Abstract
The protein expression patterns of exponentially growing, starved, and viable but nonculturable (VBNC) Enterococcus faecalis cells were analyzed to establish whether differences exist between the VBNC state and other stress responses. The results indicate that the protein profile of VBNC cells differs from that of either starved or exponentially growing bacteria. This demonstrates that the VBNC state is a distinct physiological phase within the life cycle of E. faecalis, which is activated in response to multiple environmental stresses.
It is well known that bacteria can respond to environmental stresses by activating survival mechanisms (24). While a number of microorganisms can survive by forming resistant spores, nonsporulating microorganisms are able to persist in adverse environments as vegetative cells with low metabolic activity, via the activation of the viable but nonculturable (VBNC) state (1, 4). In contrast, nutrient deprivation or exhaustion stimulates a starvation response, which allows bacterial long-term persistence in a nongrowing but culturable state (18). These different survival strategies may represent distinct metabolic responses of bacteria which are confronted with adverse environmental conditions. These two alternative adaptive responses to stress, namely, starvation and the VBNC state, have been previously described for Vibrio species (20, 21).
It has been demonstrated elsewhere (12) that the major part of the bacterial population present in the environment cannot be detected by traditional culture techniques (e.g., determination of CFU). Although they cannot be cultured, viable bacteria can be visualized when a direct viable count method is used (11). On the other hand, the monitoring of pathogenic bacteria released under unfavorable environmental conditions has demonstrated that there are significant differences between total numbers of viable cells and CFU (19). This observation suggests that the VBNC state could be a preferred survival strategy of nonsporulating bacteria in the environment.
Enterococcus faecalis is a member of the human and animal intestinal microflora which is also considered an indicator for fecal contamination when the microbiological quality of water samples is monitored (29). In addition, this microorganism can cause severe disease, being one of the most important etiologic agents of nosocomial infections (5). Previous studies have demonstrated that, when released in the environment, E. faecalis can activate the VBNC state in response to unfavorable conditions (e.g., oligotrophy and low temperatures) (15, 16). In the nonculturable state, enterococci still preserve their membrane integrity, as well as their capacity to bind and invade eukaryotic cells (16, 22). E. faecalis develops specific envelope alterations when the VBNC state is activated (27). However, macromolecular synthesis and gene expression are preserved during the VBNC state, and cellular division can be reestablished (resuscitation) upon restoration of favorable conditions (14, 16, 17). On the other hand, variations in stress conditions, such as exhaustion of the carbon source or incubation in an oligotrophic medium at permissive temperatures, activate the starvation response. This allows E. faecalis to remain culturable, even if its divisional capability is blocked (7, 10). Activation of different metabolic pathways will consequently change the protein profiles of the microorganisms confronting stress conditions. In recent years it has been demonstrated that starvation enhances the resistance of E. faecalis to environmental stresses (6). Starvation-specific alterations in the protein profiles of E. faecalis have also been reported elsewhere (8, 13).
To evaluate the differences between the VBNC state and other stress responses in E. faecalis, we analyzed the whole-protein profiles of exponentially growing, VBNC, and starved bacteria. The vancomycin-resistant (vanB) E. faecalis strain V583 was used in these studies because this is the strain which is currently being sequenced (see The Institute for Genomic Research [TIGR] website at http://www.tigr.org). Bacterial cells were grown in tryptic soy broth (Difco Laboratories, Detroit, Mich.). Cell growth was monitored with an LKB spectrophotometer by measuring the optical density at 640 nm. The number of culturable bacteria was assessed by plating them on tryptic soy agar (TSA; Difco) to which 0.2% sodium pyruvate was added (TSA-SP plates) (12). The total number of cells was established by using a Coulter Counter ZBI (Coulter Scientific, Krefeld, Germany) equipped with a 30-μm capillary, as previously described (16). For the induction of starvation and the VBNC state, exponentially growing cells were harvested at an optical density at 640 nm of 0.35, washed twice with phosphate buffer (pH 7), and resuspended in filtered and autoclaved water withdrawn from Lake Garda (Italy) at a final density of 107 cells/ml. One half of this oligotrophic microcosm was maintained at 4 ± 0.5°C without shaking and directly illuminated with four Philips Philux 100-W lamps placed at a distance of 60 cm. The other half was maintained at 21°C without direct illumination. Samples were taken every 4 days. Colony-forming ability was determined in duplicate on samples plated on TSA-SP plates. Sample size ranged from 0.1 ml for spread plate counts to 10 ml for filtered samples; the sample size was adjusted to follow the decline of bacterial culturability as previously described (16).
