2 expression in each strain harboring the mutant reporter constructs failed to block expression of the npt reporter to the levels of the wild-type sequence control (pEC80). These results indicate that the sequence 5′-AAUAAAU-3′ is necessary for class II gene-mediated regulation (Fig. 9C). Replacement of 5′-AAUAAAU-3′ (pEC80) with 5′-CCGCCCG-3′ (pEC145) abolished expression of the reporter altogether even in the Gst-expressing control strain (Fig. 9A). It appears that the position of the AU-rich sequence between the Shine-Dalgarno binding site for 16S rRNA and the AUG start codon is also essential for gene expression in Yersinia.
FIG. 9.
The sequence 5′-AAUAAAU-3′ in the 5′ UTR of yopE mRNA is necessary for Gst-YscM1- and Gst-YscM2-mediated repression. (A and B) Y. enterocolitica strain MC3 [Δ(yopQ)] harboring plasmids pEC112, pEC138 to pEC145, and pEC148 was cultured in −Ca2+ and analyzed for synthesis of Npt. Y. enterocolitica strains were transformed with pEC260 (encoding Gst), pEC347 (encoding Gst-YscM1), or pEC350 (encoding Gst-YscM2). The expression of pEC260-, pEC347-, or pEC350-carried genes was induced by the addition of 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon. Quantification of immunoreactive signals is reported as the percent amount of the Npt signal compared to the signal for bacteria overexpressing Gst. Asterisks indicate nucleotide positions in each construct where transversion substitutions were introduced. Dashed lines indicate sites of positional introduction of conserved sequence in the 5′ npt UTR of the 5′ yopE UTR (pEC148) and the −28 5′ yopQ UTR (pEC112) with substituted nucleotides indicated in boldface italics. Wild-type 5′ yopE UTR (pEC80) and 5′ npt UTR (pDA330) are included for reference. The underlined sequence CAU in pEC80 represents insertion of nucleotides at the NdeI fusion site. Small boxes represent predicted ribosome (16S rRNA) binding sites (Shine-Dalgarno box). (C and D) Graphical representation of the data generated in panels A and B, respectively, with quantified data from pEC80 and pDA330 included for reference. All chemiluminescent signals were visualized, quantified, and analyzed on a Fluorchem 8800 Imaging System (Alpha Innotech).
To examine whether the conserved sequence element is sufficient to impose yscM1- and yscM2-mediated regulation on unrelated transcripts, 5′-AUAAA-3′ was inserted into the npt UTR at −10, i.e., following the Shine-Dalgarno box as in yopE, or at −27, i.e., one of the motif locations within the 5′ yopQ UTR (Fig. 9). Positional numbers refer to nucleotide positions upstream (−) or downstream (+) of the AUG start codon. Gst-YscM1 and Gst-YscM2 expression in each strain harboring the hybrid UTR constructs was measured as expression of the npt reporter. The −10 insertion in pEC148 prevented npt expression even in the Gst-expressing control strain, again demonstrating that nucleotide sequence substitutions between the Shine-Dalgarno box and the AUG have dramatic effects on gene expression. The introduction of 5′-AUAAA-3′ at −27 in pEC112 imposed a twofold regulation on the npt reporter due to overexpression of Gst-YscM1 or Gst-YscM2 (Fig. 9D). These results suggest that introduction of the 5′-AUAAA-3′ element in the 5′ UTR of npt genes can lead to the acquisition of class II gene-mediated regulation.
