Anthrax is one of the oldest known zoonotic diseases. Natural outbreaks threaten wildlife, livestock, and human health, and Bacillus anthracis has been used as a biological weapon [1,2]. Without exception, B. anthracis strains described to date show very little sequence diversity [2–4]. This has led to the conclusion that B. anthracis represents a clonal derivative of an ancestral member of the Bacillus cereus group that obtained pathogenic potential primarily through the acquisition of two B. anthracis–specific virulence plasmids, pXO1 and pXO2 [4,5]. Two major but closely related groups of B. anthracis strains (A and B) have been described by means of characterization of six genomic and two plasmid-encoded variable number tandem repeat regions (VNTRs) [6]. Group B strains are almost exclusively restricted to Southern Africa, while group A is found throughout the world and may have been spread by the domestication and international trade of livestock [7,8].
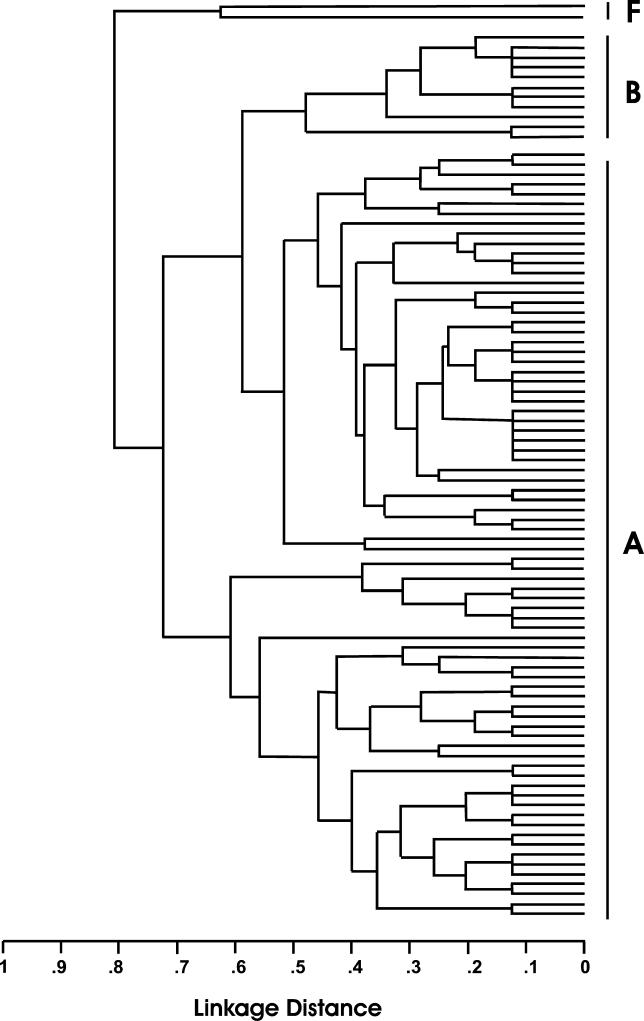
We determined that anthrax infection caused the sudden deaths of six wild chimpanzees in the Taï National Park, Côte d'Ivoire in 2001/2002 [9] and in three chimpanzees and one gorilla found dead more than 1,000 mi to the east, at the northern periphery of the Dja Reserve, Cameroon 2004/2005 [10]. In order to characterize the B. anthracis isolates from a totally new habitat that were responsible for the death of wild great apes that died in Côte d'Ivoire and Cameroon, we performed VNTR analyses on DNA isolated from tissue samples and bones of these great apes. Surprisingly, for the isolates from both outbreaks, three of the six genomic VNTRs were clearly different from those of any previously described strain, as is evident from the phylogenetic tree analysis. The strain from Côte d'Ivoire, named B. anthracis CI (Côte d'Ivoire), and the strain from Cameroon, named B. anthracis CA (Cameroon) are clearly distinct from group A and group B strains. They establish a separate branch with a new “forest anthrax cluster,” termed “F” (Figure 1) suggesting that B. anthracis is a far less homogeneous species than is currently believed. No differences were found among the six cases from Côte d'Ivoire, and also the four anthrax isolates from the three chimpanzees and the gorilla from Cameroon were identical.
