Abstract
Escherichia coli MG1655 acid-inducible genes were identified by whole-genome expression profiling. Cultures were grown to the mid-logarithmic phase on acidified glucose minimal medium, conditions that induce glutamate-dependent acid resistance (AR), while the other AR systems are either repressed or not induced. A total of 28 genes were induced in at least two of three experiments in which the gene expression profiles of cells grown in acid (pH 5.5 or 4.5) were compared to those of cells grown at pH 7.4. As expected, the genes encoding glutamate decarboxylase, gadA and gadB, were significantly induced. Interestingly, two acid-inducible genes code for small basic proteins with pIs of >10.5, and six code for small acidic proteins with pIs ranging from 5.7 to 4.0; the roles of these small basic and acidic proteins in acid resistance are unknown. The acid-induced genes represented only five functional grouping categories, including eight genes involved in metabolism, nine associated with cell envelope structures or modifications, two encoding chaperones, six regulatory genes, and six unknown genes. It is unlikely that all of these genes are involved in the glutamate-dependent AR. However, nine acid-inducible genes are clustered in the gadA region, including hdeA, which encodes a putative periplasmic chaperone, and four putative regulatory genes. One of these putative regulators, yhiE, was shown to significantly increase acid resistance when overexpressed in cells that had not been preinduced by growth at pH 5.5, and mutation of yhiE decreased acid resistance; yhiE could therefore encode an activator of AR genes. Thus, the acid-inducible genes clustered in the gadA region appear to be involved in glutatmate-dependent acid resistance, although their specific roles remain to be elucidated.
Enteric bacteria survive exposure to acidic environments, such as those encountered in the mammalian stomach, by inducing genes that provide for acid resistance and pH homeostasis (4, 14, 18). Escherichia coli has three distinct systems for acid resistance (AR): a glucose-catabolite-repressed system and two amino acid decarboxylase-dependent systems (9). These systems overlap in ways that protect both growing and nongrowing cells from acid across a broad range of environmental conditions. The mechanism of the glucose-repressed AR is not known. The amino acid decarboxylase-dependent AR systems are thought to consume protons that leak into the cell during acid stress by decarboxylation of arginine or glutamate (16), which are transported into the cell in exchange for their respective decarboxylated products (24).
The more effective E. coli AR system is dependent on glutamate and is comprised of at least three genes: gadA and gadB, encoding glutamate decarboxylase; and gadC, encoding a γ-aminobutyrate (GABA) antiporter. There are several open questions regarding the mechanism of glutamate-dependent AR. For example, it appears that the decarboxylase-dependent systems would create a futile proton cycle, since transport of glutamic acid brings a proton into the cell that is consumed by its decarboxylation. This suggests that there should be a mechanism for endogenous glutamate generation. For this reason alone, it is clear that additional genes must participate in the glutamate-dependent AR, but these genes remain to be identified (9).
Functional genomics provides a comprehensive approach for identification of additional genes involved in AR. Proteome studies revealed 13 (19) or 18 (6) E. coli proteins induced by acid, including GadA. Whole genome expression profiling of E. coli cells growing on minimal glucose medium revealed that gadA, gadB, hdeA, and hdeB were induced relative to growth on rich medium (25). The hdeA gene is thought to encode a periplasmic chaperone involved in AR (15). Other gene expression profiling experiments revealed 7 acid-inducible genes as being HN-S dependent (20), and 26 genes were induced by acetate, which increases acid survival (2). Although these array experiments identified a number of acid-induced genes, they were not designed explicitly for this purpose.
In the present study, we grew E. coli MG1655 to the mid-logarithmic phase on acidified glucose minimal medium, conditions known to induce the glutamate-dependent AR and repress the glucose-repressed AR, but which fail to induce the arginine-dependent AR. The gene expression profiles of cells grown at pH 5.5 and 4.5 were compared to those of cells grown at pH 7.4: 28 genes showing statistically significant up-regulation were identified, a number of which are clustered on the genome with gadA. Several of these genes were inactivated by allelic replacement and tested for their ability to survive acid challenge. One of them, yhiE, was found to significantly affect AR.
MATERIALS AND METHODS
Strains and culture conditions.
The wild-type E. coli strain MG1655 (CGSC 7740) was obtained from the E. coli Genetic Stock Culture Collection at Yale University. All cultures used for genomic expression profiling were grown in the minimal medium developed for E. coli proteome studies (21). Glucose (0.2%) was the sole source of carbon and energy. Morpholinepropanesulfonic acid (MOPS) was used as the buffer for pH 7.4 media, and morpholineethanesulfonic acid (MES) was used to buffer the pH 4.5 and 5.5 media. Cultures were grown aerobically with agitation at 300 rpm at 37°C in 50 ml of medium in 250-ml fleakers (Corning, Acton, Mass.). Cultures were inoculated to an initial A600 of less than 0.001 to ensure that the cells experienced at least 10 generations in the medium prior to reaching steady state (mid-logarithmic growth phase). Cell growth was monitored spectrophotometrically (DU530 spectrophotometer; Beckman Instruments, Fullerton, Calif.) at 600 nm by removing 100-μl samples without interrupting culture agitation; no more than 5% of the culture was removed prior to harvesting of cells for RNA isolation. Since both the rate of growth and total yield of biomass were affected by alterations in the medium pH, the shape of the growth curve was used to determine the midpoint in logarithmic growth for cell harvest.
RNA isolation.