To determine viability, VBNC and starved cells were stained with the Live/Dead kit (Molecular Probes), as specified by the manufacturer. This kit utilizes a mixture of the stains SYTO 9 and propidium iodide to evaluate cell membrane integrity. Green fluorescence of the intact cells was visualized under a fluorescent Leitz Orthoplan microscope. The presence of mRNA in the cells, a further marker of viability, was evaluated by reverse transcription-PCR (RT-PCR) (17) by using a primer pair specific for the pbp5 gene (23), which encodes penicillin binding protein 5 of E. faecalis, which is involved in peptidoglycan synthesis.
Proteome analysis of exponentially growing, starved, and VBNC E. faecalis cells.
To obtain proteins for proteome analysis, the VBNC cells were harvested when the number of cultivable cells was <0.1 CFU/ml. The starved population, which still contained 2 × 106 cultivable cells, was also harvested at the same time point. Two-dimensional (2D) gel electrophoresis was performed according to the method of Görg et al. (9) by using cell preparations from three different lots of each starved and VBNC microcosm, which were separately inoculated. Frozen cells (10 mg) were extracted with 1 ml of the reswelling solution for isoelectric focusing (IEF) containing 7.4 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 20 mM Tris base, 30 mM dithiothreitol, and one-half tablet of protease inhibitor (MiniComplete; Boehringer) by ultrasonication (six times for 30 s each). After 15 min of shaking at room temperature (Eppendorf mixer), the extracts were centrifuged (20 min at 4°C) to remove cell debris and DNA (16,000 × g). For further improving the sample quality for the 2D polyacrylamide gel electrophoresis (PAGE), the proteins were precipitated with ice-cold acetone (45 min at −20°C) to eliminate lipid components and salts. Then, the proteins were pelleted by centrifugation (Sorvall SS34, 5 min at 4°C, 10,000 × g). After drying, the protein pellets were dissolved in 300 μl of reswelling solution with the addition of 5% immobilized pH gradient buffer, pH 4 to 7 (Pharmacia), and applied to IEF ReadyStrips (Bio-Rad). After 100 kVh focusing on the IEF Protean instrument (Bio-Rad), the gel strips were equilibrated for the second dimension (sodium dodecyl sulfate [SDS]-PAGE) twice for 15 min each time with 5 ml of equilibration solution (6 M urea, 30% glycerol, 2% SDS, 200 μg of bromophenol blue, 50 mM Tris base [pH 8.8], 50 mg of dithiothreitol, and 0.24 g of iodacetamide). Then, the strips were applied to an SDS-10 to 15% polyacrylamide gel (1.5 mm) and developed in an IsoDALT chamber (Amersham-Pharmacia Biotech) overnight. When proteins needed to be identified by mass spectrometric analysis, the gels were stained with colloidal Coomassie blue (Roth). Otherwise, the gels were silver stained according to the method of Blum et al. (3).
To determine the N-terminal amino acid sequence (Edman degradation), the 2D-separated proteins were electroblotted onto a polyvinylidene difluoride (PVDF) membrane (tank blot, 30 V overnight, 4°C), which was stained with Coomassie brilliant blue R-250. The interesting protein spots were cut out, and N-terminal amino acid sequencing was performed with an Applied Biosystems model 494A Procise HT sequencer. To perform the mass spectrometric analysis, the protein spots were excised from colloidal Coomassie blue-stained gels and samples were prepared as previously described (30). The in-gel digest of the proteins was performed with sequencing-grade trypsin from Promega (2 μg/ml). For matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis, the peptide extracts were directly eluted from the ZipTip microcolumns (RP18 material used for desalting the samples; Millipore) with 1.5 μl of saturated α-cyano-4-hydroxycinnamic acid directly onto the MALDI target and analyzed with a MALDI-TOF Reflex apparatus (Bruker, Bremen, Germany). Samples analyzed with the Q-TOF instrument (Q-TOF II; Micromass) were also desalted with ZipTip microcolumns, but elution was performed with 10 μl of 50% methanol in water. The peptide fingerprints obtained by mass spectrometry (MALDI-TOF) were used for searches in the SwissProt/Trembl and National Center for Biotechnology Information (NCBI) protein databases with the MS-Fit tool of ProteinProspector v3.4.1. In addition, the sequence tags obtained by Edman degradation or mass spectrometric sequencing with the Q-TOF instrument were used for searches with the TIGR BLAST search engine for unfinished microbial genomes (http://www.tigr.org). To get an unambiguous identification, the resulting nucleotide sequences (contig sequence) were translated into protein sequences with the translation tool on the ExPASy home page (http://www.expasy.ch). BLAST searches were executed on the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/) for the resulting protein sequences.