We asked whether the conserved sequence elements contained in the 5′ yopQ UTR are also necessary to confer class II gene-mediated regulation. Successive truncations of yopQ UTR sequences in increments of ∼30 nucleotides were introduced in both the 5′ and 3′ directions (Fig. 10A). These fragments were fused at their respective 3′ ends to the 5′ end of the 5′ npt UTR and npt ORF. A 3′-end truncation was also constructed that simply removed the sequence containing the predicted ribosome binding site (−12 to −1) (pEC57). Plasmids were transformed into Y. enterocolitica MC3 Δ(yopQ) carrying plasmids for the overexpression of gst-yscM1, gst-yscM2, and gst. npt expression was calibrated by measuring the concentration of Npt in Yersinia cultures expressing Gst alone (100%). Appending the 178-nucleotide 5′ yopQ UTR to the npt UTR repressed the expression of npt by about fourfold (Gst-YscM1 or Gst-YscM2) compared to the Gst control (Fig. 10B). Almost all of the truncations of 5′ yopQ UTR remained sufficient to cause three- to fourfold regulation of expression of npt (Fig. 10B). One exception was construct pEC71 (−120 to −1), which failed to be synthesized even in the control strain expressing Gst, a phenotype that we do not yet understand. Truncating the yopQ UTR to 30 nucleotides at the 3′ end reduced repression to a twofold effect. Thus, the presence of multiple copies of 5′-AUAAA-3′ in the yopQ UTR has additive effects on the repression of yop genes; however, the presence of a single copy of 5′-AUAAA-3′ in the UTR appears sufficient to elicit a twofold YscM1- and YscM2-mediated repression. These observations also explain the differences in yopQ and yopE expression in the presence of calcium. As yopE harbors a single 5′-AUAAA-3′ element in the UTR, the yopE gene is much less repressed than yopQ, containing at least four copies of 5′-AUAAA-3′.
FIG. 10.
The 5′-AUAAA-3′ elements of the 5′ UTR of yopQ are sufficient to impose Gst-YscM1- and Gst-YscM2-mediated repression when fused to the 5′ UTR of npt. (A) Illustration of the truncation strategy of the 5′ yopQ UTR. Sequences highlighted in boldface are predicted to be necessary for yscM1- and yscM2-mediated regulation. Arrows indicate the inclusion of conserved sequence (bold arrow) with truncations in both the 5′ and 3′ directions. Each truncation was appended to the 5′ end of the 5′ npt UTR. (B) Y. enterocolitica strain MC3 [Δ(yopQ)] harboring plasmids pEC52 to pEC58 and pEC70 to pEC74 was cultured in −Ca2+ and analyzed for synthesis of Npt. Y. enterocolitica strains were transformed with pEC260 (encoding Gst), pEC347 (encoding Gst-YscM1), or pEC350 (encoding Gst-YscM2). The expression of pEC260-, pEC347-, or pEC350-carried genes was induced by the addition of 1 mM IPTG when shifting the temperature of bacterial cultures to induce the type III virulon. Quantification of immunoreactive signals is reported as the percent amount of the Npt signal compared to the signal for bacteria overexpressing Gst.
DISCUSSION
LcrQ (YscM1 and YscM2) has previously been hypothesized to regulate the expression of yop genes in Yersinia species at the level of transcription (7, 43, 52). This model was based on the following observations. (i) Knockout mutations in lcrQ (yscM1 and yscM2) increased the expression of yop genes compared to that in wild-type yersiniae (7, 43, 48, 52). (ii) Overexpression of yscM1 and yscM2 decreased the concentration of yop mRNA compared to that of wild-type yersiniae (48). (iii) Knockout mutations in lcrQ (yscM1 and yscM2) increased the expression of reporter genes fused to yop genes compared to wild-type yersiniae (7, 43, 52). These experiments used insertions of the luxAB gene into yopE (18) or the fusion of the chloramphenicol acetyltransferase reporter gene to the 3′ end of yopH. Although the reported data were consistent with YscM1 and YscM2 (LcrQ) acting to prevent transcription, one could not discard the possibility of a posttranscriptional regulatory mechanism (43, 52).