For RNA isolation, cells were harvested from the mid-logarithmic phase of growth by removing 5.0 ml of culture and rapidly (less than 2 s) pipetting them into 5.0 ml of ice-cold RNAlater (Ambion, Austin, Tex.). Cells were pelleted from this mixture by centrifugation (8,730 × g for 20 min). RNA was isolated and purified with an RNeasy Mini kit (Qiagen, Valencia, Calif.). After purification, genomic DNA was removed by treatment with RNase-free DNase I (Ambion), with subsequent RNA repurification with an RNeasy column. RNA samples were quantified spectrophotometrically at 260 nm (Beckman DU530 spectrophotometer).
Labeled target synthesis.
Labeled targets for hybridization were generated as described previously (25), but with slight modifications. The initial preincubation (90°C for 2 min and then 42°C for 20 min in an MJ Research MiniCycler) was performed in 0.2-ml PCR tubes (1 μg of RNA template, dATP, dTTP, and dGTP at a final concentration of 0.33 mM each), and C-terminal primers (Sigma Genosys Biotechnologies, Inc., The Woodlands, Tex.) in a final volume of 23 μl with RNase-free dH2O). After preincubation, 20 μCi of [α-33P]dCTP (2,000 to 3,000 Ci/mol; Perkin-Elmer Life Sciences, Inc., Boston, Mass.), 200 U of Superscript II RNase H− reverse transcriptase (Invitrogen Corp., Carlsbad, Calif.), 10 mM dithiothreitol (Invitrogen Corp.), and 10 U of RNase inhibitor (Invitrogen Corp.) were added, bringing the final volume to 30 μl. The cDNA reaction mixture was incubated for 2 h at 42°C. The labeled cDNA was denatured from the RNA template for 30 min at 65°C with 29 mM EDTA and 0.43 N NaOH. The denaturing reaction was neutralized with 160 mM Tris (pH 7.4) and 0.28 N HCl. Quantification of radiolabel incorporation was determined by comparing scintillation counts of 1 μl of labeled cDNA (in 5.0 ml of scintillation fluid) prior to and subsequent to removal of unincorporated nucleotide by gel filtration through a Sephadex G-50 column (Sigma-Aldrich Corp., St. Louis, Mo.). Labeling efficiencies of 70 to 90% incorporation were routinely obtained.
Hybridization of arrays and image analysis.
The DNA arrays (Panorama E. coli gene arrays; Sigma Genosys Biotechnologies, Inc.), array prehybridization, hybridization, and array washes were performed as described by Tao et al. (25), with the following modifications. Prior to prehybridization, arrays were rinsed in 50 ml of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and then prehybridized with 5.0 ml of hybridization solution (65°C for 1 to 4 h) and hybridized with 3.0 ml of hybridization solution plus 400 to 600 μl of labeled cDNA probe (65°C for 16 to 18 h). The arrays were washed as per the manufacturer's instructions and then wrapped in Saran Wrap plastic film (Dow Chemical, Midland, Mich.) and exposed to a Kodak storage phosphor screen GP (Eastman Kodak Co., Rochester, N.Y.) for 24 h. Longer exposure times were occasionally necessary to increase spot intensity and contrast. Exposed phosphor screens were scanned at a pixel density of 100 μm (10,000 dots per cm2) with a STORM 820 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The array membranes were consecutively hybridized, stripped, and rehybridized (which was repeated up to eight times). Arrays were stripped of hybridized probe by gently boiling the inverted array in a Kenmore microwave oven (Sears, Roebuck and Co., Hoffman Estates, Ill.) for 20 min at power setting 3 in 200 ml of stripping solution (10 mM Tris [pH 7.5], 1 mM EDTA, 1% sodium dodecyl sulfate). Phosphorimaging of a membrane array results in a TIFF image file that must be further processed for data analysis. The image analysis software (ArrayVision version 5.1; Imaging Research, Inc., St. Catharines, Ontario, Canada) makes use of a template that is customized to accommodate the three grid layers (3 by 1, 16 by 24, and 4 by 4) of the E. coli Panorama arrays and for the spot intensity measurements to be easily associated with the correct gene identifiers. The spot labeling protocol was edited to name each spot by its unique array coordinate. The raw spot intensities were represented in a row-column format and exported into Microsoft Excel spreadsheets for further analysis. This customized ArrayVision template file and the data analysis tools described below are available on our web site (12).
Data processing.
Raw data from each experimental replicate were analyzed in Excel workbooks containing manually executed macros written in Visual Basic (12). The first step in the analysis associates the array coordinate for each spot with a unique spot number, the gene name, and related gene annotation. On the membrane arrays, there are two spots for each gene, and these were treated as separate determinations. The raw data were normalized by expressing spot intensities as a percentage of the sum of all of the gene-specific spot intensities. The second step in the analysis applies Student's t test to determine the probability that the average of the experimental replicates (e.g., pH 5.5) is significantly different from the average of the control replicates (e.g., pH 7.4). The P values (derived from Student's t test) for the normalized and natural log-transformed data were calculated. The third step calculates relative gene expression between conditions by introducing a threshold value, chosen to be representative of the limit of detection of expressed genes, and then calculating the ratio of the experimental to control expression levels such that genes that are more highly expressed under the experimental condition are given a positive value, and genes that are more highly expressed under the control condition are given a negative value. Statistical significance was established by consideration of two metrics: the P value, which takes into consideration the uncertainty of the replicate measurements for both conditions; and the standard deviation of the mean of the log (base 10) ratios, which indicates the significance of the ratio. In the experiments reported here, we considered only those genes that were significantly up-regulated by a value greater than 3 standard deviations of the mean of the log ratios for all genes (99.9% confidence) and those that had a P value > 0.05 (95% probability that the ratio is significant).
Web site and public access to analysis tools.
