Abstract
The gene for ribonucleotide reductase from Anabaena sp. strain PCC 7120 was identified and expressed in Escherichia coli. This gene codes for a 1,172-amino-acid protein that contains a 407-amino-acid intein. The intein splices itself from the protein when it is expressed in E. coli, yielding an active ribonucleotide reductase of 765 residues. The mature enzyme was purified to homogeneity from E. coli extracts. Anabaena ribonucleotide reductase is a monomer with a molecular weight of approximately 88,000, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Superose 12 column chromatography. The enzyme reduces ribonucleotides at the triphosphate level and requires a divalent cation and a deoxyribonucleoside triphosphate effector. The enzyme is absolutely dependent on the addition of the cofactor, 5′-adenosylcobalamin. These properties are characteristic of the class II-type reductases. The cyanobacterial enzyme has limited sequence homology to other class II reductases; the greatest similarity (38%) is to the reductase from Lactobacillus leichmannii. In contrast, the Anabaena reductase shows over 90% sequence similarity to putative reductases found in genome sequences of other cyanobacteria, such as Nostoc punctiforme, Synechococcus sp. strain WH8102, and Prochlorococcus marinus MED4, suggesting that the cyanobacterial reductases form a closely related subset of the class II enzymes.
The deoxyribonucleotides required for DNA replication are produced by the reduction of the corresponding ribonucleotides. One enzyme, ribonucleotide reductase (RNR), catalyzes the reduction of the four common ribonucleotides. Although this is an essential reaction in most cells, the primary structure of the enzyme is not highly conserved. At least three different classes of RNRs that differ in amino acid sequence, substrate, and cofactor requirements have been described. Despite major differences in primary sequences, all RNRs catalyze reduction by similar mechanisms involving the generation of a cysteine thiyl radical at the active site (for a review, see reference 20).
The best-characterized RNR is the class I enzyme found in Escherichia coli and most eukaryotic cells. This enzyme is generally reported to have an α2β2 structure (20, 21). The α subunit binds substrates, effectors, and reducing agents. The β subunit is an iron-containing protein that uses oxygen to generate a stable free radical on a tyrosine residue. This radical is transferred to a cysteine residue in the α subunit, forming a transient thiyl radical during catalysis. An anaerobic RNR (class III) also occurs in E. coli and has been found in other facultative and strict anaerobes. This enzyme has an α2 structure. A second protein, referred to as the activase, is loosely associated with the α polypeptides. The activase is a dimeric iron-sulfur protein that catalyzes the formation of a stable glycyl radical in the anaerobic RNR, with S-adenosylmethionine as a cofactor. After the binding of the substrate and the effector, a transient thiyl radical is subsequently generated during catalysis (see references 10 and 21 for reviews).
The class II RNR has been found mainly in bacteria and archaea. Both monomeric and dimeric forms have been described (2, 11). The catalytic thiyl radical is formed by homolytic cleavage of the C-Co bond of the cofactor, 5′-adenosylcobalamin. This class of RNR has been reported to be common among cyanobacteria (8). The purification and characterization of a cyanobacterial RNR from the filamentous organism Anabaena sp. strain PCC 7119 have been reported previously (6). We report here the identification of the Anabaena sp. strain PCC 7120 nrdJ gene, which encodes a class II RNR, and the characterization of the protein.
MATERIALS AND METHODS
Materials.
The protease inhibitor 4-(2-aminoethyl) benzenesulfonyl fluoride, unlabeled adenosylcobalamin, dithiothreitol (DTT), HEPES, isopropyl-β-d-thiogalactopyranoside (IPTG), proteins for molecular weight determinations, Reactive Red 120 agarose, and unlabeled ribo- and deoxyribonucleoside diphosphates and triphosphates were from Sigma Chemical Co. DEAE-Sepharose fast-flow, chelating Sepharose, octyl Sepharose 4 fast-flow, and Superose 12 HR 10/30 columns and Rainbow molecular weight markers for gel electrophoresis were from Amersham Biosciences. XM-50 ultrafiltration membranes (molecular weight cutoff, 50,000) and Centricon-30 filters (molecular weight cutoff, 30,000) were obtained from Amicon. Polyethyleneimine thin-layer plates were purchased from Analtech Corporation. [5-3H]CDP and -CTP were purchased from ICN. Ecoscint A scintillation cocktail was from National Diagnostics Inc. [5′-3H2]adenosylcobalamin was synthesized by the method of Gleason and Hogenkamp (7).