Characterization of E. faecalis cells under different stress conditions.
Oligotrophic media, such as lake water, represent for E. faecalis cells a stress condition which does not sustain growth but in which bacteria are still able to survive. As shown in Table 1, depending on the conditions in which cells were maintained, E. faecalis might conserve or lose its culturability. When cells were maintained in lake water at low temperatures and directly illuminated, E. faecalis rapidly reached the VBNC state, as demonstrated by the lack of culturable cells (<0.1 CFU/ml) after 2 weeks. By using the Live/Dead staining kit and RT-PCR, we demonstrated that the majority of the VBNC cells were intact and that there was active transcription. Thus, it can be concluded that bacterial cells remained viable (Table 1). In contrast, when E. faecalis cells were maintained in an indirectly illuminated microcosm at 21°C, the starvation response was activated, keeping bacteria culturable at the time at which the VBNC state had been reached in the other experiment (2 × 106 CFU/ml, Table 1) and for at least 2 months afterward (data not shown).
TABLE 1.
Growth and viability of E. faecalis cells during starvation and VBNC states
| State and day | Measure of viability
|
|||
|---|---|---|---|---|
| No. of cell particles/mla | No. of CFU/ml | % Live cellsb | Presence of PBP5 mRNAc | |
| Starvation | ||||
| 0 | 1 × 107 | 1 × 107 | 100 | + |
| 10 | 1.5 × 107 | 3 × 106 | 91 | + |
| 20 | 9 × 106 | 2 × 106 | 83 | + |
| VBNC | ||||
| 0 | 1.5 × 107 | 1 × 107 | 100 | + |
| 10 | 9 × 106 | 1 × 102 | 86 | + |
| 20 | 8.5 × 106 | <0.1 | 70 | + |
Determined with a Coulter counter.
Determined by Live/Dead staining kit.
Determined by RT-PCR. PBP5, penicillin binding protein 5.
Comparison of protein profiles of exponentially growing, starved, and VBNC E. faecalis cells.
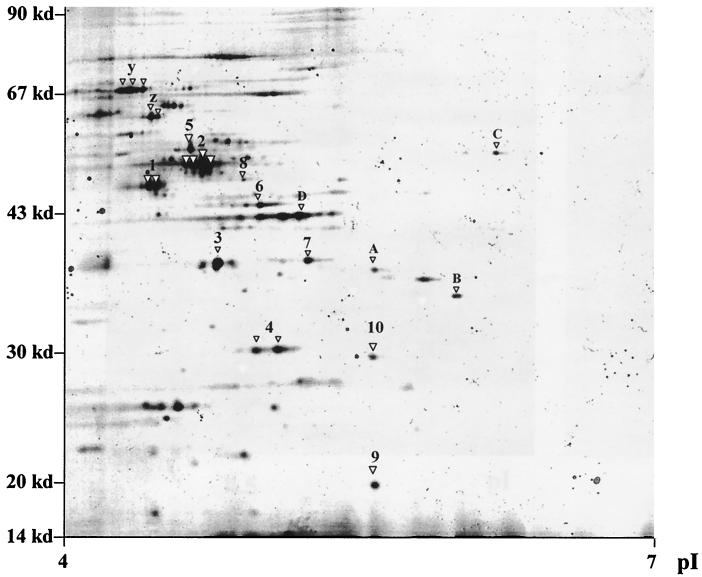
The protein profile of exponentially growing E. faecalis was initially characterized to generate a standard grid, which was used for the subsequent comparative studies with bacterial preparations obtained under stress conditions. As shown in the prototypic gel from Fig. 1, clear protein patterns were generated that enabled us to obtain sequence tags from the dominant spots (Table 2). Subsequent studies performed under a broad range of pI and molecular weight conditions with protein extracts from stressed bacteria allowed us to establish that starved and VBNC bacteria exhibited different profiles which, in turn, differed from that of exponentially growing bacteria (data not shown). These preliminary studies allowed us to select the optimal conditions to perform an analysis at higher resolution of the regions in which the main differences in the protein patterns were detected (Fig. 2 and 3).
FIG. 1.