A web site has been created for users to download these analysis tools and array protocols (http://www.ou.edu/microarray). The web site describes the step-by-step implementation of the analysis tools (12).
Characterization of acid-induced genes.
PCR was performed with Stratagene Taq2000 (La Jolla, Calif.) under standard conditions. Gene knockouts were constructed by allelic replacement. The one-step method developed by Datsenko and Wanner (12a) was used with slight modifications for optimization in E. coli MG1655: a higher concentration of the Kanr insert PCR product (≥1 μg) was employed during electroporation, and an increased concentration of kanamycin (40 μg/ml) was used during mutant screening. PCR-amplified genes were cloned by using the Stratagene PCR-Script Amp cloning kit, according to the manufacturer's instructions. Clones were selected by growth on Luria broth (LB) containing ampicillin (75 μg/ml) and verified by restriction analysis.
Acid tolerance assay.
The method used for assay of acid survival was similar to that designed by Castanie-Cornet et al. (9), except that glutamate was omitted. Strains for testing were pregrown at 37°C with shaking at 250 rpm to the stationary phase (24 h) in MOPS minimal medium (pH 7.4) and then were inoculated (optical density at 600 nm [OD600] = 0.001) into MES minimal medium (pH 5.5) for activation of the AR genes. These cultures were grown exponentially to an OD600 of 0.4 and then acid challenged at pH 2.75 for 2 h. Cultures were acidified by dilution (1:10) of the overnight culture (pH 5.5) into the same medium at pH 2.5 to give a final pH of 2.75. Acid challenge was stopped by dilution and plating on LB. Cell viability was determined by triplicate plate counts.
RESULTS
Experimental design and reproducibility.
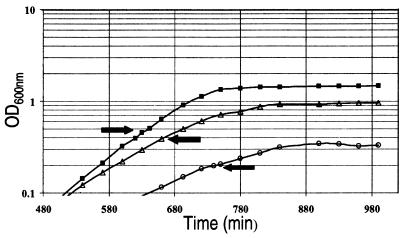
An experiment designed to favor induction of the glutamate-dependent AR system must take into consideration the mode of regulation of all three AR systems. The first AR system is catabolite repressed by glucose and induced in complex medium; the second arginine-dependent AR system is regulated by CysB and induced only in the stationary phase; and the third glutamate-dependent AR system is independently regulated by extracellular pH, HN-S, and RpoS (8, 9, 13). Thus, growth on glucose represses the first AR system, and the second AR system is not induced in mid-log-phase cells. Therefore, to favor induction of the glutamate-dependent AR system, we grew E. coli MG1655 on glucose-MOPS or MES minimal medium and compared the gene expression profiles of log-phase cells grown at pH 7.4 to cells grown at pH 4.5 and 5.5 (Fig. 1).
FIG. 1.
Growth curves of E. coli MG1655 grown in pH 7.4 MOPS minimal medium (solid squares), pH 5.5 MES minimal medium (open triangles), or pH 4.5 MES minimal medium (open circles) supplemented with 0.2% glucose. RNA was isolated at the times indicated by the arrows.
RNA samples from pH 7.4 and 5.5 cultures were labeled and hybridized in duplicate (technical replication) to matched membrane pairs of DNA arrays (Fig. 2), and the experiment was repeated (biological replication). Correlation coefficients for the technical replicates from the first experiment were 0.977 for the pH 7.4 replicates and 0.968 for the pH 5.5 replicates. The correlation coefficients for the technical replicates from the second experiment were 0.967 for the pH 7.4 replicates and 0.926 for the pH 5.5 replicates. Although the correlation coefficients of these two experiments were not significantly different, we noted that it was not possible to directly compare the two data sets, because the membranes were printed from different sets of PCR products (information provided by the manufacturer). In a third experiment, cells were grown at pH 4.5; the technical replicates had a correlation coefficient of 0.918. The expression ratios of the pH 4.5 experiment were calculated by comparison to the second pH 7.4 experiment, which was run on the same membranes.
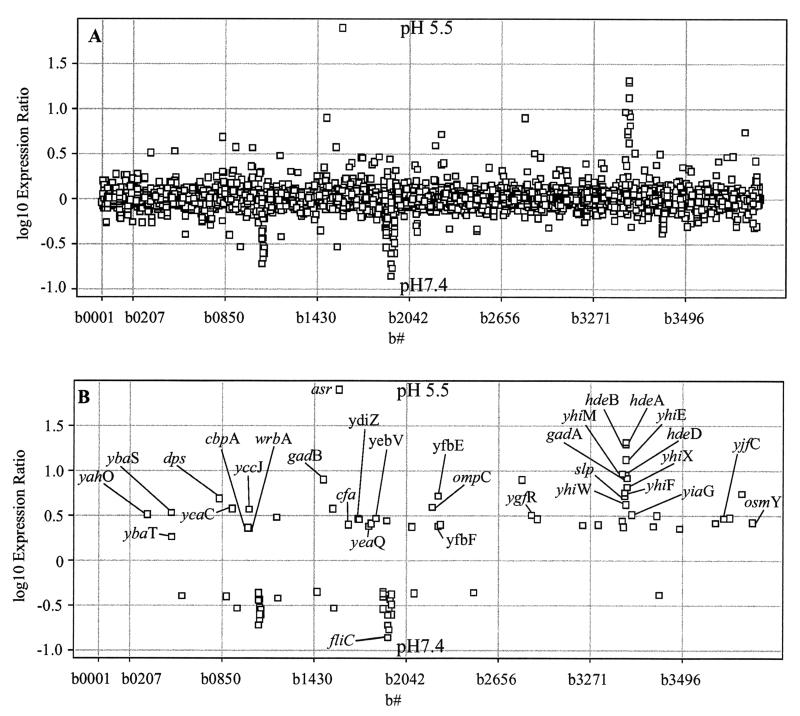
FIG. 2.