Methods. (i) Isolation of the RNR gene from genomic DNA and cloning into E. coli.
DNA was isolated from Anabaena sp. strain PCC 7120 by phenol extraction by standard procedures (12). A search of the raw Anabaena (Nostoc sp. strain PCC 7120) genomic sequences available in the Kazusa DNA Research Institute database (http://www.kazusa.or.jp/cyanobase/) identified a putative RNR gene (nrdJ). The open reading frame (ORF) of the nrdJ gene was amplified by PCR, and the resulting product was cloned into pCR2.1 by using a Topo TA cloning kit (Invitrogen) to generate plasmid pCR2.1-nrdJ. The forward (GACGGATCCTATGGTTCGTGAGCTTGAGAGAAAA) and reverse (CTCGAATTCCCAGTCTTAACAGGAAACCTGGG) oligonucleotides created BamHI and EcoRI sites, respectively, flanking the nrdJ ORF. The nrdJ ORF of pCR2.1-nrdJ was subcloned as a BamHI-to-EcoRI fragment into the corresponding sites of pRSET B to produce pRSET-nrdJ, a plasmid that expresses Anabaena RNR as a fusion protein with an N-terminal six-His tag plus an Xpress epitope, and PCR amplification of the nrdJ ORF was performed with an Expand 20kb Plus PCR system (Boehringer Mannheim) according to the manufacturer's instructions. Thermocycling conditions were 2 min at 94°C; 20 cycles of 30 s at 94°C, 30 s at 55°C, and 5 min at 68°C; and then a final incubation at 68°C for 10 min. The nrdJ ORF of pRSET-nrdJ was sequenced by the University of Minnesota Advanced Genetics Analysis Center and found to encode a protein identical to that predicted by the Anabaena genome project (accession number NP_488075.1).
(ii) Enzyme assays.
Adenosylcobalamin-dependent RNR activity can be monitored by measuring the 3H exchange between [5′-3H2]adenosylcobalamin and water (9). The assay procedure was modified from that reported previously (6). The assay mixture contained 240 mM HEPES (pH 8.2), 1.2 mM EDTA, 25 mM DTT, 2 mM CTP, 0.05 mM dATP, 1 mM CaCl2, 300 mM NaCl, 0.01 mM [5′-3H2]adenosylcobalamin (4.6 μCi/μmol), and enzyme in a final volume of 0.50 ml. Adenosylcobalamin was added in dim light, and the reaction was initiated by the addition of the enzyme fraction. The reaction tubes were incubated at 35°C in the dark. The reaction was terminated by the addition of 0.50 ml of 2.0% trichloroacetic acid containing 50 mg of activated charcoal. The reaction tubes were centrifuged, and 0.50 ml of the supernatant was added to 10 ml of the Ecoscint A scintillation cocktail. The amount of tritium was determined in a Packard Tri-Carb 2900 TR counter. Activity is reported as counts released to the water after subtraction of counts in a blank reaction without enzyme. This assay was used to monitor activity during enzyme purification. One exchange unit is defined as 1,000 cpm released per the reaction time of 15 min.
The activity of the purified enzyme was determined by measuring the conversion of 3H-labeled ribonucleotides to the corresponding deoxyribonucleotides. The reaction mixture contained 100 mM HEPES (pH 8.2), 1 mM EDTA, 40 mM DTT, 0.01 mM adenosylcobalamin, 10 mM CaCl2, 0.05 mM deoxynucleoside triphosphate effector, 0.10 mM 5-3H-labeled ribonucleotide (10 μCi/μmol), and enzyme in a final volume of 0.050 ml. Adenosylcobalamin was added in dim light, and the reaction was initiated by the addition of enzyme. Reaction mixtures were incubated at 35°C in the dark, and reactions were terminated by boiling. Labeled deoxyribonucleotide was separated from the substrate by chromatography on polyethyleneimine cellulose as described previously (8). One unit of activity corresponds to 1 nmol of deoxynucleotide produced per 15 min of incubation.
The protein concentration was estimated from the UV absorbance at 260 and 280 nm (17).
(iii) Growth of E. coli.