Proteome analysis of exponentially growing E. faecalis. Protein extracts of exponentially growing bacterial cells were separated by 2D PAGE. Spots were excised or blotted onto a PVDF membrane and further characterized by Q-TOF mass spectrometry or Edman degradation. The obtained sequence tags were compared with existing databases. The identified proteins are indicated by numbers (see also Table 2). For the spots in which sequence tags were not obtained, only peptide mass fingerprints could be compared with the available databases and the putative proteins are indicated by letters. The spots indicated with y and z were identified by comparison to the gels from the work of Giard et al. (8). Designations are as follows: 1, enolase; 2, EF-Tu; 3, EF-Ts; 4, fructose-bisphosphate aldolase; 5, ATP-synthase β-chain; 6, glycerine-3-phosphate-dehydrogenase; 7, mannose-specific PTS system component IIAB; 8, glucose-6-phosphate isomerase; 9, 50S ribosomal protein L10; 10, NADH-dependent enoyl-ACP reductase; A, catabolite regulator protein; B, ABC transporter; C, ATP binding protein of amino acid ABC transporter; D, restriction endonuclease; y, DnaK protein; and z, GroEL protein.
TABLE 2.
Main characteristics of the obtained sequence tags
| Name | Method | Sequence tag | Estimateda
|
Similarity (%)b | Function | |
|---|---|---|---|---|---|---|
| Molecular mass (kDa) | pl | |||||
| Enolase | N-terminal | SIITDIYAREVL | 46.5 | 4.5 | Enterococcus faecalis (100) | Glycolysis, gluconeogenesis |
| EF-Ts | N-terminal | ADVTAKMVKEL | 32 | 4.8 | Listeria innocua (79) | Protein synthesis |
| Fructose-bisphosphate aldolase | N-terminal | PVVSGAEFLKAA | 31 | 4.8 | Streptococcus pneumoniae (90) | Glycolysis, gluconeogenesis |
| Enoyl-ACP reductase, NADH dependent | N-terminal | FLQNKNVVIMGVAN | 26.7 | 5.3 | Lactococcus lactis (82) | Fatty acid biosynthesis |
| Glyceraldehyde-3-phosphate dehydrogenase | N-terminal | TVKVGINGFGRI | 36 | 4.9 | Lactobacillus delbrueckii (85) | Carbohydrate metabolism |
| Glucose-6-phosphate isomerase | N-terminal | HIQLDYSKLAPF | 49.7 | 4.96 | Streptococcus pneumoniae (86) | Carbohydrate metabolism |
| 50S ribosomal protein L10 | N-terminal | SEAAIAKKETLV | 17.6 | 5.4 | Streptococcus pyogenes (83) | Protein synthesis |
| Q-TOF | VSSVEQLTALAK | |||||
| ATP synthase β-chain (H+ ATPase) | Q-TOF | FTQAGSEVSALLGR | 50.8 | 4.7 | Enterococcus faecalis (87) | ATP synthesis |
| Mannose-specific PTS system, factor IIAB | Q-TOF | VLSMGQEDVDTFDLIEQAAP | 35.5 | 5.1 | Clostridium acetobutylicum (80) | Sugar active transport |
| EF-Tu | Q-TOF | DLLSEYDFPGDDVPVLAGSALK | 43.3 | 4.7 | Staphylococcus aureus (85) | Protein synthesis |
| MALDI-TOF | LLDYAEAGDNLGALLR | |||||
Calculated from sequence (TIGR).
Protein BLAST NCBI.
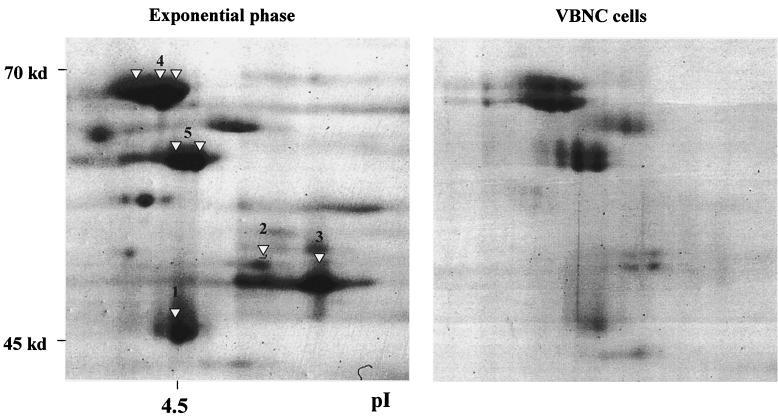
FIG. 2.
