Abstract
By primer extension inhibition assays, 70S ribosomes bound with higher affinity, or stability, than did 30S subunits to leaderless mRNAs containing AUG or GUG start codons. Addition of translation initiation factors affected ribosome binding to leaderless mRNAs. Our results suggest that translation of leaderless mRNAs might initiate through a pathway involving 70S ribosomes or 30S subunits lacking IF3.
Initiation of protein synthesis in prokaryotes involves several steps that include the binding of a 30S ribosomal subunit to the mRNA's ribosome binding site and placement of the start codon in the ribosomal P site. Translation signals within the mRNA, including the start codon (16, 20, 25) and complementary pairing between the mRNA Shine-Dalgarno (SD) and 16S rRNA anti-SD sequences (8, 9, 21), contribute to the efficiency and stability of the translation initiation complex.
The efficiency and fidelity with which the initiation complex forms is influenced also by three initiation factors (IF1, IF2, and IF3) (6). IF1 enhances the activities of IF2 and IF3 (6) and is proposed to block binding of aminoacyl-tRNA to the 30S subunit A site during initiation (2, 14). IF2 promotes binding of the initiator tRNA at the P site of the 30S subunit (6). IF3 is proposed to have several functions, including the selection of initiator tRNA and discrimination against unusual start codons (17) and binding to 30S subunits to prevent joining to a 50S subunit (13).
Much of our understanding of translation initiation results from studies of canonical mRNAs containing a 5′ untranslated leader and SD sequence. However, several leaderless mRNAs from the archaea, bacteria, and eucarya lack these features (reviewed in reference 26) but are still highly translated. It is not clear how leaderless mRNAs bind ribosomes, but several lines of evidence implicate a modified initiation pathway different from that observed with mRNAs containing a 5′ untranslated leader and SD sequence. For example, 30S subunits containing IF3 discriminate against initiation at a 5′ AUG initiation codon (4, 15, 24). Also, 30S subunits containing IF2 stimulate ribosome binding and translation of leaderless mRNAs (1, 4, 5). These data, and a report that 70S ribosomes bind leaderless mRNAs in vitro (1), suggest the possibility of a novel pathway for translation of leaderless mRNAs.
Leaderless mRNAs bind 70S ribosomes more efficiently, or more stably, than 30S ribosomal subunits.
Primer extension inhibition (toeprint) assays (7) were performed to investigate the relative ribosome binding strengths of leaderless mRNAs encoded by the Escherichia coli λ cI, Tn1721 tetR, and P2 V genes. Ribosomes (70S) and 30S subunits were prepared from E. coli MRE600 as described previously (23). Toeprint assays and construction of cI-lacZ fusions were performed as described previously (12, 16). P2 gene V codons 1 to 16 from pGCV22 (3) were fused to lacZ and used as a template to generate V-lacZ leaderless mRNAs in vitro. Tn1721 tetR codons 1 to 16 (10) were constructed from oligonucleotides, fused to lacZ, and used as a template to generate tetR-lacZ leaderless mRNAs in vitro. Ribosome binding efficiencies for cI-lacZ mRNAs were quantified, with a Molecular Dynamics (Storm 800) phosphorimager, by taking the average from three independent toeprint assays and were expressed as toeprint signal/(full-length signal + toeprint signal).
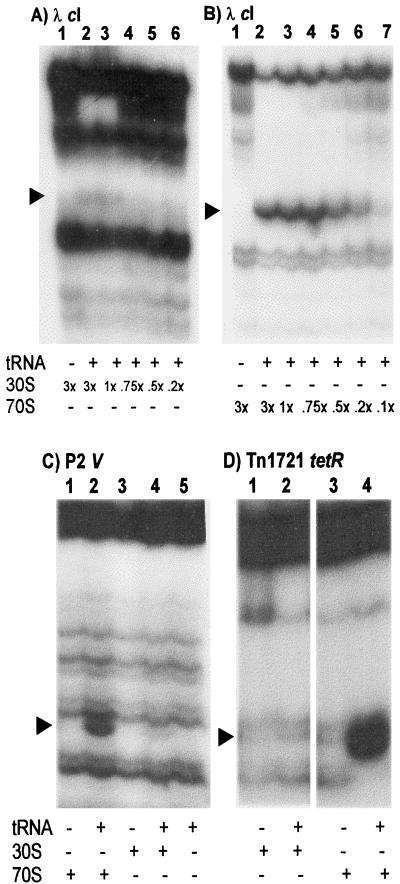
A ternary complex (i.e., mRNA, ribosome, and initiator tRNA)-dependent toeprint signal was observed at position +16 relative to the start codon of leaderless cI-lacZ mRNA (Fig. 1A and B). Phosphorimager analyses revealed that the signal with 70S ribosomes was, on average, 18-fold stronger than the signal obtained with 30S subunits. Also, maximal toeprint signal intensity was observed when the 70S ribosome/mRNA ratio was 1:1 (Fig. 1B, lanes 2 and 3), while signal intensities have been observed to continue increasing beyond a 12-fold excess of 30S subunits over mRNA (data not shown). By using gene V and tetR leaderless mRNAs, a much stronger toeprint signal was observed with 70S ribosomes (Fig. 1C, lanes 1 and 2; Fig. 1D, lanes 3 and 4) than with 30S subunits (Fig. 1C, lanes 3 and 4; Fig. 1D, lanes 1 and 2). These results are consistent with an earlier report (1) and suggest that leaderless mRNAs have a higher affinity for, or are intrinsically more stable with, 70S ribosomes than 30S subunits.