Batch cultures of E. coli containing plasmid pRSET-nrdJ were grown in 2-liter Erlenmeyer flasks containing 500 ml of Turbo Broth. Ampicillin was added to a final concentration of 100 μg/ml. The flasks were incubated at 30°C with shaking. When the cultures reached an optical density of 6.0 at 650 nm, IPTG was added to a final concentration of 0.2 mM. The cells were allowed to grow for an additional 4 h and then harvested by centrifugation at 10,000 × g for 10 min. The pellets were frozen at −15°C.
(iv) Purification of RNR.
All steps were carried out at 4°C. Purification typically started with approximately 100 g (wet weight) of packed E. coli cells. Cells were thawed and resuspended in buffer containing 10 mM Tris-HCl buffer (pH 8.2), 1.0 mM EDTA, 2.0 mM MgCl2, and 0.10 mM DTT (M-8 buffer). 4-(2-Aminoethyl) benzenesulfonyl fluoride protease inhibitor was added to a final concentration of 0.15 mM. The cells were stirred with 0.25 mg of egg white lysozyme/ml for 1 h. The partially lysed cells were further disrupted by ultrasound treatment for 20 min with a Heat Systems sonifier. The lysate was centrifuged at 18,000 × g for 15 min, and the pellets were discarded. Streptomycin sulfate was added to the supernatant to a final concentration of 1% (wt/vol). The mixture was stirred for 1 h and then centrifuged at 18,000 × g for 15 min. The pellets were discarded, and the supernatant was dialyzed for approximately 36 h against several changes of M-8 buffer (approximately 10 liters). Dialysis tubing with a molecular weight cutoff of 10,000 was used.
The dialyzed crude fraction was loaded onto a DEAE-Sepharose fast-flow column (23 by 3.5 cm) that was preequilibrated with M-8 buffer. The column was eluted with the same buffer at a flow rate of 100 ml/h, and fractions were collected. The absorbance at 280 nm (A280) was monitored. When the A280 returned to 0, the column was eluted with 0.10 M NaCl in M-8 buffer. When the A280 again returned to 0, the column was eluted with a gradient of NaCl (0.1 to 0.5 M in M-8 buffer, 1-liter total volume). Enzyme activity was determined by the 3H exchange assay. RNR activity eluted at approximately 0.4 M NaCl. Active fractions were pooled and concentrated by ultrafiltration by using an Amicon XM-50 membrane and under 70 lb of N2/in2.
The concentrated DEAE fraction was dialyzed against 1 liter of buffer containing 0.5 M NaCl in 0.02 M Na2HPO4, pH 7.2. The buffer was changed four times over 24 h. The dialyzed DEAE fraction was loaded onto a chelating Sepharose column (9 by 3 cm) which had been charged with 0.10 M CoSO4, washed extensively, and prerun with eluting buffer according to the manufacturer's directions. The column was preequilibrated with the above-mentioned buffer. After the dialyzed fractions were loaded, the column was eluted with the same buffer and the absorbance was monitored at 280 nm. When the A280 returned to 0, the column was eluted with 0.10 M imidazole in 0.02 M Na2HPO4, pH 7.2. Enzyme activity was determined by the 3H exchange assay. Active fractions were pooled and concentrated by ultrafiltration as described above.
The volume of the chelating fraction was measured, and 4 M NaCl was added to a final concentration of approximately 2 M NaCl. The fraction was loaded onto an octyl Sepharose column (10 by 3 cm) which had been preequilibrated with 2 M NaCl in M-8 buffer. The column was eluted with the same buffer at a flow rate of approximately 100 ml per h. When the A280 returned to 0, the column was eluted with a gradient of NaCl (2 M NaCl to zero NaCl in M-8 buffer; 400-ml total volume). RNR activity eluted near the end of the gradient. Active fractions were pooled and concentrated by ultrafiltration as described above.
The octyl fraction was loaded onto a Reactive Red agarose column (35 by 1 cm) which was preequilibrated with 0.10 M NaCl in M-8 buffer. The column was eluted with the same buffer at a flow rate of 25 ml/h. When the A280 returned to 0, the column was eluted with a gradient of 0.10 to 1.0 M NaCl in M-8 buffer (total volume, 400 ml). RNR activity elutes approximately halfway through the gradient. Active fractions were pooled and concentrated to a protein concentration of approximately 1 mg/ml. The preparation was frozen at −15°C.
(v) Purity and molecular weight determination.
The progress of the purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Copeland (3). The samples were denatured by boiling for 2 min in 1% SDS containing 10 mM DTT. Proteins were separated on alkaline SDS-10% polyacrylamide gels. Rainbow high-molecular-weight standards were used to estimate relative masses. Gels were stained with Coomassie blue.