Figure 1. Multilocus VNTR-Analysis–Based Dendrogram.
Eight VNTR loci were analyzed following standard protocols [6]. The amplification products were analyzed by DNA sequencing and compared with 89 B. anthracis reference strains [4] processed with Sequence Type Analysis and Recombinational Tests (START) software (K. Jolley, University of Oxford). A, B. anthracis phylogenetic group A; B, B. anthracis phylogenetic group B [7]; F, the newly discovered B. anthracis group from sub-Saharan rainforests including the strains B. anthracis CI and CA.
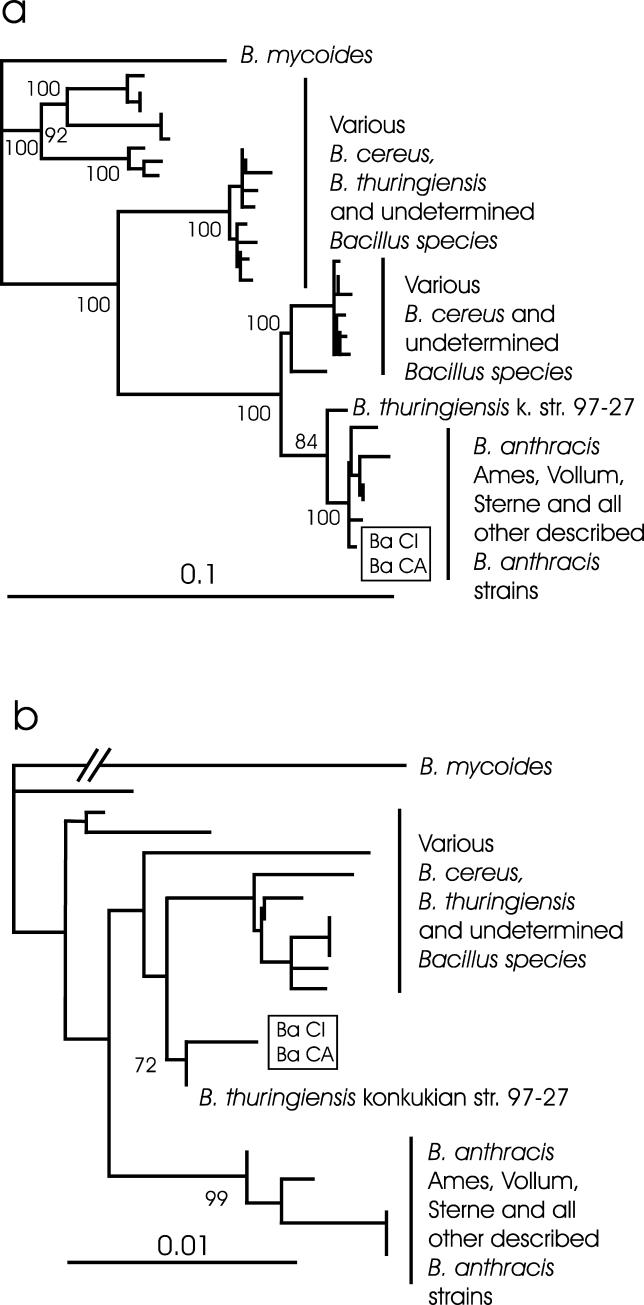
Since these new B. anthracis strains were so clearly distinct from “classic” B. anthracis, we next examined the relationship of the new strains to other members of the B. cereus group. Therefore, we compared the sequences of two chromosomal genes, gyrB and rpoB, that typically differentiate B. anthracis from other closely related Bacillus species [11,12]. The gyrB sequences of the B. anthracis CI and CA were identical and in this analysis fell in a cluster of published B. anthracis strains that were well separated from all other members of the B. cereus group and supported by a 100% bootstrap value (Figure 2A). While isolates CI and CA were also identical in the rpoB sequences, they fell outside the well-supported cluster of classic B. anthracis strains and instead clustered with B. cereus and Bacillus thuringiensis strains and most closely with a recently described atypical and pathogenic B. thuringiensis (72% bootstrap value) (Figure 2B).