FIG. 1.
Relative binding efficiencies of 30S subunits and 70S ribosomes for leaderless mRNA. (A) Toeprint assays with 30S subunits in a threefold molar excess over mRNA without initiator tRNA (lane 1) or with 30S subunits in a 3-fold (lane 2), 1-fold (lane 3), 0.75-fold (lane 4), 0.5-fold (lane 5), or 0.2-fold (lane 6) excess over mRNA with initiator tRNA. (B) Toeprint assays with 70S ribosomes in a threefold molar excess over mRNA without initiator tRNA (lane 1) or 70S ribosomes in a 3-fold (lane 2), 1-fold (lane 3), 0.75-fold (lane 4), 0.5-fold (lane 5), 0.2-fold (lane 6), or 0.1-fold excess over mRNA with initiator tRNA. (C) Toeprint assays of V-lacZ leaderless mRNA without ribosomes (lane 5) or with 70S ribosomes (lane 2) or 30S subunits (lane 4) in a 10-fold molar excess over mRNA or a 10-fold molar excess of 70S ribosomes (lane 1) or 30S subunits (lane 3) in the absence of initiator tRNA. (D) Toeprint assays of tetR-lacZ leaderless mRNA with 30S subunits (lane 2) or 70S ribosomes (lane 4) in a 10-fold molar excess over mRNA or a 10-fold molar excess of 30S subunits (lane 1) or 70S subunits (lane 3) in the absence of initiator tRNA. The arrowheads indicate the positions of the toeprint signal.
70S ribosomes bind to leaderless cI mRNA with either an AUG or a GUG start codon.
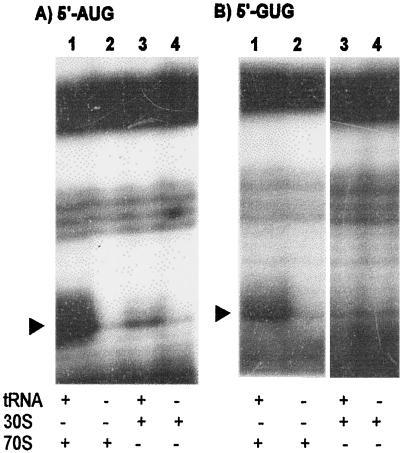
Previous reports (16, 25) indicate that leaderless mRNAs with an AUG start codon are most efficiently expressed but that mRNAs with a 5′-terminal GUG are also translated. Because 30S subunits have been reported previously not to bind leaderless cI mRNA with a GUG start codon (16), we investigated the ability of 70S ribosomes to bind this mRNA. With the use of cI-lacZ leaderless mRNAs with AUG or GUG start codons (16), 30S subunits yielded the expected toeprint signal from mRNA with an AUG start codon (Fig. 2A, lanes 3 and 4) but not from mRNA with a GUG start codon (Fig. 2B, lanes 3 and 4); assays with 70S ribosomes revealed a ternary complex-dependent signal for leaderless mRNA with an AUG (Fig. 2A, lanes 1 and 2) or GUG (Fig. 2B, lanes 1 and 2) start codon. Toeprint reactions with cI-lacZ leaderless mRNAs containing a UUG or CUG initiation codon did not reveal a toeprint signal with 30S subunits (16) or 70S ribosomes (data not presented). These results suggest that leaderless GUG-initiated mRNAs have substantially reduced affinity for, or do not associate stably with, 30S subunits.
FIG. 2.