The Mr of the native protein was determined by fast protein liquid chromatography (FPLC) on a Superose 12 HR 10/30 column. The column was preequilibrated with 0.10 M NaCl in M-8 buffer. A sample size of 0.10 ml (containing approximately 0.5 mg of protein) was loaded on the column, which was run at 0.10 ml per min by using a Pharmacia FPLC system. Molecular weight standards were prepared by dissolving commercial preparations of the following proteins: sweet potato β-amylase (Mr = 200,000), yeast alcohol dehydrogenase (Mr = 150,000), bovine serum albumin (Mr = 66,000), and egg albumin (Mr = 45,000) at a final concentration of 5 mg/ml. The column was eluted with the above-mentioned buffer alone or in buffer containing 0.050 or 0.10 mM dATP.
RESULTS
Cloning and sequence.
The RNR gene (nrdJ) was discovered during a search of the genome database for Anabaena sp. strain PCC 7120 (Nostoc sp. strain PCC 7120) (Kazusa DNA Research Institute [http://www.kazusa.or.jp/cyanobase/]). The nrdJ ORF encoded a protein of 1,172 amino acids, with a molecular weight of approximately 120,000. This was surprising since the relative molecular weight for the enzyme from Anabaena sp. strain PCC 7119 was reported to be 72,000 (6). However, the protein was predicted to contain an intein of 407 amino acids, as shown in Fig. 1.
FIG. 1.
(A) Alignment of the Anabaena strain PCC 7120 RNR with the reductase from L. leichmannii. The five cysteines corresponding to those in the active site of the Lactobacillus enzyme are indicated with a ▾. The site of the intein insertion in the Anabaena enzyme is indicated by a filled arrow. The alignment was generated with the GAP program (Genetics Computer Group, University of Wisconsin). (B) Intein sequence expressed by the Anabaena nrdJ gene. The putative endonuclease active site is underlined.
The nrdJ ORF was ligated into a pRSET B vector to create a gene expressing a fusion protein consisting of an N-terminal Xpress epitope fused to the reductase. Using the Anti-Xpress antibodies, Western blots of crude extracts of E. coli which express the Anabaena RNR gene were run. These blots weakly detected a protein at a high molecular weight and strongly detected a protein at an Mr of 88,000 (data not shown). Presumably these proteins corresponded to the expressed protein with and without the intein, respectively. Adenosylcobalamin-dependent reductase was readily detected by the tritium exchange assay in extracts of transformed E. coli cells, confirming that the intein processing occurred in the foreign bacterium.
Purification and molecular weight.
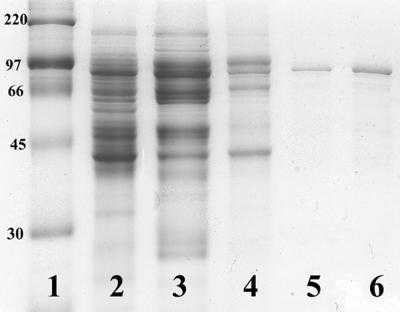
The cloning vector adds a sequence of 6 histidine residues to the N terminus of the expressed protein to facilitate purification by metal affinity chromatography. Attempts to purify the cyanobacterial reductase by standard techniques on Ni- or Cu-chelated columns were unsuccessful. Active enzyme could not be recovered from these columns. Most likely, any free metal ions formed complexes with the cysteines of the enzyme, resulting in a progressive loss of activity. Approximately 12-fold purification was possible on a chelating column charged with cobalt (Table 1). The column was washed extensively after being charged, and active enzyme was eluted with 0.10 M imidazole. However, several proteins in the extract bind under these conditions, and the reductase was not homogenous after chelating chromatography, as seen upon SDS-PAGE (Fig. 2).
TABLE 1.
Purification of Anabaena sp. strain PCC 7120 RNR from E. coli
| Fractiona | Vol (ml) | Protein (mg/ml) | Total protein | Sp act (U/mg) | Purification factor | Total activity (U) | % Recovery |
|---|---|---|---|---|---|---|---|
| Crude extract | 320 | 36 | 11,520 | 2.4 | 27,650 | ||
| DEAE pool | 73 | 46 | 3,219 | 7.5 | 3.1 | 24,970 | 90 |
| Chelate pool | 32 | 8.7 | 278 | 30.3 | 12.6 | 8,436 | 31 |
| Octyl pool | 24 | 4.4 | 106 | 46.4 | 19.3 | 4,918 | 18 |
| Red agarose | 5 | 2.0 | 10 | 120.2 | 50.1 | 1,202 | 4 |
Fractions were assayed by 3H exchange to the solvent from [5′-3H2]adenosylcobalamin as described in the text under “Methods.” CTP was the substrate, and dATP was used as the effector.