Figure 2. Neighbour-Joining Phylogenetic Tree of the (A) gyrB (1,159-bp) and (B) rpoB (700-bp) Genes.
The B. anthracis strains CI and CA are marked by the box. Analyses were carried out following the procedure described by others [11,12]. The trees were statistically evaluated with a bootstrap analysis with 1,000 bootstraps. Only relevant bootstrap values above 70% are shown for major branches.
The nucleotide differences found in the rpoB sequence from both strains prevent sensitive detection in B. anthracis specific real-time PCR assays designed for rapid detection and differentiation of B. anthracis from closely related B. cereus and B. thuringiensis strains [13– 16]. Other PCR assays target segments of each of four recently identified B. anthracis–specific genomic insertions of 16.5-kb–45.5-kb regions (termed A, C, D, and E) not found in B. cereus strains [13,14]. All four PCR assays for these regions were negative for the CI and CA strains, while B. anthracis control isolates yielded the expected amplification products. We confirmed that the assay failure was due to the absence of the expected B. anthracis insertions in the CI and CA strains by designing specific primer pairs for each of the four loci, with a primer located 5′ to the expected insert, either combined with a reverse primer specific for the insert sequence, resulting in an amplification product of approximately 200 bp in case the B. anthracis–specific insert was present, or with a reverse primer located 3′ of the expected insert, resulting in a product of approximately 700 bp in case of absence of the B. anthracis–specific insert. In the event that the insertion was present, the latter primer combination would not have resulted in a product since the fragment was too long to be amplified under the conditions used. As expected, the CI and CA strains yielded products of about 700 bp for regions A, C, and D, and sequencing confirmed the absence of three insertions thought to be specific for B. anthracis. Repeated attempts with different primer sets to amplify segments of the E region from the CI and CA strains failed, although they produced products of expected sizes from B. cereus. This suggests that for the CI and CA strains, the sequence of this region differs from that of both previously described (“classic”) B. anthracis as well as from B. cereus. These data demonstrate that use of molecular assays relying upon chromosomal sequences for the identification of pathogenic B. anthracis may lead to misdiagnosis, and in the light of the potential use of B. anthracis as a biological weapon, it is important that diagnostic tests target various chromosomal sequences as well as both virulence plasmids pXO1 and pXO2 [16].
Differences were also found in the sequence data from two genes (pag and cya) located on the B. anthracis plasmid pXO1. Previously, pag was shown to display limited sequence variation, with a total of only six single-nucleotide differences (three causing amino acid changes) found among 26 pag sequences [3]. Our sequence analysis of the pag toxin gene from the CI and CA strain identified three additional mutations, which result in unique amino acid changes (Table 1) in both new strains. For the strain B. anthracis CA, one additional but conservative mutation, not resulting in an amino acid change, was found. Analysis of 2,231 nucleotides of the cya toxin gene on plasmid pXO1 was only possible for the CI strain but identified four new nucleotide changes (three causing amino acid changes), compared with the six cya sequences from other B. anthracis strains.
Table 1.
Mutations Identified in the pag and cya Toxin Genes
The evergreen rainforest of Central and West Africa is a refuge for a number of animals and plant species and a high biodiversity has developed [17]. Thus there is a possibility that the B. anthracis detected in chimpanzees and gorilla is a “forest strain,” hidden for centuries or millennia in the rainforest of sub-Saharan Africa. Although it is tempting to conclude that modern human activities have caused anthrax emergence in wild apes, there are no data to support such a conclusion. Rather the description of this new group of B. anthracis underscores the need to collect baseline data on pathogens found in remote areas, such as the rainforests of Africa. In this respect, monitoring wild great apes, our closest relatives, for various pathogens may give important hints toward the presence of pathogens of importance for human health. In the light of increasing deforestation, disturbance of ecosystems, and bush-meat handling and consumption, these baseline data will contribute to the assessment of new pathogens which may spread to humans, such as shown for Ebola [18] and monkey pox [19] or other zoonotic diseases.