Ribosome binding (toeprint) assays for cI-lacZ leaderless mRNAs containing an AUG or GUG start codon. (A) Toeprint assays with an AUG start codon and 70S ribosomes (lane 1) or 30S subunits (lane 3) in a 12-fold molar excess over mRNA or a 12-fold molar excess of 70S ribosomes (lane 2) or 30S subunits (lane 4) in the absence of initiator tRNA. (B) Toeprint assays with a GUG start codon and 70S ribosomes (lane 1) or 30S subunits (lane 3) in a 12-fold molar excess over mRNA or a 12-fold molar excess of 70S ribosomes (lane 2) or 30S subunits (lane 4) in the absence of initiator tRNA. The arrowheads indicate the positions of the toeprint signal.
IFs affect 30S subunit binding to leaderless mRNAs.
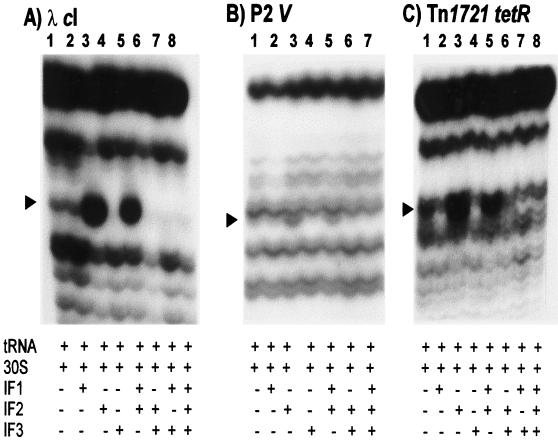
The contribution of IFs in 30S subunit binding to leaderless mRNAs was assessed by toeprint assays with cI, gene V, and tetR leaderless mRNAs. IF1, IF2, and IF3 were isolated essentially as described previously (22) from IF-overproducing strains of E. coli (kindly provided by C. Gualerzi). IFs were present in a twofold molar excess over 30S subunits and were prebound to 30S subunits (37°C, 5 min) prior to addition of mRNA. Initiator tRNA was charged and formylated, as described previously (19). Addition of IF1 reduced the toeprint signal for tetR (Fig. 3C, lanes 1 and 2) but did not significantly affect signal intensities for cI (Fig. 3A, lanes 1 and 2) or gene V (Fig. 3B, lanes 1 and 2); however, addition of IF2 increased binding to each leaderless mRNA (compare lanes 1 and 3 of Fig. 3A to C, respectively). IF2 stimulation of 30S subunit binding to leaderless mRNA is consistent with previous reports (1, 4, 5). Addition of IF3 decreased toeprint signal intensity (compare lanes 1 and 4 of Fig. 3A to C), consistent with an earlier report (24). Addition of IF1 and IF2 did not significantly affect toeprint intensity relative to that with reactions with IF2 alone (compare lanes 3 and 5 of Fig. 3A to C). Addition of IF2 and IF3 (lane 6 of Fig. 3A to C) or IF1 and IF3 (lane 7 of Fig. 3A and B) reduced the toeprint signal, similar to that seen with IF3 alone, suggesting that the inhibitory effect of IF3 dominates the stimulatory effect of IF2. Interestingly, addition of IF1, IF2, and IF3 to 30S subunits eliminated the toeprint signal from all three leaderless mRNAs (Fig. 3A, lane 8; Fig. 3B, lane 7; and Fig. 3C, lane 8), suggesting that the IFs disallow 30S initiation complex formation on leaderless mRNAs; alternatively, IFs might destabilize the ternary complex for detection by reverse transcription.
FIG. 3.
30S subunit binding (toeprint) assays for λ cI, P2 gene V, and Tn1721 tetR leaderless mRNAs in the presence or absence of IFs. Toeprint assays were done with 30S subunits in a sixfold molar excess over mRNA without IFs (lanes 1) or with IF1 (lanes 2), IF2 (lanes 3), IF3 (lanes 4), IF1 and IF2 (lanes 5), IF2 and IF3 (lanes 6), and IF1 and IF3 (lanes 7, panels A and C) or with IF1, IF2, and IF3 (lanes 8, panels A and C, and lane 7, panel B) and fMet-tRNA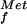 . The arrowheads indicate the positions of the toeprint signal.
. The arrowheads indicate the positions of the toeprint signal.