FIG. 2.
SDS-PAGE of an Anabaena RNR preparation. Lane 1, molecular weight standards for myosin (220,000), phosphorylase b (97,000), bovine serum album (66,000), ovalbumin (45,000), and carbonic anhydrase (30,000); lane 2, DEAE pool; lane 3, chelating pool; lane 4, octyl pool; lane 5, Reactive Red 120 agarose pool, 3 μg of protein; lane 6, Reactive Red 120 agarose pool, 6 μg of protein.
A further purification was carried out by using affinity chromatography. As reported earlier (6), the cyanobacterial reductase will bind to Reactive Red 120 agarose, presumably by its nucleotide binding site. The column has a limited capacity, and some enzyme activity was generally found in the fraction that passes through the column. However, most of the RNR binds to the column and elutes with a NaCl gradient-yielded enzyme that is at least 90% as pure as that seen in Fig. 2. The relative molecular weight on the denaturing gel was estimated to be 88,000. The mass as calculated from the amino acid composition of the fusion protein is 90,882 Da (87,148 Da for the native enzyme).
The Mr of the native enzyme was determined by chromatography on a Superose 12 column. The molecular weight calculated from the elution profile is 88,000, as shown in Fig. 3. The column was also run with the allosteric effector dATP (see Tables 2 and 3) in the equilibration buffer. It has been reported that some adenosylcobalamin-dependent reductases readily dimerize in the presence of effectors (4). This does not appear to be the case with the cyanobacterial enzyme, since the elution profile was unchanged in the presence of either 0.05 or 0.10 mM dATP (data not shown).
FIG. 3.
Estimation of the Mr of native Anabaena RNR by FPLC on a Superose 12 column. The column was calibrated with four standards (λ): sweet potato β-amylase (Mr, 200,000), yeast alcohol dehydrogenase (Mr, 150,000), bovine serum albumin (Mr, 66,000), and egg albumin (Mr, 45,000). The column was equilibrated and run as described in the text under “Methods.” Anabaena RNR elutes at the position indicated by *, with a calculated molecular weight of 88,000.
TABLE 2.
Requirements for ribonucleotide reductiona
| Reaction conditions | Amt of deoxynucleotide produced (U/mg) |
|---|---|
| Complete with 3H-CTP | 400 |
| Complete with 3H-CDP | 19 |
| Minus adenosylcobalamin | 0 |
| Minus DTT | 0 |
| Minus dATP | 4.4 |
| Minus CaCl2 | 11.8 |
Reduction of CTP or CDP was determined as described in the text under “Methods.” Reaction conditions were identical except for the substrate.
TABLE 3.
Substrate and effector specificities as determined by 3H-exchange reaction from [5′-3H2]adenosylcobalamina
| Substrate | Effector | Amt of tritium exchanged (U/mg) |
|---|---|---|
| ATP | 1.4 | |
| ATP | dGTP | 52.0 |
| dGTP | 0 | |
| GTP | 13.8 | |
| GTP | dTTP | 40.2 |
| dTTP | 0 | |
| CTP | 14.4 | |
| CTP | dATP | 126.4 |
| dATP | 0 | |
| UTP | 4.6 | |
| UTP | dCTP | 10.6 |
| dCTP | 0 |
Reaction conditions are described in the text under “Methods.” Units are reported after subtraction of the value for a blank reaction without protein.
Enzyme activity.
As seen in Table 2, the Anabaena reductase is most active with ribonucleoside triphosphate substrates. A small amount of activity was seen with CDP, but this may be due to contamination of the diphosphate with triphosphate. No reduction occurs in the absence of adenosylcobalamin or a reducing agent. As reported earlier for the enzyme from Anabaena strain PCC 7119 (6), little activity is detected in the absence of dATP, which is the only deoxyribonucleoside triphosphate that activates CTP reduction. The enzyme also seems to require a divalent ion for activity. Previous results with the partially purified reductase from Anabaena strain PCC 7119 (6) showed that the enzyme required a divalent cation for maximal activity; the Ca2+ ion was the most effective, but MgCl2 or MnCl2 also stimulated reduction. The residual activity seen here in the absence of a divalent cation is probably due to Mg2+ in the enzyme buffer.