Stainier et al. observed that the regulatory effect of yscM1 and yscM2 on yop expression did not involve type III secretion but required additional genes that were located on the Yersinia virulence plasmid (52). Rimpilainen et al. (48) as well as Williams and Straley (55) observed a requirement of lcrH and yopD for lcrQ-mediated regulation. Further evidence for functional cooperation among yscM1, yscM2, yopD, and lcrH was provided by the observation that knockout mutations in all four genes could bypass the Yersinia requirement for an environmental amino acid signal (glutamate) to induce type III secretion (37). Recently, Anderson et al. reported that yopD and lcrH regulate the expression of yopQ at a posttranscriptional step that involves binding of YopD and LcrH to yopQ mRNA (2). It is reported here that yscM1 and yscM2 regulated the expression of yopQ and yopE at the same posttranscriptional step as did yopD and lcrH. The target of regulation was the 5′ UTR of yop mRNAs, which presumably contained a specific binding site for YopD and LcrH. The 5′-AUAAA-3′ sequence represents the target element, as mutations in the conserved sequence motif abolish class II gene-mediated regulation. Further, introduction of a single 5′-AUAAA-3′ into the 5′ npt UTR is sufficient to impose at least a twofold repression by the class II-mediated mechanism. Fusions of truncations of the 178-nucleotide 5′ yopQ UTR to the 5′ npt UTR and npt reporter gene, all containing at least one of the four aforementioned conserved sequence elements, are also sufficient for class II gene-mediated regulation. Future work will provide further characterization of this sequence element. Because expression of yopQ is more tightly regulated by the class II genes than by other type III substrates, it is likely that the presence of multiple conserved sequence elements in the relatively long 5′ yopQ UTR provides for the engagement of several repressor complexes compared to other substrates which are not as tightly regulated. It is also possible that the conserved sequence element found in other parts of the transcript, i.e., the coding region, could contribute to class II gene-mediated regulation. There is evidence of yopD-sensitive nucleotide substitution mutations in one of two conserved 5′-AUAAA-3′ sequences in the yopQ coding sequence (2, 46). This may explain why repression of the npt reporter by YscM1 or YscM2 overexpression is maximal only when npt is appended to the full-length coding sequence of yopQ (Fig. 5C).
It is yet unclear whether YscM1 and YscM2 are capable of binding yop mRNA or whether they contribute to the binding of YopD and LcrH. Alternatively, YscM1- and YscM2-mediated control of yop gene expression could be indirect. Wulff-Strobel et al. proposed a model whereby LcrQ may regulate the activity of the type III secretion machinery (56). The observations in Fig. 2 are consistent with this model. It is, however, also plausible that class II genes control the expression of a regulatory factor for the type III pathway in addition to regulating yop genes, as YscM1 or YscM2 overexpression lowers the expression of other factors in the yop virulon (7, 52). Our data are consistent with the notion that binding of YopD and LcrH to yop mRNA alone is not sufficient to prevent Yop synthesis but requires at least one additional factor, YscM1 or YscM2. Further, YscM1 and YscM2 can be viewed as negative regulators of yop expression while SycH functions as an activator.
We believe that the simplest model for the data presented here and elsewhere is the formation of a protein complex composed of YopD, LcrH, YscM1, and YscM2 on yop mRNA. The assembly of the complex likely prevents translation of the transcript and may even target mRNAs for degradation (2). Multiple events contribute to relieving the posttranscriptional repression of the yop virulon. Binding of SycH to YscM1 and YscM2 certainly suffices to induce yop expression. However, depletion of YopD, YscM1, and YscM2 from the bacterial cytoplasm via the type III transport is another possible mechanism of regulation. We reason that the assembly of a YopD, LcrH, YscM1, and YscM2 repressor complex may represent the setting of a switch. The switch may be set once Yersinia enters the host and the temperature is raised to 37°C. Flipping the switch, i.e., causing a large increase in yop expression, requires the environmental signals glutamate (amino acid), albumin (serum protein), and calcium. Three regulatory response mechanisms seem coupled to these signals: (i) the secretion of YopD in the presence of calcium and albumin; (ii) the binding of SycH to YscM1 and YscM2 in the presence of the glutamate, albumin, and calcium signals; and (iii) the targeting of YscM1 and YscM2 into the cytoplasm of tissue culture cells. Available evidence suggests that all three mechanisms are reversible and perhaps not essential to the flipping of the switch. Other regulatory mechanisms could play a role. For example, temporal posttranslational modification of any one class II gene product, YopD, LcrH, or YscM1 and YscM2, could inactivate the entire complex, a possibility that will need to be addressed in future studies. The drawing in Fig. 11 summarizes our present knowledge and views of the events that regulate yop expression by the class II gene products YopD, LcrH, YscM1, and YscM2.