The detection of two closely related strains forming an unrecognized highly divergent group of B. anthracis at two different locations more than 1,000 mi apart was highly unexpected and implies distribution through Western and Central Africa. The presented genetic data suggest that both “classic” and the “forest anthrax” strains have a common ancestor with B. cereus, and that the forest strains diverged before the other B. anthracis strains acquired the characteristic B. anthracis–specific chromosomal insertions. From the data obtained, it is not possible to determine whether the plasmid pXO1 was already present in the CI and CA strains prior to divergence from the common B. anthracis ancestor, or whether the pXO1 plasmid was acquired independently in B. anthracis group F and “classic” B. anthracis. However, the recent description of a pathogenic B. cereus strain containing a circular plasmid showing 99.6% similarity to the B. anthracis pXO1 [20] demonstrates that plasmid acquisition has occurred more than once within B. cereus.
Even though the anthrax described here shows several important differences from “classic” anthrax strains, the pathological and cytological findings reported in the anthrax victims of Taï National Park, Côte d'Ivoire, are in agreement with those reported in experimental infections of nonhuman primates with anthrax [9]. Unfortunately, no pathological and histological evaluation could be performed on the chimpanzees and gorilla carcasses found near the Dja Reserve in Cameroon since the carcasses were in an advanced stage of decomposition. However, in bone marrow, dental pulpa, and muscle tissue of these animals a high copy number of B. anthracis–specific pag and capC genes encoded by plasmids pXO1 and pXO2, respectively, and the chromosomal rpoB gene were detected, leading to the conclusion that these great apes also died of an acute B. anthracis infection [10]. So far we have found only great apes suffering from this new variant of the pathogen. However, this may be due to the fact that our research was focused only on great apes. In general, much mortality in wildlife but also in domestic animals and in humans in sub-Saharan Africa remains undiagnosed, and the pathogens responsible are frequently not found. The findings presented here show that a variety of new pathogens or variants of known pathogens have to be expected in these areas. It will be important to elucidate the capacity of classic microbiological tests to detect this new B. anthracis, and it would not be much of a surprise if anthrax cases caused by this pathogen go unrecognized. Therefore, studies will have to be undertaken to describe the distribution, biological features, and the pathology of these new B. anthracis forest strains.
Acknowledgments
We thank authorities in Côte d'Ivoire for long-term support, especially the Ministry of the Environment and Forests as well as the Ministry of Research, the directorship of the Taï National Park, and the Swiss Research Center in Abidjan. In Cameroon we thank the MINFoF, MINRESI, the Service de la Conservation de la Réserve du Dja, and various organizations, especially Last Great Ape, the Limbe Wildlife Center, and Projet Grand Singe. For skillful technical support we thank T. Deschner, Y. Moebius, H. Emmel, and also C. Hedemann and J. B. Glanville for copyediting. We are grateful to P. Walsh for helpful discussions. This work was supported by the Robert Koch-Institut and the Max Planck Society.
Abbreviations
- CA
Cameroon
- CI
Côte d'Ivoire
- VNTR
variable number tandem repeat regions
Footnotes
Citation: Leendertz FH, Yumlu S, Pauli G, Boesch C, Couacy-Hymann E, et al. (2006) A new Bacillus anthracis found in wild chimpanzees and a gorilla from West and Central Africa. PLoS Pathog 2(1): e8.