All four common ribonucleoside triphosphates are substrates for the Anabaena reductase, as determined by the tritium exchange assay (Table 3). As noted above, there is little activity in the absence of effectors. The dNTPs shown in Table 3 are the only ones that substantially enhance activity. These positive effectors are the same as the specificity activators found for other RNRs (10). In contrast to the reductase from Lactobacillus leichmannii, the Anabaena enzyme does not catalyze exchange in the presence of the effectors alone (23). UTP is a poor substrate, and none of the common deoxynucleoside triphosphates substantially enhance activity.
DISCUSSION
The RNR from Anabaena strain PCC 7120 joins a growing list of RNRs in all classes that contain inteins. Other intein-containing class II reductases include enzymes from Pyrococcus furiosus, Pyrodictium abyssi, and Deinococcus radiodurans (16). Inteins have been reported to occur only in the proteins of unicellular organisms and are generally associated with enzymes of DNA metabolism (14). For example, in the complete genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803, inteins are found in the DNA helicase, polymerase, and gyrase but not in the class I RNR found in this cyanobacterium (InBase, the New England Biolabs intein database, http://www.neb.com/inteins). The Anabaena strain PCC 7120 intein has features which are common to many other homing endonucleases (Fig. 1B). It has an N-terminal cysteine residue and a C-terminal sequence, His-Asn-Ser, where cleavage occurs after the Asn residue and the extein sequence resumes with a serine, which forms a peptide bond to arginine 275. The intein has the conserved endonuclease active-site motif L-A-G-L-M-D-S-D-G (Fig. 1B). It shows some homology to the intein found in the RNR of P. abyssi.
The Anabaena RNR shows limited sequence similarity to other reductases in the GenBank database, including most class II reductases. The greatest homology is to the RNR from L. leichmannii, and a suggested alignment is presented in Fig. 1. Some of the same sequence homology is found in putative RNRs from mycobacteriophages D29 and L5 and roseophage SI01. The sequence alignment in Fig. 1A shows that the five cysteine residues which are involved in catalysis in the L. leichmannii reductase (2) are conserved in the cyanobacterial reductase. Cysteine 408 in the Lactobacillus enzyme is the site of the thiyl radical, and this corresponds with cysteine 408 in the cyanobacterial reductase. Similarly, cysteine 419 in the Lactobacillus reductase is one of an active redox pair of cysteines which reduce the substrate. This residue corresponds to cysteine 418 in the Anabaena RNR. A glutamic acid at 410 and asparagine 406, which H bonds to Glu 410, are also conserved. Together, these serve as an acid-base pair during catalysis. The other cysteine in the redox pair is residue 119 in the Lactobacillus reductase, which aligns with cysteine 102 in the cyanobacterial enzyme, although there is little conservation of sequence in this part of the primary structure. A similar lack of homology in this region of RNRs was previously noted by Tauer and Benner (22). The assignment of this cysteine to the active site seems reasonable based on the recently published three-dimensional structure of the L. leichmannii reductase (19).
Cysteines 731 and 736 in the L. leichmannii reductase also function as a redox pair and accept electrons from the H donor, thioredoxin or glutaredoxin. These electrons are then transferred to the active-site pair, cysteines 119 and 419, during reduction. The Anabaena sequence has a similar C terminus, with cysteine 752 separated from cysteine 757 by 4 amino acid residues. Based on homology, it seems likely that these residues also serve to interact with cyanobacterial thioredoxins (1). The three-dimensional structure of the Lactobacillus enzyme shows a bound cobalamin analog interacting with a cleft in the protein formed by residues 565 to 626 and 685 to 724. The alignment proposed in Fig. 1 shows little sequence similarity to the Anabaena reductase in this area of the protein. Since adenosylcobalamin is loosely bound to RNRs, tertiary rather than primary structure conservation may be critical in the cofactor binding region. Alternatively, the cyanobacterial reductase may bind the cofactor more tightly than the L. leichmannii RNR because binding of adenosylcobalamin in Anabaena RNR requires both a substrate and an effector.