(A) Spotfire scatter plot analysis of E. coli MG1655 gene expression under acidic (pH 5.5) and neutral (pH 7.4) growth conditions. Gene expression was determined with RNA isolated from pH 5.5 MES minimal or pH 7.4 MOPS minimal medium supplemented with 0.2% glucose. All 4,290 E. coli genes are represented in the scatter plot, with each square indicating the log10 expression ratio of an individual gene. Genes more highly expressed at pH 5.5 are shown above the centerline, while those below the centerline are more highly expressed at pH 7.4. (B) Statistically significant genes from panel A. Statistical significance was determined as induction ≥3 standard deviations from the mean (no change in expression) and a P value of ≤ 0.05 as determined by Student's t test. Induced genes discussed in the text and shown in Table 1 are labeled on the plot.
Acid-induced genes.
Genes with statistically significant differential expression had to meet two criteria: a log expression ratio of ≥3 standard deviations of the mean of the log ratios (99.9% significance) and a P value of ≤0.05 (95% probability that the ratio is significant). This statistical approach revealed genes with a reasonable probability of induction (or repression) by acid, when compared to the control at pH 7.4 (12). In the first experiment, 49 genes were expressed at significantly higher levels at pH 5.5, and 34 genes were expressed at significantly higher levels at pH 7.4 (Fig. 2). In the second experiment, 24 genes were expressed at significantly higher levels at pH 5.5, and 10 were expressed at significantly higher levels at pH 7.4 (data not shown). In the third experiment, 35 genes were expressed at significantly higher levels at pH 4.5, and 23 were expressed at significantly higher levels at pH 7.4 (data not shown). Interestingly, only one gene, fliC, was expressed at significantly higher levels at pH 7.4 in all three experiments. Since we were primarily interested in genes induced by acid, we did not give any further consideration to genes repressed by acid.
The 28 genes that were induced by acid in at least two of the three experiments were considered in detail (Table 1). There were 21 genes with significant induction in both pH 5.5 experiments, 22 genes were significantly induced at pH 4.5 and in the first pH 5.5 experiment, and 16 genes were significantly induced at pH 4.5 and in the second pH 5.5 experiment. In sum, 15 genes were significantly induced in all three experiments. Three other genes, ybaT, yhiF, and yhiW, were added to Table 1 because they were induced in at least one of the experiments and appeared to be located in operons with other significantly induced genes. The annotations for the 31 genes listed in Table 1 were updated by BLAST and Domain Structure (ProDom) analysis by using the Colibri, EcoGene, NCBI, and SwissProt databases (as of 15 February 2002); the genes were assigned to functional groups accordingly.
TABLE 1.
Expression ratios and statistical significance of acid-induced gene comparisons
| b no. | Gene | Gene product | Functional group | Size (amino acids) | Log10 expression ratioa
|
Pb
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| pH 5.5
|
pH 4.5 | pH 5.5
|
pH 4.5 | |||||||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | |||||||
| b1661 | cfa | Cyclopropane fatty acyl phospholipid synthase | Cell envelope | 382 | 0.387 | 0.094e | 0.470 | 9.47E-06 | 8.09E-02 | 2.35E-08 |
| b1795 | yeaQ | Putative transglycosylase-associated membrane protein | Cell envelope | 82 | 0.366 | 0.254 | 0.254e | 3.82E-04 | 7.18E-03 | 3.60E-03 |
| b2215 | ompC | Outer membrane protein 1b (lb;c) | Cell envelope | 367 | 0.583 | 0.405 | 0.410e | 1.27E-04 | 1.38E-04 | 3.71E-04 |
| b2253 | yfbE | Putative aminotransferase; modification of lipid A with aminoarabinose | Cell envelope | 379 | 0.704 | 0.352 | 0.300e | 3.57E-04 | 5.24E-04 | 1.90E-03 |
| b2254 | yfbF | Putative glycosyl transferase; LPS biosynthesis | Cell envelope | 254 | 0.371 | 0.263 | 0.180e | 2.83E-04 | 2.09E-05 | 1.06E-03 |
| b3506 | slpc | Outer membrane lipoprotein; induced by stationary phase and starvation | Cell envelope | 188 | 0.710 | 0.462 | 0.976 | 8.54E-05 | 6.01E-05 | 6.88E-10 |
| b3509 | hdeBc | Putative periplasmic protein; unknown function | Cell envelope | 108 | 1.287 | 0.625 | 1.113 | 1.60E-09 | 2.38E-04 | 4.70E-06 |
| b3511 | hdeDc | Putative membrane protein; unknown function | Cell envelope | 190 | 0.942 | 0.342 | 0.584 | 4.50E-06 | 8.59E-05 | 6.93E-07 |
| b4376 | osmY | Periplasmic protein; hyperosmotically inducible | Cell envelope | 201 | 0.414 | 0.259 | 0.107e | 6.33E-03 | 3.32E-02 | 9.68E-02 |