Comparison of the protein profiles of exponentially growing and VBNC E. faecalis cells within the 45- to 70-kDa region. Protein extracts of exponentially growing bacteria or VBNC cells were separated by 2D PAGE. Spots were excised or blotted onto a PVDF membrane and further characterized by Q-TOF mass spectrometry or Edman degradation. The obtained sequence tags were compared with existing databases. The putative proteins are indicated by numbers (see also Table 2). Designations are as follows: 1, enolase; 2, ATP-synthase β-chain; 3, EF-Tu; 4, DnaK; and 5, GroEL.
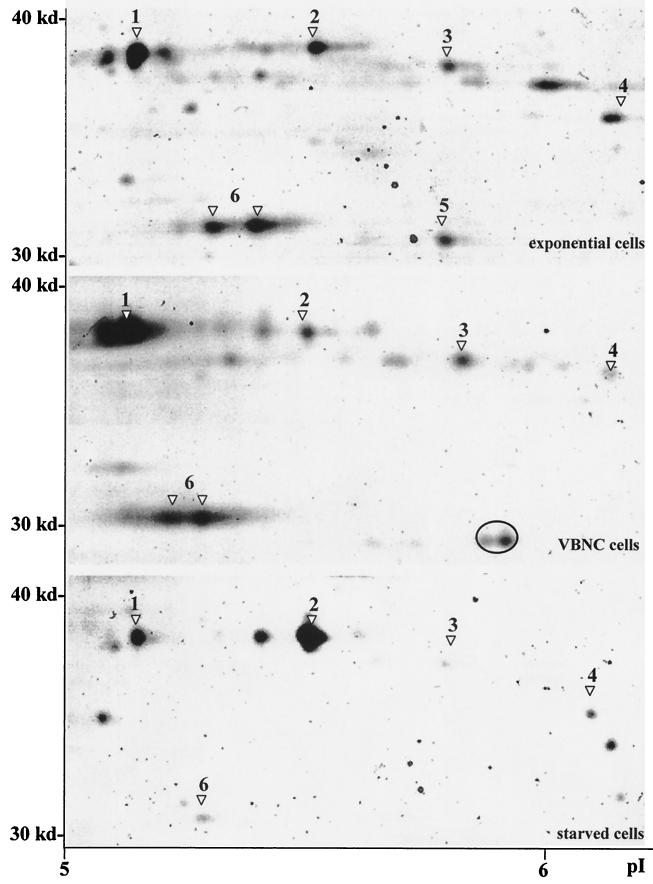
FIG. 3.
Comparison of the protein profiles of exponentially growing, VBNC, and starved E. faecalis cells within the 30- to 40-kDa region. Protein extracts of exponentially growing bacteria, VBNC cells, or starved cells were separated by 2D PAGE. Spots were excised or blotted onto a PVDF membrane and further characterized by Q-TOF mass spectrometry or Edman degradation. The obtained sequence tags were compared with existing databases. The identified proteins are indicated by numbers (see also Table 2). A putative unknown VBNC-specific protein spot is enclosed by a circle. Designations are as follows: 1, EF-Ts; 2, homologue of mannose-specific PTS system component IIAB (EC 2.7.1.69) of L. lactis (73% similarity); 3, putative catabolite regulator protein; 4, putative ABC transporter; 5, NADH-dependent enoyl-ACP reductase; and 6, fructose-bisphosphate aldolase.
Figure 2 shows the expression of proteins from exponentially growing and VBNC cells in the higher-molecular-weight region. The two general stress proteins GroEL and DnaK (8) seem to be expressed at slightly lower levels in the VBNC state than in exponential growth. Interestingly, other high-molecular-weight proteins, such as the putative proteins enolase and ATP-synthase β-chain, or the homologue of the elongation factor Tu (EF-Tu) of Staphylococcus aureus, are dramatically reduced in VBNC cells.
Figure 3 shows the protein spots with molecular masses between 30 and 40 kDa and pIs in the range of 5 to 6, a zone of the gels in which significant differences were observed among the protein profiles of exponentially growing, starved, and VBNC cells. Three proteins exhibiting homology to the elongation factor Ts (EF-Ts) of Listeria innocua, the fructose-bisphosphate aldolase of Streptococcus pneumoniae, and a putative catabolite regulator protein of E. faecalis (CAA09491) were overexpressed in VBNC cells compared with exponentially growing cells. Interestingly, these three proteins were in turn down-regulated in starved bacteria. A pH shift of fructose-bisphosphate aldolase was also observed in VBNC cells, the more basic spot present in exponentially growing cells being replaced by an acidic variant. In contrast, a homologue of the mannose-specific phosphotransferase (PTS) system component IIAB of Clostridium acetobutylicum seemed to be activated under starvation but not under VBNC conditions. The homologue of an enoyl-acyl carrier protein (ACP) reductase of Lactococcus lactis was down-regulated in both starved and VBNC bacteria. Finally, a protein spot with an approximate molecular mass of 30 kDa at pI 6 was observed only in VBNC cells. Our efforts to identify this protein proved unsuccessful. However, we cannot rule out the possibility that it is a modified form of the enoyl-ACP reductase homologue.