Based on the intracellular concentrations of ribosomes (∼10 μM) and IFs (∼1 μM each), and IF Kas for 30S subunits, Gualerzi and Pon (6) have speculated that most IFs in vivo are bound to 30S subunits. Based on our toeprint results, this would suggest that 30S subunits containing all three IFs are biased against initiation complex formation with leaderless mRNAs. These data further support the suggestion that 70S ribosomes, or 30S subunits lacking a full complement of IFs, might bind and initiate translation of leaderless mRNAs and explain the way by which leaderless mRNAs compete with leadered mRNAs for initiating ribosomes in vivo.
IF2 affects 70S ribosome binding to leaderless mRNAs.
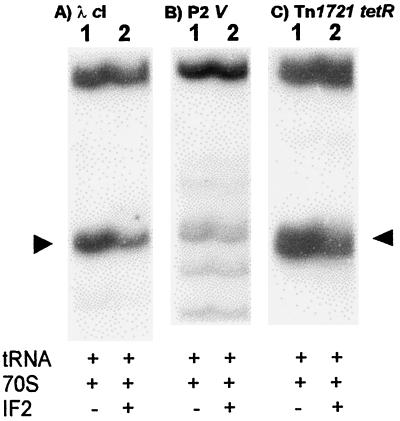
Because of a recent report that IF2 protects the 23S rRNA of 70S ribosomes (11), toeprint assays were performed to investigate the effect of IF2 on 70S ribosome binding to leaderless mRNA. IF2 was present in a twofold molar excess over ribosomes and was prebound to ribosomes (37°C, 5 min) prior to addition of mRNA. Initiator tRNA was charged and formylated, as described previously (19). Addition of IF2 to 70S ribosomes reduced the toeprint signal observed for cI (Fig. 4A) and tetR (Fig. 4C) but had minimal effect on binding the gene V (Fig. 4B) leaderless mRNA. Because IF2 has a higher affinity for 30S subunits than for 70S ribosomes (18), the slight inhibition observed for IF2 in binding leaderless mRNA to 70S ribosomes might be minimal in vivo.
FIG. 4.
70S ribosome binding (toeprint) assays for λ cI, P2 gene V, and Tn1721 tetR leaderless mRNAs in the presence or absence of IF2. Toeprint assays were done with a sixfold molar excess of 70S ribosomes over mRNA without IF2 (lanes 1) or with IF2 (lanes 2) and 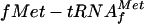 . Arrowheads indicate the positions of the toeprint signal.
. Arrowheads indicate the positions of the toeprint signal.
Concluding remarks.
The results reported here suggest that the conventional pathway for translation initiation with leadered mRNAs (6), involving 30S subunits bound by IFs, might not contribute significantly to translation of leaderless mRNA. Given the in vivo competition for 30S subunits by the multitude of SD sequence-containing leadered mRNAs, it is reasonable to speculate that initiation complexes of leaderless mRNA and 30S subunits might assemble only through interaction with subunits lacking IF3. Based on our in vitro ribosome binding assays, more cI, gene V, and tetR leaderless mRNA was bound by 70S ribosomes than by 30S subunits, consistent with an earlier report for cI mRNA (1). The higher binding affinity, or increased stability, suggests that leaderless mRNAs with AUG or GUG start codons might more readily form initiation complexes with 70S ribosomes, thereby relieving competition for 30S subunits; expression levels from leaderless mRNAs might then reflect the abundance, or available pool, of free 70S ribosomes. Translation initiation via 70S ribosomes would represent a novel initiation pathway and might contain mRNA-ribosome interactions not present in the conventional pathway that initiates with 30S subunits.
Acknowledgments
We thank Richard Calendar for providing plasmid pGCV22; Uttam RajBhandary for providing plasmids pQE16-MTF and pQE60-MetRS; Claudio Gualerzi for providing plasmids encoding IF1, IF2, and IF3 and protocols for IF purification; and Anton Vila for providing helpful advice on tRNA charging reactions. Inita Forti is acknowledged for inspiring discussions.
This work was supported by grant GM45923 from the National Institutes of Health and, in part, by funds received through the Research Challenge program of the Ohio Board of Regents. S.M.O. thanks the Miami University Graduate School for a Graduate Student Achievement Award.