| b1000 | cbpA | Curved-DNA binding protein; functions closely related to DnaJ | Chaperones | 306 | 0.352 | 0.244e | 0.473 | 2.07E-03 | 1.70E-03 | 1.92E-05 |
| b3510 | hdeAc | Periplasmic chaperone; acid resistance phenotype | Chaperones | 110 | 1.309 | 0.677 | 1.187 | 1.18E-07 | 1.61E-03 | 1.57E-05 |
| b0329 | yahO | ORF, hypothetical protein; induced in stationary phase | Hypothetical | 91 | 0.498 | 0.285 | 0.383e | 4.37E-05 | 1.37E-02 | 1.13E-03 |
| b1003 | yccJc | ORF, hypothetical protein | Hypothetical | 75 | 0.555 | 0.274 | 0.461 | 3.56E-08 | 7.87E-03 | 2.17E-05 |
| b1597 | asrc | Acid shock RNA; ORF, hypothetical protein | Hypothetical | 102 | 1.889 | 1.828 | 2.363 | 2.68E-08 | 9.31E-15 | 3.86E-06 |
| b1724 | ydiZ | ORF, hypothetical protein | Hypothetical | 96 | 0.448 | 0.152e | 0.514 | 6.61E-05 | 2.84E-01 | 7.78E-06 |
| b1836 | yebV | ORF, hypothetical protein | Hypothetical | 78 | 0.456 | 0.204e | 0.833 | 8.38E-04 | 7.82E-02 | 3.10E-07 |
| b3491 | yhiMc | ORF, hypothetical protein | Hypothetical | 364 | 0.956 | 0.559 | 1.030 | 7.35E-06 | 9.45E-04 | 1.33E-07 |
| b1004 | wrbA | Putative flavodoxin protein | Metabolism | 198 | 0.344 | 0.193e | 0.463 | 4.18E-04 | 7.97E-02 | 8.23E-06 |
| b2885 | ygfRc | Putative xanthine permease | Metabolism | 455 | 0.496 | 0.421 | 0.722 | 1.47E-02 | 4.38E-03 | 8.54E-04 |
| b4186 | yjfC | Putative synthetase/amidase | Metabolism | 387 | 0.455 | 0.151e | 0.545 | 1.84E-04 | 7.28E-02 | 7.15E-07 |
| b0485 | ybaSc | Putative glutaminase | Metabolism | 310 | 0.520 | 0.355 | 0.755 | 1.97E-04 | 2.31E-06 | 2.72E-10 |
| b0486 | ybaTd | Putative amino acid/amine transporter | Metabolism | 430 | 0.186a | 0.147e | 0.404e | 1.11E-02 | 5.55E-03 | 1.05E-07 |
| b0897 | ycaC | Putative isochorismatase | Metabolism | 208 | 0.565 | 0.239e | 0.828 | 2.55E-06 | 1.11E-02 | 1.33E-07 |
| b1493 | gadBc | Glutamate decarboxylase isozyme | Metabolism | 466 | 0.884 | 0.637 | 1.135 | 3.51E-05 | 3.01E-03 | 5.27E-10 |
| b3517 | gadAc | Glutamate decarboxylase isozyme | Metabolism | 466 | 0.908 | 0.805 | 1.425 | 7.36E-04 | 3.90E-04 | 1.12E-10 |
| b3507 | yhiFd | Putative luxR transcriptional regulator | Regulatory | 176 | 0.746 | 0.079e | 0.379e | 3.05E-08 | 3.52E-01 | 4.66E-07 |
| b3512 | yhiEc | Putative luxR transcriptional regulator | Regulatory | 175 | 1.114 | 0.602 | 1.242 | 2.40E-05 | 4.54E-05 | 1.05E-07 |
| b3515 | yhiWd | Putative ARAC-type (family 2) regulatory protein | Regulatory | 242 | 0.610 | 0.071e | 0.073e | 6.24E-06 | 2.54E-02 | 1.10E-01 |
| b3516 | yhiXc | Putative ARAC-type (family 2) regulatory protein | Regulatory | 274 | 0.804 | 0.430 | 0.836 | 6.71E-07 | 6.29E-04 | 5.69E-06 |
| b3555 | yiaGc | Putative transcriptional regulator; helix-turn-helix motif | Regulatory | 96 | 0.498 | 0.264 | 0.586 | 1.07E-03 | 8.75E-03 | 9.34E-05 |
| b0812 | dpsc | DNA binding protein; global regulator, starvation conditions | Regulatory | 167 | 0.675 | 0.401 | 0.780 | 2.97E-05 | 2.47E-04 | 1.79E-06 |
Log induction ratio of experimental pH versus pH 7.4.
P value calculated by application of Student’s t test for replicates of natural log-transformed, normalized spot intensities.
Significantly induced in all three experiments.
Significantly induced in only one experiment.
Induction ratio not significantly different.
The 31 acid-induced genes listed in Table 1 fit into only five categories when grouped by function. Interestingly, 9 of the 31 acid-induced genes code for small proteins of 110 amino acids or less. Of these genes, asr and yeaQ code for small basic proteins with pIs of 11.1 and 10.5, respectively. With the exception of yiaG, which codes for a putative transcription factor with a pI of 8.1, the remainder of these genes, hdeB, hdeA, yahO, yccJ, ydiZ, and yebV, code for small acid proteins with pIs ranging from 5.7 to 4.0.
Nine acid-induced genes are associated with cell envelope structures or modifications: cfa encodes cyclopropane fatty acyl phospholipid synthase; yfbE and yfbF, which apparently form an operon, as well as yeaQ, are putatively involved in glycosyl transferase reactions that modify lipopolysaccharide; ompC encodes an outer membrane protein, slp encodes a stationary-phase-inducible outer membrane lipoprotein of unknown function (22); hdeD encodes an integral membrane protein of unknown function; osmY encodes a periplasmic protein that is induced by membrane perturbations and osmotic and other stresses (5), and hdeB encodes a putative periplasmic protein.
Two of the acid-induced genes encode chaperones. One of these is cbpA, which encodes a DnaJ paralog that appears to be involved in folding of cytoplasmic proteins (26). Acid induction of hdeA is consistent with its role as a periplasmic chaperone that suppresses the aggregation of acid-denatured proteins (15).