The proteome data were further confirmed by a transcriptional analysis of the fba (fructose bisphosphate aldolase), tuf (EF-Tu), and tsf (EF-Ts) genes of E. faecalis cells which had entered the VBNC state, performed by RT-PCR as previously described (17). As shown in Table 3, the mRNAs corresponding to the two proteins overexpressed in the VBNC state, namely, fructose-bisphosphate aldolase and EF-Ts, were detected in E. faecalis for at least 14 days after cells entered the VBNC state. In contrast, although the EF-Tu mRNA was present when bacteria reached the VBNC state, it was not detected in later samples. The ftsz gene, which is involved in septum formation during cell division, and the pbp5 gene, previously shown to be expressed for 3 months in VBNC cells of E. faecalis (17), were included as negative and positive controls, respectively.
TABLE 3.
RT-PCR analysis of VBNC E. faecalis cells
| Gene | Protein | Primers | Amplicon (bp) | mRNA detected in VBNC cells
|
||
|---|---|---|---|---|---|---|
| Day 0 | Day 7 | Day 14 | ||||
| fba | Fructose-bisphosphate aldolase | 5′ GCAGCAGAAGCTAA 3′ | 270 | + | + | ± |
| 5′ AGATACCTTTAGCGTG 3′ | ||||||
| tsf | EF-Ts | 5′ TCTTCCCCTTGGTTACC 3′ | 180 | + | + | + |
| 5′ CGTAAAGAACGCATTGC 3′ | ||||||
| tuf | EF-Tu | 5′ TGGCCCTATGCCTCAAAC 3′ | 400 | + | − | − |
| 5′ ACCAGTTGCAACAGTACC 3′ | ||||||
| pbp5 | Penicillin binding protein 5 | 5′ CATGCGCAATTAATCGG 3′ | 444 | + | + | + |
| 5′ CATAGCCTGTCGCAAAAC 3′ | ||||||
| ftsz | FTSZ | 5′ AAGTTATCGGTGTAG 3′ | 400 | ± | − | − |
| 5′ GCGGCAAAACGACCA 3′ | ||||||
It is well known that the EF-Ts protein mediates the regeneration of EF-Tu-GDP complex into the active form, EF-Tu-GTP. The active form of EF-Tu facilitates the entry of aminoacyl-tRNA into the ribosome, enabling protein synthesis (26). Besides its essential role in protein biosynthesis, EF-Tu may play a role in cell growth regulation and stress responses (32). EF-Tu can regulate translation through its interaction with tRNA and ribosomes, stopping the translation of unnecessary proteins and triggering the synthesis of stress-induced proteins (32). Nutrient deprivation in the environment might result in EF-Tu hypermethylation and membrane release (31). Thus, considering its central regulatory role, the observed down-regulation of the EF-Tu in VBNC cells is intriguing. The up-regulation of EF-Ts may represent a compensatory mechanism to ensure protein synthesis and EF-Tu-mediated regulatory activities in conditions in which EF-Tu is no longer expressed. The overproduction of EF-Ts may also allow the regeneration of EF-Tu-GTP from the EF-Tu-GDP pool, in order to allow the protein synthesis to continue for a limited period.
As regards the other proteins differing in their levels of expression among exponentially growing, starved, and VBNC cells, three of them are involved in catabolic pathways. Enolase is involved in glycolysis, ATP synthase is involved in oxidative phosphorylation, and enoyl-ACP reductase is involved in the biosynthesis of phospholipids. This seems to be in agreement with a shutdown of the metabolism in VBNC cells. It can be hypothesized that VBNC cells would require alternative metabolic pathways to obtain energy in order to survive under unfavorable conditions. This function can be provided by the overexpressed homologue of the fructose-bisphosphate aldolase. The expression of this enzyme was demonstrated to be activated when bacteria are grown under nitrogen-limiting conditions (25). The fructose-bisphosphate aldolase plays a crucial role in central carbohydrate metabolic pathways such as glycolysis, gluconeogenesis, the pentose phosphate pathway, fructose and mannose metabolism, and carbon fixation (http://www.genome.ad.jp/dbget-bin/www_bget?enzyme + 4.1.2.13).