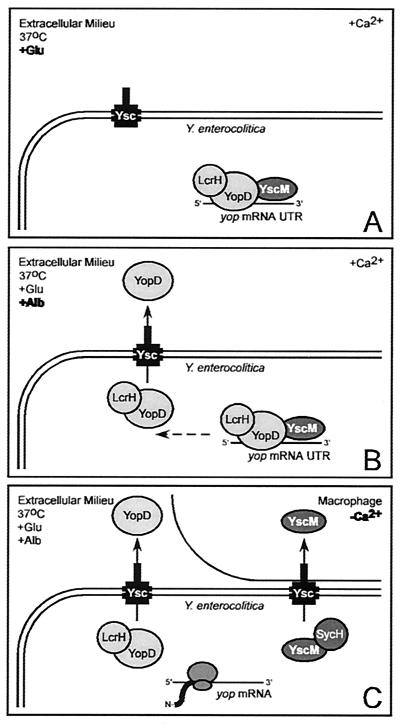
FIG. 11.
Model for the regulation of yop genes by class II gene products. (A) Upon host entry and exposure to 37°C, glutamate (serum amino acid), and 1.2 mM calcium, YopD, LcrH, YscM1, and YscM2 prevent the translation of yop mRNAs by recognizing a feature in the 5′ UTR. (B) Albumin (serum protein) activates the type III secretion of YopD. (C) Insertion of type III needles into the plasma membrane of macrophages triggers a calcium signal that activates the type III targeting of YscM1 and YscM2 via binding to the SycH chaperone. Secretion of YopD and targeting of YscM1 and YscM2 relieve the inhibition of yop mRNA translation and activate Yersinia type III injection of effector Yops.
Acknowledgments
We thank Deborah Anderson, Vincent Lee, Kumaran Ramamurthi, and Christina Tam for help and reagents.
E.D.C. was supported in part by the Cellular and Molecular Biology Training Grant at UCLA, US-PHS National Research Award GM07185. This work was supported by NIAID-NIH grant AI42797 to O.S.
REFERENCES
- 1.Allaoui, A., R. Schulte, and G. R. Cornelis. 1995. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol. Microbiol. 18:343-355. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. M., K. S. Ramamurthi, C. Tam, and O. Schneewind. 2002. YopD and LcrH regulate the expression of Yersinia enterocolitica YopQ at a posttranscriptional step and bind to yopQ mRNA. J. Bacteriol. 184:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. M., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31:1139-1148. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, T., S. Hakansson, A. Forsberg, L. Norlander, A. Macellaro, A. Backman, I. Bolin, and H. Wolf-Watz. 1991. Analysis of V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of lcrH and lcrV. J. Bacteriol. 173:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland, A., M.-P. Sory, M. Iriarte, C. Kerbourch, P. Wattiau, and G. R. Cornelis. 1996. Status of YopM and YopN in the Yersinia yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 15:5191-5201. [PMC free article] [PubMed] [Google Scholar]
- 6.Bolin, I., and H. Wolf-Watz. 1988. The plasmid-encoded Yop2b protein of Yersinia pseudotuberculosis is a virulence determinant regulated by calcium and temperature at the level of transcription. Mol. Microbiol. 2:237-245. [DOI] [PubMed] [Google Scholar]
- 7.Cambronne, E. D., L. W. Cheng, and O. Schneewind. 2000. LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone-dependent mechanism. Mol. Microbiol. 37:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, L. W., O. Kay, and O. Schneewind. 2001. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183:5293-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, L. W., and O. Schneewind. 2000. Type III machines of pathogenic Yersinia deliver the goods. Trends Microbiol. 8:214-220. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, L. W., and O. Schneewind. 2000. Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for the specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J. Bacteriol. 182:3183-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis, G., C. Sluiters, C. Lambert de Rouvroit, and T. Michiels. 1989. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M.-P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelis, G. R., and C. Colson. 1975. Restriction of DNA in Yersinia enterocolitica detected by the recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87:285-291. [DOI] [PubMed] [Google Scholar]