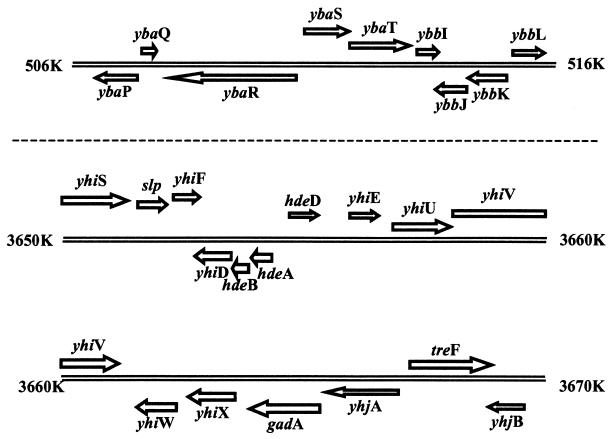
Eight acid-induced metabolism genes were identified. Two of them, gadA and gadB, encode isoforms of glutamate decarboxylase, which plays a primary role in glutamate-dependent AR (9). The glutamate/γ-aminobutyrate transporter, encoded by gadC, the third known component of glutamate-dependent AR, was not induced (data not shown). Other acid-inducible metabolism genes included wrbA, which encodes a flavodoxin protein (wrbA apparently forms an operon with yccJ, an acid-inducible unknown gene); ygfQ, which encodes a putative xanthine permease; ycaC, which encodes a putative isochorismatase; yjfC, which encodes a putative bifunctional glutathionylspermidine synthetase/amidase; ybaS, which encodes a putative glutaminase; and ybaT, which encodes a putative amino acid/amine transporter and is predicted to form an operon (see Fig. 4) with ybaS (regulon DB).
FIG. 4.
Genome location and orientation of acid-induced genes examined in this study.
The remaining six acid-inducible genes encode putative regulatory proteins. A general stress response is indicated by induction of dps, which encodes a general DNA binding protein that functions to protect DNA during stress (1). Another acid-inducible gene is yiaG, which encodes a putative transcription factor. The remaining four regulatory proteins are all located in the hdeA-gadA region. Two are luxR family members, yhiF and yhiE, and two, yhiX and yhiW, are putative transcription factor genes of the araC family.
Acid tolerance of knockout mutants.
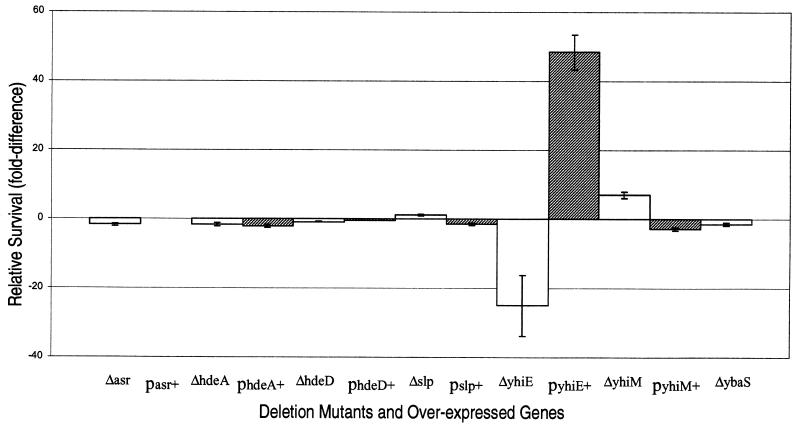
To determine whether some of the acid-inducible genes are directly involved in AR, we made knockout mutations and tested them for survival in an acid shock test. We focused on the genes in the hdeA-gadA region that were significantly induced by acid. Since several of these genes had no obvious role in amino acid decarboxylation and could be involved in endogenous glutamate formation, we tested survival at pH 2.75 for 2 h in the absence of glutamate. Of the seven genes tested, only mutation of yhiE resulted in significantly decreased AR; there was a 25-fold decrease in survival relative to that of the wild type (Fig. 3). This decrease in survival of the yhiE mutant during acid challenge is similar to that of a gadA gadB double mutant (9). Mutation of hdeA, which would be expected to have a polar effect on hdeB transcription (if they form an operon), had no effect on AR. Likewise, mutations of slp or ybaS, which could have a polar effect on yhiF and ybaT, respectively, did not affect AR. Nor was AR affected by mutations of hdeD, yhiM, or even asr, the most significantly induced of all the genes.
FIG. 3.
Relative survival of acid challenge at pH 2.75 for 2 h. Isogenic deletion strains (Δ; open columns) were compared to wild-type E. coli strain MG1655. Strains overexpressing cloned genes (p indicates gene overexpression; shaded columns) were compared to E. coli MG1655 (pPCRScriptAmp). Increased survival compared to the controls is indicated by positive values, and decreased survival is indicated by negative values. Each value reported is the mean of at least nine replicates, and the standard deviations are indicated by error bars.
Acid tolerance conferred by overexpression.
Genes that negatively affect acid tolerance when mutated might confer improved acid tolerance when overexpressed. To test this possibility, we cloned several of the acid-inducible genes from the hdeA-gadA region, grew the wild-type strain overexpressing these genes at pH 7.4 (conditions that do not cause induction of AR), and measured survival in the acid shock test. Under these conditions, only overexpression of yhiE significantly improved AR; there was a 50-fold increase in survival relative to the parent-vector control (Fig. 3). Interestingly, while overexpression of asr had no effect on AR when grown at pH 7.4, the strain carrying the asr plasmid was unable to grow at pH 5.5 (data not shown).