Although it has limited similarity to most known class II reductases, the Anabaena RNR shows over 90% sequence homology to putative reductases of other cyanobacteria and cyanophage P60 (accession number NP_570337). The partial sequences from Synechococcus sp. strain WH8102, Nostoc punctiforme, Prochlorococcus marinus MED4, and Prochlorococcus strain MIT9313 in the U.S. Department of Energy Joint Genome Institute database (http://spider.jgi-psf.org) are easily aligned over the entire sequence. This finding is in agreement with previous data showing that the adenosylcobalamin-dependent reductases from the cyanobacteria have similar properties in most common strains (8). The class II reductases seem to be confined to organisms that synthesize cobalamins. The class I reductase found in the genome of Synechocystis sp. strain PCC 6308 is an anomaly and may have been acquired by gene transfer (13).
Functionally, the Anabaena RNR is more similar to the reductase from Lactobacillus than to any other characterized enzyme in class II. The Anabaena strain PCC 7120 RNR described here is very similar to the enzyme previously isolated from cells of Anabaena strain PCC 7119 (6). It reduces nucleosides at the triphosphate level rather than diphosphates, as reported for enzymes from Pyrococcus (16) and Thermotoga (4). Like the Lactobacillus reductase, it seems to function as a single polypeptide. There is no evidence that Anabaena reductase forms dimers under the conditions used in these assays, which is consistent with the recent analysis of the tertiary structure of the L. leichmannii RNR. Sintchak and coworkers (19) reported that the effector binding region of the L. leichmannii enzyme (amino acids 168 to 320 in Fig. 1) forms a four-helix bundle and is structurally similar to the dimer interface in the class I α2 structure. However, this four-helix bundle prevents the formation of a dimer in the Lactobacillus reductase. Residues 130 to 275 in the Anabaena enzyme are approximately 50% similar, including a conserved aspartic acid at 180 (223 in the Lactobacillus sequence) and an arginine at 215 (258 in the Lactobacillus sequence), which are implicated in effector binding (5). Assuming that the cyanobacterial reductase also has a helical structure in this part of the protein, it may not be able to dimerize. It is interesting that this region in the Anabaena reductase terminates at the intein insertion site. This may suggest some role for mobile genetic elements in the evolution of the monomeric versus the dimeric type of class II reductases.
Like many other class II reductases, the Anabaena enzyme does not seem to have an overall activity site. Deoxynucleoside triphosphates are required for activity, and none of these inhibits reduction as dATP does in the E. coli reductase. UTP is not a particularly good substrate for the Anabaena RNR, as reported earlier (6). Although some stimulation is seen on addition of dCTP, it seems more likely that most of the dTTP required for DNA synthesis is generated by deoxycytidine deaminase and subsequent methylation as proposed for other microorganisms. This assumption correlates with the higher reductase activity observed in vitro with CTP.
The lack of sequence similarity among RNRs from different organisms and among the different enzyme classes presents an evolutionary puzzle. It is expected that an enzyme that performs an essential function will be highly conserved. Given the similarity in the catalytic mechanisms and three-dimensional structures of all known classes of RNR, it has been proposed that a single enzyme arose in the evolutionary transition from the RNA to the DNA world (15). This original RNR then diverged into multiple forms in response to environmental challenges or genomic changes, such as the loss or gain of genes for cobalamin synthesis. This divergence led to a major loss of primary structural homology among reductases over time, while those features necessary for catalysis and regulation were preserved. The class II enzyme found in cyanobacteria may be closely related to the original RNR. It is monomeric and has a simple method of regulation. It is also widespread among cyanobacteria, which are among some of the most ancient eubacteria known from the fossil record (18). These findings indicate an early evolution of the monomeric class II reductases in the eubacteria. The cloning of the gene for the Anabaena enzyme provides a tool for further investigation of a relatively simple RNR. We have recently made a construct in the pET3a vector without the His tag and the epitope which expresses native Anabaena RNR in E. coli. Further structural and kinetic analysis will be done when homogenous protein is available.
Acknowledgments
We thank Tina Thornton for help with the Western blots.
This research was supported by a grant from the Graduate School, University of Minnesota.
REFERENCES
- 1.Alam, J., S. Curtis, F. K. Gleason, M. Gerami-Nejad, and J. A. Fuchs. 1989. Isolation, sequence, and expression in Escherichia coli of an unusual thioredoxin gene from the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 171:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booker, S., S. Licht, J. Broderick, and J. A. Stubbe. 1994. Coenzyme B12-dependent ribonucleotide reductase: evidence for the participation of five cysteine residues in ribonucleotide reduction. Biochemistry 33:12676-12685. [DOI] [PubMed] [Google Scholar]
- 3.Copeland, R. A. 1994. Methods for protein analysis. Chapman & Hall, New York, N.Y.