REFERENCES
- 1.Balakin, A. G., E. A. Skripkin, I. N. Shatsky, and A. A. Bogdanov. 1992. Unusual ribosome binding properties of mRNA encoding bacteriophage λ repressor. Nucleic Acids Res. 20:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, T. Hartsch, B. T. Wimberly, and V. Ramakrishnan. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291:498-501. [DOI] [PubMed] [Google Scholar]
- 3.Christie, G. E., and R. Calendar. 1985. Bacteriophage P2 late promoters II. Comparison of the four late promoter sequences. J. Mol. Biol. 181:373-382. [DOI] [PubMed] [Google Scholar]
- 4.Grill, S., C. O. Gualerzi, P. Londei, and U. Balsi. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 19:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grill, S., I. Moll, D. Hasenohrl, C. O. Gualerzi, and U. Blasi. 2001. Modulation of ribosomal recruitment to 5′-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett. 495:167-171. [DOI] [PubMed] [Google Scholar]
- 6.Gualerzi, C. O., and C. L. Pon. 1990. Initiation of mRNA translation in prokaryotes. Biochemistry 29:5881-5889. [DOI] [PubMed] [Google Scholar]
- 7.Hartz, D., D. S. McPheeters, R. Traut, and L. Gold. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164:419-425. [DOI] [PubMed] [Google Scholar]
- 8.Hui, A. J., and H. A. deBoer. 1987. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 84:4762-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob, W. F., M. Santer, and A. E. Dahlberg. 1987. A single base change in the Shine-Dalgarno region of the 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl. Acad. Sci. USA 84:4757-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klock, G., and W. Hillen. 1986. Expression, purification, and operator binding of the transposon Tn1721-encoded Tet repressor. J. Mol. Biol. 189:633-641. [DOI] [PubMed] [Google Scholar]
- 11.LaTeana, A., C. O. Gualerzi, and A. E. Dahlberg. 2001. Initiation factor IF2 binds to the α-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7:1173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Farmer, J., and G. R. Janssen. 1999. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 31:1025-1038. [DOI] [PubMed] [Google Scholar]
- 13.McCutcheon, J. P., R. K. Agrawal, S. M. Phillios, R. A. Grassucci, S. E. Gerchman, W. M. Clemons, V. Ramakrishnan, and J. Frank. 1999. Location of translation initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. USA 96:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moazed, D., R. R. Samaha, C. Gualerzi, and H. F. Noller. 1995. Specific protection of 16S rRNA by translation initiation factors. J. Mol. Biol. 248:207-210. [DOI] [PubMed] [Google Scholar]
- 15.Moll, I., A. Resch, and U. Blasi. 1998. Discrimination of 5′-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett. 436:213-217. [DOI] [PubMed] [Google Scholar]
- 16.O'Donnell, S. M., and G. R. Janssen. 2001. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 183:1277-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrelli, D., A. LaTeana, C. Garofalo, R. Spurio, C. L. Pon, and C. O. Gualerzi. 2001. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 20:4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pon, C. L., M. Paci, R. T. Pawlik, and C. O. Gualerzi. 1985. Structure-function relationship in Escherichia coli initiation factors. J. Biol. Chem. 260:8918-8924. [PubMed] [Google Scholar]
- 19.Ramesh, V., S. Gite, and U. L. RajBhandary. 1998. Functional interaction of an arginine conserved in the sixteen amino acid insertion module of Escherichia coli methionyl-tRNA formyltransferase with determinants for formylation in the initiator tRNA. Biochemistry 37:15925-15932. [DOI] [PubMed] [Google Scholar]
- 20.Ringquist, S., S. Shinedling, D. Barrick, L. Green, J. Binkley, G. D. Stormo, and L. Gold. 1992. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6:1219-1229. [DOI] [PubMed] [Google Scholar]
- 21.Shine, J., and L. Dalgarno. 1974. The 3′ terminal of Escherichia coli 16S ribosomal RNA: complementarity to non-sense triplets and ribosomal binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soffientini, A., R. Lorenzetti, L. Gastaldo, J. H. Parlett, R. Spurio, A. LaTeana, and K. Islam. 1994. Purification procedure for bacterial translational initiation factors IF2 and IF3. Protein Expr. Purif. 5:118-124. [DOI] [PubMed] [Google Scholar]
- 23.Spedding, G. 1990. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles, p. 1-27. In G. Spedding (ed.), Ribosomes and protein synthesis: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 24.Tedin, K., I. Moll, S. Grill, A. Resch, A. Graschopf, C. O. Gualerzi, and U. Blasi. 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 31:67-77. [DOI] [PubMed] [Google Scholar]
- 25.Van Etten, W. J., and G. R. Janssen. 1998. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol. Microbiol. 27:987-1001. [DOI] [PubMed] [Google Scholar]
- 26.Wu, C.-J., and G. R. Janssen. 1996. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5′ untranslated leader. Mol. Microbiol. 22:339-355. [DOI] [PubMed] [Google Scholar]