DISCUSSION
The DNA array experiments presented here revealed 28 genes that were significantly induced during the logarithmic phase in acidified growth medium, conditions that favor induction of the glutamate-dependent AR. It is unlikely that all 28 of these acid-induced genes are involved in the glutamate-dependent AR, but the clustering of 9 of them in the gadA region suggests there are several genes in this system (Fig. 4). The gadA gene encodes one of two glutamate decarboxylase enzymes that are critically important for glutamate-dependent AR (9). The gadA region also contains the hdeA gene, which is necessary for AR in Shigella flexneri (29). HdeA is a periplasmic chaperone that is presumably important for suppressing aggregation of periplasmic proteins in acid conditions (15). Our experiments extend these results by demonstrating for the first time that hdeA is induced by acid in E. coli. However, unlike the situation in S. flexneri, our results show that mutation of the E. coli hdeA gene does not affect AR. Apparently this periplasmic chaperone is not essential for AR in E. coli, or there is a redundant function in the absence of HdeA. None of the other seven acid-inducible genes in the gadA region have been previously implicated as being involved in AR. Our results demonstrate that yhiE, encoding a putative transcription factor of the luxR family, is important for AR; mutation of this gene decreases AR, whereas overexpression of yhiE from a high-copy-number plasmid increases AR in cells grown under neutral conditions (Fig. 3). No other mutations of genes in the gadA region affected AR, nor were all genes in the gadA region acid inducible. Nonetheless, the results indicate that this region is important for AR.
The fact that four of the nine acid-inducible genes in the gadA region encode transcription factors suggests that the AR genes are subject to complex regulation. We previously identified a conserved 20-bp sequence element upstream of gadA, gadB, and hdeA (25). This 20-bp sequence was recently shown to function in pH control of transcription (8). We and others have suggested that this 20-bp sequence might be the target of binding by the transcription factor YhiX (8, 25). In fact, overexpression of YhiX causes elevated expression of gadA, gadB, and hdeD, suggesting that YhiX is an activator of these genes (20, 23). Future work planned in this laboratory will shed light on regulation by YhiX. However, it is important not to ignore other regulatory genes in the gadA region during studies of acid regulation (e.g., yhiW, yhiF, and yhiE). yhiF is thought to encode a regulator of dctA, a gene involved in dicarboxylate transport (7). The effects of the yhiE mutation and its overexpression, described above, suggest that YhiE acts as a positive transcription factor for AR genes; the identities of these genes await further investigation.
Additional factors are involved in regulation of the acid-inducible genes in the gadA region. The mnemonic “hde” stands for “HN-S dependent,” and the hdeA, hdeB, and hdeD genes were originally identified as being regulated by HN-S (30). Recently, whole-genome expression profiling provided a comprehensive list of HN-S-dependent genes (20); interestingly, nine of the acid-inducible genes in the gadA region are on that list (the exception is yhiD). Thus, our results further support the hypothesis that HN-S plays a central role in regulating glutamate-dependent AR. Negative control of gadA and gadB by cyclic AMP (cAMP) receptor protein and positive control by the stationary-phase sigma factor, RpoS, points to further complexity in the regulation of acid-inducible genes (8).
There are three genes in the gadA region that encode proteins associated with the cell envelope. slp encodes a stationary-phase-inducible outer membrane lipoprotein of unknown function (22). hdeD encodes an integral membrane protein of unknown function. hdeB encodes a putative periplasmic protein. A fourth cell envelope-associated protein, HdeA, is the only one whose function is known. Its role as a chaperone implies that the cell must combat the deleterious effects of acid on periplasmic proteins. The results further imply that modification of the cell envelope is important for AR. However, mutation of slp, hdeA, and hdeD had no effect on AR. Perhaps the individual mutation of these genes had no effect on AR because of the presence of other acid-induced cell envelope genes with redundant functions.
In contrast to the clustering of acid-induced genes in the gadA region, the remainder are scattered throughout the genome. Of the six small acidic or basic proteins located outside of the gadA region, five of them are unknown genes. Acid induction of small acidic or basic proteins was not previously reported (6), although there is evidence that several of these genes do in fact encode protein products (28). Perhaps these highly charged proteins serve as proton sinks for cytoplasmic pH homeostasis. Alternatively, these proteins could be acting as acid chaperones, as does HdeA. However, the fact that overexpression of asr prevented growth at pH 5.5 indicates that production of these proteins must be strictly controlled.
Outside of the gadA region there are six acid-induced genes that are associated with cell envelope modifications that can lead to improved stress resistance. cfa encodes cyclopropane fatty acyl phospholipid synthase, which is involved in a modification of the phospholipid profile that makes cells more resistant to acid (10). yeaQ, yfbE, and yfbF are putatively involved in glycosyl transferase reactions that modify lipopolysaccharide to make cells more resistant to antibiotics and other stresses (17). ompC encodes an outer membrane protein. Finally, osmY encodes a periplasmic protein that is induced by membrane perturbations and osmotic and other stresses (5). Acid induction of these cell envelope genes further emphasizes the importance of the outer membrane, periplasm, and cytoplasmic membrane in AR.
Another acid-inducible gene outside of the gadA region is cbpA. CbpA was first identified as a curved-DNA binding protein (3), but cbpA was also shown to be a paralog of dnaJ, which encodes a molecular chaperone (26). CbpA appears to be induced in the stationary phase, and dnaJ cbpA double mutants have a severe growth phenotype (27). The involvement of CbpA in AR was not reported previously, but acid induction of cbpA implies that refolding of cytoplasmic proteins is important for AR.