- 15.DeBord, K., V. T. Lee, and O. Schneewind. 2001. On the role of LcrG and LcrV during the type III targeting of effector Yops by Yersinia enterocolitica. J. Bacteriol. 183:4588-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lorenzo, V., E. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 17.Fallman, M., K. Andersson, S. Hakansson, K. E. Magnusson, O. Stendahl, and H. Wolf-Watz. 1995. Yersinia pseudotuberculosis inhibits Fc receptor-mediated phagocytosis in J774 cells. Infect. Immun. 63:3117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg, A., and R. Rosqvist. 1993. In vivo expression of virulence genes of Yersinia pseudotuberculosis. Infect. Agents Dis. 2:275-278. [PubMed] [Google Scholar]
- 19.Forsberg, A., A.-M. Viitanen, M. Skunik, and H. Wolf-Watz. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5:977-986. [DOI] [PubMed] [Google Scholar]
- 20.Forsberg, A., and H. Wolf-Watz. 1988. The virulence protein Yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-pIB1-encoded trans-acting elements controlled by temperature and calcium. Mol. Microbiol. 2:121-133. [DOI] [PubMed] [Google Scholar]
- 21.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075-1093. [DOI] [PubMed] [Google Scholar]
- 22.Francis, M. S., and H. Wolf-Watz. 2000. A study of the YopD-LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol. Microbiol. 38:85-102. [DOI] [PubMed] [Google Scholar]
- 23.Francis, M. S., H. Wolf-Watz, and A. Forsberg. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5:166-172. [DOI] [PubMed] [Google Scholar]
- 24.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1333. [DOI] [PubMed] [Google Scholar]
- 25.Galyov, E. E., S. Hakansson, A. Forsberg, and H. Wolf-Watz. 1993. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature 361:730-732. [DOI] [PubMed] [Google Scholar]
- 26.Hakansson, S., T. Bergman, J.-C. Vanooteghem, G. Cornelis, and H. Wolf-Watz. 1993. YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect. Immun. 61:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakansson, S., E. Gaylov, R. Rosqvist, and H. Wolf-Watz. 1996. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol. Microbiol. 20:593-603. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-572. [DOI] [PubMed] [Google Scholar]
- 29.Hoe, N. P., and J. D. Goguen. 1993. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175:7901-7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoe, N. P., F. C. Minion, and J. D. Goguen. 1992. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J. Bacteriol. 174:4275-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoiczyk, E., and G. Blobel. 2001. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. USA 98:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmstrom, A., J. Petterson, R. Rosqvist, S. Hakansson, F. Tafazoli, M. Fallman, K.-E. Magnusson, H. Wolf-Watz, and A. Forsberg. 1997. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol. Microbiol. 24:73-91. [DOI] [PubMed] [Google Scholar]
- 33.Hueck, C. J. 1998. Type III protein secretion in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 35.Iriarte, M., and G. R. Cornelis. 1998. YopT, a new Yersinia effector protein, affects the cytoskeleton of host cells. Mol. Microbiol. 29:915-929. [DOI] [PubMed] [Google Scholar]
- 36.Lee, V. T., D. M. Anderson, and O. Schneewind. 1998. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28:593-601. [DOI] [PubMed] [Google Scholar]
- 37.Lee, V. T., S. K. Mazmanian, and O. Schneewind. 2001. A program of Yersinia enterocolitica type III secretion reactions is triggered by specific host signals. J. Bacteriol. 183:4970-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, V. T., and O. Schneewind. 1999. Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol. Microbiol. 31:1619-1629. [DOI] [PubMed] [Google Scholar]