- 4.Eliasson, R., E. Pontis, A. Jordan, and P. Reichard. 1999. Allosteric control of three B12-dependent (class II) ribonucleotide reductases. Implications for the evolution of ribonucleotide reduction. J. Biol. Chem. 274:7182-7189. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson, M., U. Uhlin, S. Ramaswamy, M. Ekberg, K. Regnström, B.-M. Sjöberg, and H. Eklund. 1997. Binding of allosteric effectors to ribonucleotide reductase system from Escherichia coli. J. Biol. Chem. 267:25541-25547. [DOI] [PubMed] [Google Scholar]
- 6.Gleason, F. K., and T. D. Frick. 1980. Adenosylcobalamin-dependent ribonucleotide reductase from the blue-green alga, Anabaena sp. J. Biol. Chem. 255:7728-7733. [PubMed] [Google Scholar]
- 7.Gleason, F. K., and H. P. C. Hogenkamp. 1971. Preparation of 5′-deoxyadenosylcobalamin-5′-3H2. Methods Enzymol. 18C:65-71. [Google Scholar]
- 8.Gleason, F. K., and J. M. Wood. 1976. Ribonucleotide reductase in blue-green algae: dependence on adenosylcobalamin. Science 192:1343-1344. [DOI] [PubMed] [Google Scholar]
- 9.Hogenkamp, H. P. C., R. K. Ghambeer, C. Brownson, R. L. Blakley, and E. Vitols. 1968. Cobamides and ribonucleotide reduction VI. J. Biol. Chem. 243:799-808. [PubMed] [Google Scholar]
- 10.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 11.Jordan, A., E. Torrents, C. Jeanthon, R. Eliasson, U. Hellman, C. Wernstedt, J. Barbe, I. Gibert, and P. Reichard. 1997. B12-dependent ribonucleotide reductases from deeply rooted eubacteria are structurally related to the aerobic enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 94:13487-13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Ochman, H., J. G. Lawrence, and E. A. Grossman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 14.Paulus, H. 2000. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69:447-496. [DOI] [PubMed] [Google Scholar]
- 15.Reichard, P. 1993. From RNA to DNA, why so many ribonucleotide reductases? Science 260:1773-1777. [DOI] [PubMed] [Google Scholar]
- 16.Riera, J., F. T. Robb, R. Weiss, and M. Fontecave. 1997. Ribonucleotide reductase in the archaeon Pyrococcus furiosus: a critical enzyme in the evolution of DNA genomes? Proc. Natl. Acad. Sci. USA 94:475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schleif, R. F., and P. C. Wensink. 1981. Practical methods in molecular biology. Springer-Verlag, New York, N.Y.
- 18.Schopf, J. W., A. B. Kudryavtsev, D. G. Agresti, T. J. Wdowiak, and A. D. Czaja. 2002. Laser-Raman imagery of Earth's earliest fossils. Nature 416:73-76. [DOI] [PubMed] [Google Scholar]
- 19.Sintchak, M. D., G. Arjara, B. A. Kellogg, J. A. Stubbe, and C. L. Drennan. 2002. The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer. Nat. Struct. Biol. 9:293-300. [DOI] [PubMed] [Google Scholar]
- 20.Sjöberg, B.-M. 1997. Ribonucleotide reductases—a group of enzymes with different metallosites and a similar reaction mechanism. Struct. Bonding 8:139-173. [Google Scholar]
- 21.Stubbe, J. A., J. Ge, and C. S. Yee. 2001. The evolution of ribonucleotide reduction revisited. Trends Biochem. Sci. 26:93-99. [DOI] [PubMed] [Google Scholar]
- 22.Tauer, A., and S. A. Benner. 1997. The B12-dependent ribonucleotide reductase from the archaebacterium Thermoplasma acidophila: an evolutionary solution to the ribonucleotide reductase conundrum. Proc. Natl. Acad. Sci. USA 94:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada, R., Y. Tamao, and R. L. Blakley. 1971. Degradation of 5′-deoxyadenosylcobalamin by ribonucleoside triphosphate reductase and binding of degradation products to the active center. Biochemistry 10:3959-3968. [DOI] [PubMed] [Google Scholar]