The alpha enolase of S. pneumoniae has a plasminogen-binding function and seems to increase bacterial virulence in invasive infections (2). The enolase of E. faecalis shows 80% homology with the one from S. pneumoniae; thus, a similar function can be proposed for the former. Its down-regulation during the VBNC state does not exclude the maintenance of other virulence factors during the unculturable state, as suggested by previous reports (22). Alternatively, the enolase of E. faecalis may not be involved in the pathogenesis process, its function being restricted to the metabolism of carbohydrates. Finally, the overexpression of the mannose-specific PTS-system component IIAB in starved cells is in agreement with the requirement forcarbon sources under certain stress conditions. Although the PTS system appears to play a vital role under different stress conditions (28), this does not seem to apply for the VBNC stress. In fact, the level of expression of this protein in the VBNC state was similar to that of growing cells.
The genetic pathways underlying the VBNC response appear to be in part the same as those leading to the starvation response, as indicated by the presence of similar expression profiles for certain proteins (e.g., enoyl-ACP reductase). However, major differences in the expression patterns of other proteins (e.g., EF-Ts, fructose-bisphosphate aldolase, and mannose-specific PTS system) were observed. This suggests that the underlying pathways in the starvation and VBNC states are not overlapping but rather share a few crossing points. Moreover, the VBNC state also differs from other general stress responses, as indicated by the down-regulation of a general stress protein such as the mannose-specific PTS system. In conclusion, our results indicate that the protein profile of VBNC cells is different from those of cells which are either starved or exponentially growing. This demonstrates that the VBNC state constitutes a physiologically distinct state within the life cycle of E. faecalis, which is activated in response to multiple environmental stresses.
Acknowledgments
We thank D. Bludau and M. C. Tafi for outstanding technical help and K. N. Timmis for generous support and encouragement during this study.
This study was in part supported by the grant 01.00255.PF 49 from the Consiglio Nazionale delle Ricerche (CNR), by Cofin2000 from the Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR), and by a fund from the European Commission (AQUA-CHIP project, QLK4-2000-00764).
The authors are solely responsible for the content of this publication. It does not represent the opinion of the European Commission. The European Commission is not responsible for any use that might be made of data appearing therein.
REFERENCES
- 1.Barcina, I., P. Lebaron, and J. Vives-Rego. 1997. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol. Ecol. 23:1-9. [Google Scholar]
- 2.Bergmann, S., M. Rohde, S. Gursharan S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 3.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 4.Colwell, R. R. 2000. Bacterial death revisited, p. 325-342. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 5.Edmond, M. B., J. F. Ober, J. D. Dawson, D. L. Weinbaum, and R. P. Wenzel. 1996. Vancomycin-resistant enterococcal bacteremia: natural history and attributable mortality. Clin. Infect. Dis. 23:1234-1239. [DOI] [PubMed] [Google Scholar]
- 6.Giard, J. C., A. Hartke, S. Flahaut, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. Starvation-induced multiresistance in Enterococcus faecalis JH2-2. Curr. Microbiol. 32:264-271. [DOI] [PubMed] [Google Scholar]
- 7.Giard, J. C., A. Hartke, S. Flahaut, P. Boutibonnes, and Y. Auffray. 1997. Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res. Microbiol. 148:27-35. [DOI] [PubMed] [Google Scholar]
- 8.Giard, J. C., J. M. Laplace, A. Rincé, V. Pichereau, A. Benachour, C. Leboeuf, S. Flahaut, Y. Auffray, and A. Hartke. 2001. The stress proteome of Enterococcus faecalis. Electrophoresis 22:2947-2954. [DOI] [PubMed] [Google Scholar]
- 9.Görg, A., G. Boguth, C. Obermaier, A. Posch, and W. Weiss. 1995. Two-dimensional polyacrylamide gel electrophoresis with immobilized pH gradients in the first dimension (IPG-DALT): the state of the art and the controversy of vertical versus horizontal systems. Electrophoresis 16:1079-1086. [DOI] [PubMed] [Google Scholar]
- 10.Hartke, A., J. C. Giard, J. M. Laplace, and Y. Auffray. 1998. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl. Environ. Microbiol. 64:4238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kogure, K., U. Simidu, and N. Taga. 1979. A tentative direct microscopic method for counting living bacteria. Can. J. Microbiol. 25:415-420. [DOI] [PubMed] [Google Scholar]