References
- Smith KL, De Vos V, Bryden HB, Hugh-Jones ME, Klevytska A, et al. Meso-scale ecology of anthrax in southern Africa: A pilot study of diversity and clustering. J Appl Microbiol. 1999;87:204–207. doi: 10.1046/j.1365-2672.1999.00871.x. [DOI] [PubMed] [Google Scholar]
- Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- Price LB, Hugh-Jones M, Jackson PJ, Keim P. Genetic diversity in the protective antigen gene of Bacillus anthracis. J Bacteriol. 1999;181:2358–2362. doi: 10.1128/jb.181.8.2358-2362.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KK, Ticknor LO, Okinaka RT, Asay M, Blair H, et al. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl Environ Microbiol. 2004;70:1068–1080. doi: 10.1128/AEM.70.2.1068-1080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler TM. Bacillus anthracis genetics and virulence gene regulation. Curr Top Microbiol Immunol. 2002;271:143–164. doi: 10.1007/978-3-662-05767-4_7. [DOI] [PubMed] [Google Scholar]
- Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, et al. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Klevytska AM, Price LB, Schupp JM, Zinser G, et al. Molecular diversity in Bacillus anthracis. J Appl Microbiol. 1999;87:215–217. doi: 10.1046/j.1365-2672.1999.00873.x. [DOI] [PubMed] [Google Scholar]
- Smith KL, DeVos V, Bryden H, Price LB, Hugh-Jones ME et al. Bacillus anthracis diversity in Kruger National Park. J Clin Microbiol. 2000;38:3780–3784. doi: 10.1128/jcm.38.10.3780-3784.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leendertz FH, Ellerbrok H, Boesch C, Couacy-Hymann E, Matz-Rensing K, et al. Anthrax kills wild chimpanzees in a tropical rainforest. Nature. 2004;430:451–452. doi: 10.1038/nature02722. [DOI] [PubMed] [Google Scholar]
- Leendertz FH, Lankester F, Guislain P, Néel C, Drori O, et al. Anthrax in western and central African great apes. Am J Primatol. 2006. [DOI] [PubMed]
- Ko KS, Kim JM, Kim JW, Jung BY, Kim W, et al. Identification of Bacillus anthracis by rpoB sequence analysis and multiplex PCR. J Clin Microbiol. 2003;41:2908–2914. doi: 10.1128/JCM.41.7.2908-2914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Duc MT, Satomi M, Agata N, Venkateswaran K. gyrB as a phylogenetic discriminator for members of the Bacillus anthracis-cereus-thuringiensis group. J Microbiol Methods. 2004;56:383–394. doi: 10.1016/j.mimet.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Radnedge L, Agron PG, Hill KK, Jackson PJ, Ticknor LO, et al. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl Environ Microbiol. 2003;69:2755–2764. doi: 10.1128/AEM.69.5.2755-2764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer KG, Lamonica JM, Schumacher JA, Williams LE, Bishara J, et al. Identification of Bacillus anthracis specific chromosomal sequences by suppressive subtractive hybridization. BMC Genomics. 2004;5:15. doi: 10.1186/1471-2164-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Patra G, Liang X, Williams LE, Rose S, et al. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl Environ Microbiol. 2001;67:3720–3727. doi: 10.1128/AEM.67.8.3720-3727.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbrok H, Nattermann H, Ozel M, Beutin L, Appel B, et al. Rapid and sensitive identification of pathogenic and apathogenic Bacillus anthracis by real-time PCR. FEMS Microbiol Lett. 2002;214:51–59. doi: 10.1111/j.1574-6968.2002.tb11324.x. [DOI] [PubMed] [Google Scholar]
- Martin C. Die Regenwälder West-Afrikas. Basel: Birkhäuser-Verlag; 1989. pp. 38–41. [Google Scholar]
- Leroy EM, Rouquet P, Formenty P, Souquiere S, Kilbourne A, et al. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303:387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- Reed KD, Melski JW, Graham MB, Regnery RL, Sotir MJ, et al. The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci U S A. 2004;101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]