Since Dps is a DNA binding protein that plays a role in protecting DNA during stress (1), induction of dps by acid indicates a general stress response. Mutation of dps is known to decrease acid tolerance of E. coli O157:H7 (11). Thus, protection of DNA may be an important aspect of AR.
It has been suspected for some time that there are additional genes involved in glutamate-dependent AR (9). In particular, since the decarboxylase-dependent systems could create a futile proton cycle by consuming the proton that is transported into the cell together with glutamic acid, there should be a mechanism for endogenous glutamate generation. Thus, there are three acid-inducible metabolism genes that could play a role in AR. ybaS encodes a putative glutaminase, ybaT encodes a putative amino acid/amine transporter, and yjfC encodes a putative bifunctional glutathionylspermidine synthetase/amidase. The acid induction of a gene involved in formation of glutamate and ammonia from glutamine and another involved in ammonia transport points to a possible mechanism for endogenous formation of glutamate in conjunction with the glutamate-dependent AR. However, mutation of ybaS does not affect acid shock survival in the absence of glutamate. Still, this result does not rule out endogenous formation of glutamate as being involved in AR, although ybaS is clearly not essential for AR. The remaining acid-inducible metabolism genes, wrbA, ygfQ, and ycaC, have no obvious role in acid resistance and therefore pose interesting questions.
It is disappointing that gadA was the only gene identified by expression profiling whose product was also identified in two-dimensional gels as being induced by acid (6, 19). On the other hand, gene expression profiling demonstrated that the glutamate-dependent AR genes were induced by 100 mM acetate in the mid-logarithmic phase on minimal glucose medium (2). Acetate induced 13 of the 26 genes induced in our experiments (cfa, dps, gadA, gadB, hdeA, hdeB, hdeD, ompC, osmY, slp, yccJ, yeaQ, and yhiX), including 5 genes in the gadA region. Of the 13 genes induced by both acid and acetate, 4 encode small basic or acidic proteins, and 7 are in the cell envelope functional group. Acetate also induced several genes involved in the oxidative stress response, but this did not occur in cells grown on acid. Comparison of these results further supports the importance of genes encoding small basic or acidic proteins and genes in the cell envelope and chaperone functional groups. Further characterization of the gadA region will be important for understanding the complex mechanisms of acid tolerance.
Acknowledgments
We thank Simon Sims for helpful discussions.
This work was supported by a grant from the NIH (RO1-AI48945-01).
REFERENCES
- 1.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam, T. A., and A. Ishihama. 1999. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 274:33105-33113. [DOI] [PubMed] [Google Scholar]
- 4.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, C., H. Bernstein, C. M. Payne, S. E. Beard, and J. Schneider. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 39:68-72. [DOI] [PubMed] [Google Scholar]
- 6.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boogerd, F. C., L. Boe, O. Michelsen, and P. R. Jensen. 1998. atp mutants of Escherichia coli fail to grow on succinate due to a transport deficiency. J. Bacteriol. 180:5855-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 9.Castanie-Cornet, M.-P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. Y., and J. E. Cronan, Jr. 1999. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33:249-259. [DOI] [PubMed] [Google Scholar]
- 11.Choi, S. H., D. J. Baumler, and C. W. Kaspar. 2000. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 66:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conway, T., B. Kraus, D. L. Tucker, D. J. Smalley, A. F. Dorman, and L. McKibben. 2002. DNA array analysis in a Microsoft Windows environment. BioTechniques 32:110-119. [DOI] [PubMed] [Google Scholar]
- 12a.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 14.Foster, J. W. 1999. When protons attack: microbial strategies of acid adaptation. Curr. Opin. Microbiol. 2:170-174. [DOI] [PubMed] [Google Scholar]
- 15.Gajiwala, K. S., and S. K. Burley. 2000. HdeA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 16.Gale, E. F., and H. M. R. Epps. 1942. The effect of the pH of the medium during growth on the enzymic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem. J. 36:600-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, H. K., K. L. Karem, and J. W. Foster. 1995. Molecular responses of microbes to environmental pH stress. Adv. Microb. Physiol. 37:229-272. [DOI] [PubMed] [Google Scholar]
- 19.Hickey, E. W., and I. N. Hirshfield. 1990. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl. Environ. Microbiol. 56:1038-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 21.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price, G. P., and A. C. St. John. 2000. Purification and analysis of expression of the stationary phase-inducible slp lipoprotein in Escherichia coli: role of the Mar system. FEMS Microbiol. Lett. 193:51-56. [DOI] [PubMed] [Google Scholar]
- 23.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 24.Small, P. L., and S. R. Waterman. 1998. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 6:214-216. [DOI] [PubMed] [Google Scholar]
- 25.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueguchi, C., M. Kakeda, H. Yamada, and T. Mizuno. 1994. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc. Natl. Acad. Sci. USA 91:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueguchi, C., T. Shiozawa, M. Kakeda, H. Yamada, and T. Mizuno. 1995. A study of the double mutation of dnaJ and cbpA, whose gene products function as molecular chaperones in Escherichia coli. J. Bacteriol. 177:3894-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasinger, V. C., and I. Humphery-Smith. 1998. Small genes/gene-products in Escherichia coli K-12. FEMS Microbiol. Lett. 169:375-382. [DOI] [PubMed] [Google Scholar]
- 29.Waterman, S. R., and P. L. Small. 1996. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol. 21:925-940. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida, T., C. Ueguchi, and T. Mizuno. 1993. Physical map location of a set of Escherichia coli genes (hde) whose expression is affected by the nucleoid protein H-NS. J. Bacteriol. 175:7747-7748. [DOI] [PMC free article] [PubMed] [Google Scholar]