- 12.Kurath, G., and Y. Morita. 1983. Starvation-survival physiological studies of a marine Pseudomonas sp. Appl. Environ. Microbiol. 45:1206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leboeuf, C., L. Leblanc, Y. Auffray, and A. Hartke. 2000. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by CcpA. J. Bacteriol. 182:5799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lleo, M. D. M., B. Bonato, M. C. Tafi, C. Signoretto, M. Boaretti, and P. Canepari. 2001. Resuscitation rate in different enterococcal species in the viable but non-culturable state. J. Appl. Microbiol. 91:1095-1102. [DOI] [PubMed] [Google Scholar]
- 15.Lleo, M. D. M., M. C. Tafi, C. Signoretto, C. Dal Cero, and P. Canepari. 1999. Competitive polymerase chain reaction for quantification of nonculturable Enterococcus faecalis cells in lake water. FEMS Microbiol. Ecol. 30:345-353. [DOI] [PubMed] [Google Scholar]
- 16.Lleo, M. D. M., M. C. Tafi, and P. Canepari. 1998. Nonculturable Enterococcus faecalis cells are metabolically active and capable of resuming active growth. Syst. Appl. Microbiol. 21:333-339. [DOI] [PubMed] [Google Scholar]
- 17.Lleò, M. D. M., S. Pierobon, M. C. Tafi, C. Signoretto, and P. Canepari. 2000. mRNA detection by reverse transcription-PCR for monitoring viability over time in an Enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl. Environ. Microbiol. 66:4564-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita, R. Y. 1993. Bioavailability of energy and the starvation state, p. 8-16. In S. Kjelleberg (ed.), Starvation in bacteria. Plenum Press, Inc., New York, N.Y.
- 19.Oliver, J. D. 2000. The public health significance of viable but nonculturable bacteria, p. 277-300. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 20.Oliver, J. D., L. Nilsson, and S. Kjelleberg. 1991. Formation of nonculturable Vibrio vulnificus cells and its relationship to the starvation state. Appl. Environ. Microbiol. 57:2640-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paludan-Muller, C., D. Weichart, D. McDougald, and S. Kjelleberg. 1996. Analysis of starvation conditions that allow for prolonged culturability of Vibrio vulnificus at low temperature. Microbiology 142:1675-1684. [DOI] [PubMed] [Google Scholar]
- 22.Pruzzo, C., R. Tarsi, M. M. Lleó, C. Signoretto, M. Zampini, R. R. Colwell, and P. Canepari. 2002. In vitro adhesion to human cells by viable but nonculturable Enterococcus faecalis. Curr. Microbiol. 45:105-110. [DOI] [PubMed] [Google Scholar]
- 23.Robbi, C., C. Signoretto, M. Boaretti, and P. Canepari. 1996. The gene encoding for penicillin binding protein 5 of Enterococcus faecalis is useful for development of a species-specific DNA probe. Microb. Drug Resist. 2:215-218. [DOI] [PubMed] [Google Scholar]
- 24.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmi, R., E. M. Uhlemann, L. Nolden, G. Wersch, R. Hecker, T. Hermann, A. Marx, and A. Burkovski. 2000. Response to nitrogen starvation in Corynebacterium glutamicum. FEMS Microbiol. Lett. 187:83-88. [DOI] [PubMed] [Google Scholar]
- 26.Seshadri, R., L. R. Hendrix, and J. E. Samuel. 1999. Differential expression of translational elements by life cycle variants of Coxiella burnetii. Infect. Immun. 67:6026-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Signoretto, C., M. D. M. Lleò, M. C. Tafi, and P. Canepari. 2000. Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl. Environ. Microbiol. 66:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stülke, J., and W. Hillen. 2000. Regulation of carbon metabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 29.Toranzos, G. A., and G. A. McFeters. 1997. Detection of indicator microorganisms in environmental freshwaters and drinking waters, p. 184-194. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 30.Wissing, J., S. Heim, L. Flohè, U. Bilitewski, and R. Frank. 2000. Enrichment of hydrophobic proteins via Triton X-114 phase partitioning and hydroxyapatite column chromatography for mass spectrometry. Electrophoresis 21:2589-2593. [DOI] [PubMed] [Google Scholar]
- 31.Wong, D. K., B. Lee, M. A. Horwitz, and B. W. Gibson. 1999. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect. Immun. 67:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young, C. C., and R. W. Bernlohr. 1991. Elongation factor Tu is methylated in response to nutrient deprivation in Escherichia coli. J. Bacteriol. 173:3096-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]