- 39.Lee, V. T., C. Tam, and O. Schneewind. 2000. Yersinia enterocolitica type III secretion. LcrV, a substrate for type III secretion, is required for toxin-targeting into the cytosol of HeLa cells. J. Biol. Chem. 275:36869-36875. [DOI] [PubMed] [Google Scholar]
- 40.Michiels, T., P. Wattiau, R. Brasseur, J.-M. Ruysschaert, and G. Cornelis. 1990. Secretion of Yop proteins by yersiniae. Infect. Immun. 58:2840-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills, S. D., A. Boland, M.-P. Sory, P. van der Smissen, C. Kerbouch, B. B. Finlay, and G. R. Cornelis. 1997. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc. Natl. Acad. Sci. USA 94:12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Persson, C., R. Nordfelth, A. Holmstrom, S. Hakansson, R. Rosqvist, and H. Wolf-Watz. 1995. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol. Microbiol. 18:135-150. [DOI] [PubMed] [Google Scholar]
- 43.Petterson, J., R. Nordfelth, E. Dubinina, T. Bergman, M. Gustafsson, K. E. Magnusson, and H. Wolf-Watz. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231-1233. [DOI] [PubMed] [Google Scholar]
- 44.Price, S. B., C. Cowan, R. D. Perry, and S. C. Straley. 1991. The Yersinia pestis V antigen is a regulatory protein necessary for Ca2+-dependent growth and maximal expression of low-Ca2+ response virulence genes. J. Bacteriol. 173:2649-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramamurthi, K. S., and O. Schneewind. 2002. Type III protein secretion in Yersinia species. Annu. Rev. Cell Dev. Biol. 18:107-133. [DOI] [PubMed] [Google Scholar]
- 46.Ramamurthi, K. S., and O. Schneewind. 2002. Yersinia enterocolitica type III secretion: mutational analysis of the yopQ secretion signal. J. Bacteriol. 184:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reiss, B., R. Sprengel, and H. Schaller. 1984. Protein fusions with the kanamycin resistance gene from transposon Tn5. EMBO J. 3:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rimpilainen, M., A. Forsberg, and H. Wolf-Watz. 1992. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J. Bacteriol. 174:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosqvist, R., A. Forsberg, M. Rimpilainen, T. Bergman, and H. Wolf-Watz. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary host defense. Mol. Microbiol. 4:657-667. [DOI] [PubMed] [Google Scholar]
- 50.Rosqvist, R., A. Forsberg, and H. Wolf-Watz. 1991. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun. 59:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosqvist, R., K.-E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stainier, I., M. Iriarte, and G. R. Cornelis. 1997. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol. Microbiol. 26:833-843. [DOI] [PubMed] [Google Scholar]
- 53.Wattiau, P., B. Bernier, P. Deslee, T. Michiels, and G. R. Cornelis. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. USA 91:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wattiau, P., and G. R. Cornelis. 1994. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 176:3878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams, A. W., and S. C. Straley. 1998. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol. 180:350-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wulff-Strobel, C. R., A. W. Williams, and S. C. Straley. 2002. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 43:411-423. [DOI] [PubMed] [Google Scholar]
- 57.Yother, J., T. W. Chamness, and J. D. Goguen. 1986. Temperature-controlled plasmid regulon associated with low calcium response in Yersinia pestis. J. Bacteriol. 165:443-447. [DOI] [PMC free article] [PubMed] [Google Scholar]